Abstract
DNA damage by agents crosslinking the strands presents a formidable challenge to the cell to repair for survival and to repair accurately for maintenance of genetic information. It appears that repair of DNA crosslinks occurs in a path involving double strand breaks in the DNA. Mammalian cells have multiple systems involved in the repair response to such damage, including the Fanconi anemia pathway that appears to be directly involved, although the mechanisms and site of action remain elusive. A particular finding relating to deficiency of the Fanconi anemia pathway is the observation of chromosomal radial formations. The basis of formation of such chromosomal aberrations is unknown although they appear secondarily to double strand breaks. Here we review the processes involved in response to DNA interstrand crosslinks which might lead to radial formation and the role of the nucleotide excision repair gene, ERCC1, which is required for a normal response, not just to DNA crosslinks, but also for double strand breaks at collapsed replication forks caused by substrate depletion.
Interstrand crosslinks (ICLs) are a potent form of DNA damage in which the strands are covalently linked by a bifunctional chemical. The result is a block of normal DNA replication and transcription. The most readily available template for repair, the opposing strand, is also involved with the damage, complicating error-free repair. Straight forward excision and gap filling, as with nucleotide excision repair (NER), seems to be precluded. The many steps involved in IC makes ICL- inducing agents attractive as chemotheraputic drugs (e.g. cisplatin). However, not all ICLs are products of exogenous chemicals; ICLs can also be created by byproducts of metabolism, including the lipid peroxidation product malondialdehyde (Minko et al., 2008). Thus, we find ICL repair mechanisms in organisms from bacteria through humans.
In E. coli, the ICL repair pathway utilizes both nucleotide excision repair (NER) and homologous recombination (HR) (Cole, 1973), apparently acting in a single pathway. UvrABC incises the DNA 5′- and 3′- of the ICL on one strand, and the 5′-exonuclease activity of DNA polymerase I creates a single stranded DNA (ssDNA) region required for RecA-mediated recombination. Strand invasion creates a structure on which UvrABC can act, removing the ICL-containing DNA fragment (Dronkert and Kanaar, 2001). The gap is filled by DNA polymerase I and covalently bonded by polynucleotide ligase.
In Sacchromyces cerevisiae, studies have shown many genes are involved in ICL repair. Genetic evidence indicates there are three distinct ICL repair pathways in S. cerevisiae, representing ‘NER’, post-replication repair, and HR represented by SNM1, REV3, and RAD51 epistasis groups respectively (Grossmann et al., 2001). Apparently early in ICL repair double strand breaks (DSB) are formed as intermediates (Jachymczyk et al., 1981). Subsequently, a recombination-dependent step utilizing RAD51 may act in completing ICL repair, through repair of the DSB (Jachymczyk et al., 1981). This occurs only when a homologous sequence is available.
Post-replication/translesion synthesis utilizing error-prone polymerases such as ζ (REV3/Rev7) or η might replicate past the ICL following DSB formation (Jachymczyk et al., 1981). While rev3 mutants are sensitive to ICL damage, yeast DNA polymerase η mutants show normal sensitivity to ICL, suggesting no role for this bypass polymerase in repair (Grossmann et al., 2001). Less is known about this pathway than NER and HR repair; however, it appears that the pathway allows cells to bypass an ICL as opposed to actually repairing the lesion (Dronkert and Kanaar, 2001).
Yeast snm1 Δ mutants are specifically sensitive to ICL (Henriques et al., 1997) but display normal incision (Li and Moses, 2003). They do not, however, resolve the DSB and restore high molecular weight DNA after cross-links (Magana-Schwencke et al., 1982) (Li et al., 2005). Apparently the SNM1 protein, known to be a 5’-exonuclease (Li et al., 2005), acts to modify intermediates of DSB repair. Interestingly, snm1 mutants show normal processing of mating type, indicating that the DSBs occurring in that pathway are processed normally. This leads to the conclusion that the DSBs arising during the ICL repair process have a specific structure, different from DSBs created in mating type switching, thus requiring different processing.
As noted, ICL repair appears to involve DSB intermediates; the processing of the DSB resulting from ICL repair requires specific activities peculiar to the process. Therefore, