Short abstract
Automated use of phylogenetic trees to deduce orthology relationships in proteins.
Abstract
Reliable orthology prediction is central to comparative genomics. Although orthology is defined by phylogenetic criteria, most automated prediction methods are based on pairwise sequence comparisons. Recently, automated phylogeny-based orthology prediction has emerged as a feasible alternative for genome-wide studies.
Homologous sequences - that is, those derived from a common ancestral sequence - can be further divided into two different classes according to the mode in which they diverged from their last common ancestor [1]. The divergence of two homologous sequences by a speciation event gives rise to orthologous sequences, whereas a duplication event will define a paralogous relationship between the duplicates. Although such straightforward definitions could suggest that distinguishing paralogs and orthologs is simple, it is definitely not. For example, it is not unusual for multiple lineage-specific gene loss or duplication events, as well as other evolutionary processes, to result in intricate scenarios that are difficult to interpret. Far from being a simple curiosity, the establishment of correct orthology and paralogy relationships is crucial in many biological studies. For instance, phylogenetic analyses that aim to infer correct evolutionary relationships between several species should be based on orthologous sets of sequences [2]. Moreover, as orthologs are, relative to paralogs, more likely to share a common function, the correct determination of orthology has deep implications for the transfer of functional information across organisms [3]. Finally, the establishment of equivalences among genes in different genomes is a prerequisite for comparative analyses of genome-wide data to detect evolutionarily conserved traits [4,5].
Originally defined on an evolutionary basis, orthology relationships are best established through phylogenetic analysis. This usually involves the reconstruction of a phylogenetic tree describing the evolutionary relationships among the sequences and species involved, so that speciation and duplication events can then be mapped on the nodes of the tree. This is the classical procedure for establishing orthology relationships. However, the availability of whole sequenced genomes means the need to detect orthology at a genomic scale, a task for which the, mostly manual, phylogeny-based approach is not suited. Automated approaches were soon developed that inferred orthology relationships from pairwise sequence comparisons. Although these methods perform reasonably well, they have many drawbacks that can lead to annotation errors or misinterpretation of data [6,7]. To avoid such pitfalls, and in an attempt to approximate the classical approach for detecting orthology, several automatic methods have been proposed that delineate orthology relationships from phylogenetic trees. Despite the greater accuracy of such methods compared with pairwise approaches, the large demands of time and computing power needed to generate reliable trees have limited their use to datasets of moderate size. Recently, however, the combination of automated large-scale phylogenetic reconstruction with newer algorithms is paving the way for the use of phylogeny-based methods for orthology detection at genomic scales [8,9]. This progress is likely to have a deep impact on future comparative studies.
Homology, orthology and paralogy
Homology is defined as the relationship that exists between two biological entities - for example, two sequences or two anatomic characters - that are derived from a common ancestor. In 1970, Walter Fitch coined the concepts of orthology and paralogy to distinguish two types of homology relationships between biological sequences [1]. Orthologous sequences are those that derive by a speciation event from their common ancestor, whereas the origin of paralogous sequences can be traced back to a gene-duplication event. Despite this clear definition, orthology and paralogy are often misinterpreted by biologists. This is partly due to the fact that what may seem simple when comparing pairs of closely related species, easily gets complicated when wider groups of distantly related species are involved. It is sometimes wrongly claimed, for example, that only two sequences from the same species can be regarded as paralogs, or that two sequences from different species are orthologous to each other only if they perform the same biological function. I will briefly summarize here the main misunderstandings that can arise when dealing with properties of orthologous sequences (see [7] for a more thorough discussion), which are key to understanding why some of the methods discussed later would be more appropriate than others.
The first clarification is that orthology is a purely evolutionary concept, certainly related to, but not based on, the functionality of the sequences involved. All homologous proteins have a common ancestry and thus are expected to have similar three-dimensional structures and to perform related functions. But changes in functionality within a homologous family of proteins caused by sequence variation or context-dependency are not rare [10]. This is especially true in the case of paralogs, because processes of neo- or subfunctionalization may favor the retention of duplicate genes [11]. Orthologous sequences derived by speciation are, therefore, less prone to functional shifts but are definitely not free from them.
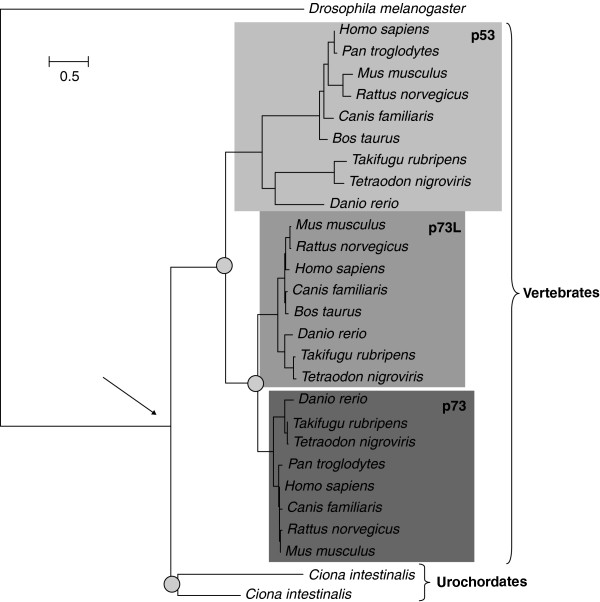
A second important point to note is that the orthology or paralogy relationship between two genes will extend to their descendants as they disperse by further speciation or duplication events. Thus, groups of orthologs, and not just pairs, may more adequately represent the ancestral relationships of the genes in a set of organisms. An important corollary of its definition is that orthology, in contrast to homology, is not transitive. If a gene A is orthologous to B and B to C, A and C are not necessarily orthologous to each other. For instance, if A and C are related by a duplication event, they will be paralogous to each other while both being co-orthologous to B. This is best explained with a graphical example (Figure 1). The human tumor suppressor protein p53 belongs to a wider family of proteins that also includes p73 and p73L. The tree shown in Figure 1 depicts the evolutionary relationships among several metazoan members of the family, ranging from insects to mammals. As can be inferred from the tree, several duplications (nodes marked with gray circles) occurred at different periods. Most significantly, two consecutive duplications at the base of the vertebrates originated three sister groups (shadowed regions in the tree) that correspond to the p53, p73 and p73L subfamilies. Human p53 can be considered orthologous to the sequences in other vertebrates that cluster within the same shadowed region, because they all derive by speciation events. Paralogous relationships can be drawn between human p53 and human p73 and p73L, because their common ancestral node always corresponds to a duplication node. The same reasoning can be used to infer paralogous relationships between any sequence within the p53 subfamily and those in the p73 and p73L subfamilies, even though they might not be encoded in the same genome, such as human p53 and mouse p73L. The only criteria to mark them as paralogs is the fact that they derived by the duplication of an ancestral gene. Human p53 is also orthologous to any of the two Ciona intestinalis sequences, because they diverged from a speciation node (marked with an arrow). Note that this is the only node that is important in defining their orthology relationship, and we do not consider the fact that, subsequent to that speciation, both lineages experienced duplication events. These later duplication events are, however, important to define other proteins at the same orthology level. In fact, human p53, p73 and p73L all are orthologous to any of the sequences in C. intestinalis because they diverged at the same speciation node. To accurately define the orthology relationships between human and C. intestinalis members of this family one should say that human p53, p73L and p73 are all co-orthologous to the two C. intestinalis proteins.
Figure 1.
p53 phylogeny. Phylogenetic tree representing the evolutionary relationships among p53 and related proteins. Sequences were obtained from the p53 tree at phylomeDB [35] (entry code Hsa0012331). After selecting a group of representative sequences, a maximum likelihood tree was reconstructed using the same parameters used for the JTT tree in PhylomeDB. Shaded boxes indicate vertebrate members of the p53, p73 and p73L subfamilies. Duplication nodes are marked with a gray circle. The arrow indicates the speciation node that marks the bifurcation between urochordates and vertebrates.
Yet another complication in defining orthology relationships among proteins is that they often comprise distinct domains that may have followed different evolutionary histories [12]. Such evolutionary chimeras can be created by fusion and recombination events between different genes and may lead to situations in which, for example, a single member of a given protein family has recently acquired a new domain through recombination with another family. In such cases the different domains should, in principle, be treated as independent evolutionary units and orthology relationships be delineated accordingly. Thus, in multidomain families, orthology relationships should be first established among core domains and then extended, where possible, to adjacent regions.
Pairwise methods for orthology inference
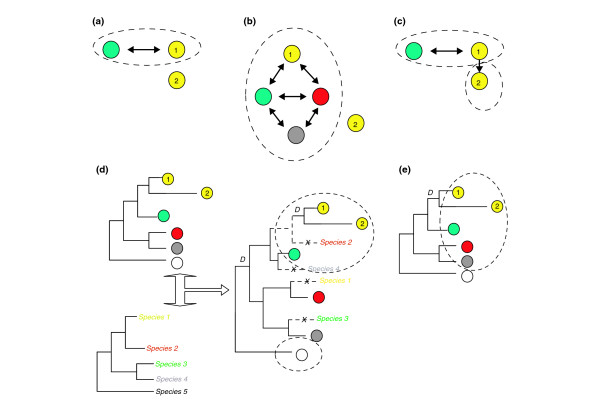
The need to compare sets of genomic sequences has prompted the development of several automatic methods that infer orthology relationships from pairwise sequence comparisons. The first, and still most widely used, method for automatically establishing orthology relationships is based on the detection of best bi-directional best hits (BBH), also known as best reciprocal hits (BRH), which consists of the detection of pairs of sequences from different species that are, reciprocally, the best hit of each other in a sequence search [13] (Figure 2a). This operational definition of orthology is fairly adequate when comparing two closely related genomes. At larger evolutionary distances, however, the scenario becomes more complicated. By definition, the BBH approach can only account for one-to-one orthology relationships. Therefore, if gene duplications have taken place in any of the two compared lineages after their divergence, a one-to-many or a many-to-many relationship will be necessary to properly describe their orthology relationships. In such cases the BBH approach will miss many true orthologs.
Figure 2.
Orthology prediction methods. (a-c) Pairwise-based and (d, e) phylogeny-based methods. Circles of different colors indicate proteins encoded in genomes from different species. Black arrows represent reciprocal BLAST hits. Proteins within dashed ovals are predicted by the method to belong to the same orthologous group. (a) Best bi-directional hit (BBH). All pairs of proteins with reciprocal best hits are considered orthologs. Note that this method is unable to predict the othology with the yellow protein 2. (b) COG-like approach. Proteins in the nodes of triangular networks of BBHs are considered as orthologs (green, red and yellow protein 1 in the example). New proteins are added to the orthologous group if they are present in BBH triangles that share an edge with a given cluster; for example, the gray protein will be added to the orthologous group because it forms a BBH triangle with the red and green proteins. Note that a BBH link with yellow protein 1 is not required. The COG-like approach can add additional proteins from the same genome if they are more similar to each other than to proteins in other genomes, or if they form BBH triangles with members of the cluster. This is not the case for yellow protein 2, which is, again, misclassified. (c) Inparanoid approach. This is similar to (a), but other proteins within a proteome (yellow protein 2 in this example) are included as 'in-paralogs' if they are more similar to each other than to their corresponding hits in the other species. (d) Tree-reconciliation phylogenetic approach. Duplication nodes (marked with a D) are defined by comparing the gene tree (small tree at the top) with the species tree (small tree at the bottom) to derive a reconciled tree (big tree on the right) in which the minimal number of duplication and gene loss (dashed lines) events necessary to explain the gene tree are included. In this case, both the yellow proteins are included in the orthologous group but the red and gray proteins are excluded. (e) Species-overlap phylogenetic approach. All proteins that derive from a common ancestor by speciation are considered members of the same orthologous group. Duplication nodes are detected when they define partitions with at least one shared species. A one-to-many orthology relationship emerges because of a recent duplication in the lineage leading to the yellow proteome.
To avoid these pitfalls and extend the procedure to multiple genome comparisons, Tatusov and colleagues introduced the concept of clusters of orthologous groups (COGs) [14] (Figure 2b). COGs are derived from the search for 'triangular' BBH relationships across a minimum of three species, and their subsequent combination into larger groups. This strategy has been followed by many groups and is the operational definition of orthology used by many databases such as EGO [15] and STRING [16].
Other extensions of the BBH approach include recent implementations such as Inparanoid [17] (Figure 2c) or OrthoMCL [18], which achieve higher sensitivity through sequence-clustering techniques that consider a range of BLAST scores beyond the absolute best hits. For instance, Inparanoid predicts paralogs resulting from lineage-specific duplications, which it calls 'in-paralogs', by including intraspecific BLAST hits that are reciprocally better than between-species BLAST hits. So, to a certain level, Inparanoid is able to include one-to-many and many-to-many relationships. Its limitation is that it is designed for comparing pairs of genomes only. OrthoMCL expands the procedure to comparisons of multiple genomes. It first uses a similar strategy to Inparanoid to define orthologous relationships between each pair of genomes. The comparisons of all possible pairs of genomes are represented as a graph in which the nodes represent genes and the edges represent orthology relationships. A Markov clustering algorithm (MCL) is then applied. In brief, OrthoMCL simulates random walks on the graph of orthology predictions to determine the transition probabilities among the nodes, that is, the probabilities that two nodes are connected in a random walk. The graph is partitioned into different orthologous groups on the basis of these probabilities.
Yet another type of method that cannot be strictly considered pairwise-based but that does not specifically build phylogenetic trees to define orthology, aims to refine previously made COGs. Generally, these methods organize clusters of orthologous genes into a hierarchical structure by using some evolutionary information. For instance, COCO-CL subdivides a given orthologous group on the basis of the correlation coefficient between their sequences, as inferred from a multiple sequence alignment [19]. In contrast, OrthoDB uses the information regarding the species to which a given sequence belongs to organize an orthologous group in a hierarchy that is guided by the species tree [20].
Phylogeny-based orthology inference in tree reconciliation
In the classical procedure for determining orthology relationships, a phylogenetic tree is constructed from an alignment of homologous sequences and subsequently compared to a species tree. This comparison allows the geneticist to infer the events of gene loss and duplication that have occurred along the evolution of the sequence family considered. The first strategy for inferring such relationships automatically was proposed by Goodman and colleagues [21], who developed an algorithm for fitting a given gene tree to its corresponding species tree and inferring the minimum set of duplications needed to explain the data. This problem came to be known as 'tree reconciliation' (Figure 2d), and several other algorithms have been implemented that solve it efficiently [22-24]. These tree-based algorithms for orthology detection are very intuitive, as they simply implement automatically what an expert would do manually and, provided that correct species and gene trees are given, the algorithm will infer the correct orthology relationships. A number of databases have been developed that use such algorithms to derive orthology relationships from automatically reconstructed trees [25-27].
The main limitation of the tree-reconciliation method is that for many scenarios the species tree is not known with confidence. Moreover, it has been shown that another assumption of the tree-reconciliation problem, the correctness of the gene tree, is frequently violated [28]. In such cases, erroneous gene trees will inevitably lead to incorrect orthology and paralogy assignments and the inference of many extraneous duplications and gene losses. As a result, these methods are very sensitive to slight variations in the topology or the rooting of the gene tree and, when applied at a large scale they perform similarly to and even worse than standard pairwise methods [29] and need manual curation [30]. Even if the gene tree is correctly reconstructed, it may not conform to the species tree in cases where horizontal gene transfer events have occurred. Such gene trees are hard to reconcile with the species tree and are often confused by apparent events of massive gene loss.
One possible solution to cope with the existing ambiguity in gene and species trees is to account for this uncertainty during the process of tree reconciliation. Some approaches consider the uncertainty of the different nodes of the gene tree as inferred from their bootstrap, or equivalent, values, and weight the gene loss and duplication events accordingly [31,32]. Another approach that tackles the uncertainty of both the gene and the species tree was recently proposed by the group of David Liberles [33]. This algorithm, called 'soft parsimony', modifies uncertain or poorly supported branches by minimizing the number of gene duplication and loss events implied by the tree. It starts by generating all possible rooted trees that can be derived from a given gene tree. Then the edges that have a support value under a given threshold are collapsed. Each tree is subsequently reconciled with the species tree, which can include multifurcations at unresolved nodes, and the number of duplications is computed. If more than one tree minimizes the necessary duplications, these are compared in terms of the number of gene losses implied. Finally, the collapsed nodes are reconstituted.
Soft parsimony is able to solve the most obvious errors arising from tree reconciliation, which normally implies a multitude of gene losses and duplications. It also allows the use of species trees with unresolved nodes, which usually better represent what we really know about relationships within most phylogenetic groups. Nevertheless, these algorithms still need a certain level of resolution in the species trees and have a number of underlying assumptions that should be taken into account. For instance, the scenario with the minimal number of losses and gene duplications is not necessarily the real one, as losses and duplications can be rampant in some cases [34]. Furthermore, the number of iterations and tree-reconciliation steps that these methods involve may limit its use in large-scale datasets.
Species-overlap methods
Yet another way out of the problem of ambiguity in species and gene trees is to consider the gene tree topology in a very relaxed way and minimize the need to know the true evolutionary relationships of species. This approach is followed in recent algorithms that are based on the level of overlap between the species encountered within a tree. Basically, these algorithms examine the level of overlap in the species connected to two related nodes to decide whether their parental node represents a duplication or speciation event (Figure 2e). They assume that a node represents a duplication event if it is ancestral to two tree-partitions that contain sets of species that overlap to some degree. Conversely, if the two partitions contain sets of species that are mutually exclusive, the node is considered to represent a speciation event. The only evolutionary information that such algorithms require is that needed to root the tree so that a polarity (ancestors to descendants) between the internal nodes is defined.
One such algorithm has been used in the prediction of all orthology and paralogy relationships for all human genes and their homologs in 38 other eukaryotic species [8]. The reason for using this type of algorithm was its speed and the high degree of topological diversity observed in the human phylome, something that would have resulted in many wrong assignments if a reconciliation algorithm had been used. This orthology-prediction methodology is now implemented in all phylomes deposited at PhylomeDB [35]. Van der Heijden and colleagues implemented a species-overlap algorithm in a program called LOFT (Levels of Orthology From Trees) [36]. Besides predicting orthology relationships between genes in a phylogenetic tree, LOFT assigns a hierarchy to the orthology relationships. Similar to the Enzyme Classification (EC) numbers, each gene of a family is given a code that indicates its level within the orthology hierarchy. In this way orthologous groups can be defined at different levels and the orthology and paralogy relationships can be readily inferred from the code.
In conclusion, the prediction of orthology, rather than just homology, relationships among genes in sequenced genomes is a necessary task that often needs to be performed in an automated way. Most automatic strategies to derive such orthology relationships still use rough approximations that are far away from the original definition of orthology. Nowadays, however, the increasing speed at which computer programs can generate phylogenetic trees, as well as the availability of new algorithms, allows the possibility of actually predicting orthology by mapping the speciation and duplication events on a tree, thus following the formal definition of orthology. It is likely that soon this strategy will become the most commonly used in genome-wide searches for orthology. The expected increase in the accuracy of the predicted relationships will result in a higher reliability of transfer of information across species. Recent analyses show that phylogeny-based methods are less prone to error than similarity-based approaches. The same analyses show, however, that there is still room for improvement and that future algorithms will need to take into account the inherent topological variability that is expected in any genome-wide phylogenetic analysis.
Acknowledgments
Acknowledgements
This work was partly funded by grants from the Spanish Ministries of Health (FIS06-213) and Science and Innovation (GEN2006-27784-E/PAT) to TG.
References
- Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–113. doi: 10.2307/2412448. [DOI] [PubMed] [Google Scholar]
- Moreira D, Philippe H. Molecular phylogeny: pitfalls and progress. Int Microbiol. 2000;3:9–16. [PubMed] [Google Scholar]
- Gabaldón T. Evolution of proteins and proteomes, a phylogenetics approach. Evol Bioinf Online. 2005;1:51–56. [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T, Huynen MA. Prediction of protein function and pathways in the genome era. Cell Mol Life Sci. 2004;61:930–944. doi: 10.1007/s00018-003-3387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen MA, Gabaldón T, Snel B. Variation and evolution of biomolecular systems: searching for functional relevance. FEBS Lett. 2005;579:1839–1845. doi: 10.1016/j.febslet.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Eisen JA. Phylogenomics: improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 1998;8:163–167. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Dopazo H, Dopazo J, Gabaldón T. The human phylome. Genome Biol. 2007;8:R109. doi: 10.1186/gb-2007-8-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. Automatic genome-wide reconstruction of phylogenetic gene trees. Bioinformatics. 2007;23:i549–i558. doi: 10.1093/bioinformatics/btm193. [DOI] [PubMed] [Google Scholar]
- Thornton JW, DeSalle R. Gene family evolution and homology: genomics meets phylogenetics. Annu Rev Genomics Hum Genet. 2000;1:41–73. doi: 10.1146/annurev.genom.1.1.41. [DOI] [PubMed] [Google Scholar]
- Roth C, Rastogi S, Arvestad L, Dittmar K, Light S, Ekman D, Liberles DA. Evolution after gene duplication: models, mechanisms, sequences, systems, and organisms. J Exp Zool B Mol Dev Evol. 2007;308:58–73. doi: 10.1002/jez.b.21124. [DOI] [PubMed] [Google Scholar]
- Doolittle RF. The multiplicity of domains in proteins. Annu Rev Biochem. 1995;64:287–314. doi: 10.1146/annurev.bi.64.070195.001443. [DOI] [PubMed] [Google Scholar]
- Huynen MA, Bork P. Measuring genome evolution. Proc Natl Acad Sci USA. 1998;95:5849–5856. doi: 10.1073/pnas.95.11.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Lee Y, Sultana R, Pertea G, Cho J, Karamycheva S, Tsai J, Parvizi B, Cheung F, Antonescu V, White J, Holt I, Liang F, Quackenbush J. Cross-referencing eukaryotic genomes: TIGR Orthologous Gene Alignments (TOGA). Genome Res. 2002;12:493–502. doi: 10.1101/gr.212002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Kruger B, Snel B, Bork P. STRING 7 - recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;(35 Database):D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien KP, Remm M, Sonnhammer EL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;(33 Database):D476–D480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothi R, Zotenko E, Tasneem A, Przytycka TM. COCO-CL: hierarchical clustering of homology relations based on evolutionary correlations. Bioinformatics. 2006;22:779–788. doi: 10.1093/bioinformatics/btl009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriventseva EV, Rahman N, Espinosa O, Zdobnov EM. OrthoDB: the hierarchical catalog of eukaryotic orthologs. Nucleic Acids Res. 2008;(36 Database):D271–D275. doi: 10.1093/nar/gkm845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M, Czelusniak J, Moore GM, Romero-Herrera AE, Matsuda G. Fitting the gene lineage into its species lineage, a parsimony strategy illustrated by cladograms constructed from globin sequences. Syst Zool. 1979;28:132–163. doi: 10.2307/2412519. [DOI] [Google Scholar]
- Zmasek CM, Eddy SR. A simple algorithm to infer gene duplication and speciation events on a gene tree. Bioinformatics. 2001;17:821–828. doi: 10.1093/bioinformatics/17.9.821. [DOI] [PubMed] [Google Scholar]
- Page RD, Charleston MA. From gene to organismal phylogeny: reconciled trees and the gene tree/species tree problem. Mol Phylogenet Evol. 1997;7:231–240. doi: 10.1006/mpev.1996.0390. [DOI] [PubMed] [Google Scholar]
- Dufayard JF, Duret L, Penel S, Gouy M, Rechenmann F, Perriere G. Tree pattern matching in phylogenetic trees: automatic search for orthologs or paralogs in homologous gene sequence databases. Bioinformatics. 2005;21:2596–2603. doi: 10.1093/bioinformatics/bti325. [DOI] [PubMed] [Google Scholar]
- Zmasek CM, Eddy SR. RIO: analyzing proteomes by automated phylogenomics using resampled inference of orthologs. BMC Bioinformatics. 2002;3:14. doi: 10.1186/1471-2105-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal PS, Boore JL. A phylogenomic gene cluster resource: the Phylogenetically Inferred Groups (PhIGs) database. BMC Bioinformatics. 2006;7:201. doi: 10.1186/1471-2105-7-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu JC, Lee EK, Egan MG, Sarkar IN, Coruzzi GM, DeSalle R. OrthologID: automation of genome-scale ortholog identification within a parsimony framework. Bioinformatics. 2006;22:699–707. doi: 10.1093/bioinformatics/btk040. [DOI] [PubMed] [Google Scholar]
- Rasmussen MD, Kellis M. Accurate gene-tree reconstruction by learning gene- and species-specific substitution rates across multiple complete genomes. Genome Res. 2007;17:1932–1942. doi: 10.1101/gr.7105007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsen T, Huynen MA, de Vlieg J, Groenen PM. Benchmarking ortholog identification methods using functional genomics data. Genome Biol. 2006;7:R31. doi: 10.1186/gb-2006-7-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Coghlan A, Ruan J, Coin LJ, Hériché JK, Osmotherly L, Li R, Liu T, Zhang Z, Bolund L, Wong GK, Zheng W, Dehal P, Wang J, Durbin R. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;(34 Database):D572–D580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand D, Halldorsson BV, Vernot B. A hybrid micro-macroevolutionary approach to gene tree reconstruction. J Comput Biol. 2006;13:320–335. doi: 10.1089/cmb.2006.13.320. [DOI] [PubMed] [Google Scholar]
- Chen K, Durand D, Farach-Colton M. NOTUNG: a program for dating gene duplications and optimizing gene family trees. J Comput Biol. 2000;7:429–447. doi: 10.1089/106652700750050871. [DOI] [PubMed] [Google Scholar]
- Berglund-Sonnhammer AC, Steffansson P, Betts MJ, Liberles DA. Optimal gene trees from sequences and species trees using a soft interpretation of parsimony. J Mol Evol. 2006;63:240–250. doi: 10.1007/s00239-005-0096-1. [DOI] [PubMed] [Google Scholar]
- Gabaldón T, Huynen MA. Lineage-specific gene loss following mito-chondrial endosymbiosis and its potential for function prediction in eukaryotes. Bioinformatics. 2005;21(Suppl 2):ii144–ii150. doi: 10.1093/bioinformatics/bti1124. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Bueno A, Dopazo J, Gabaldón T. PhylomeDB: a database for genome-wide collections of gene phylogenies. Nucleic Acids Res. 2008;(36 Database):D491–D496. doi: 10.1093/nar/gkm899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijden RT van der, Snel B, van Noort V, Huynen MA. Orthology prediction at scalable resolution by phylogenetic tree analysis. BMC Bioinformatics. 2007;8:83. doi: 10.1186/1471-2105-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]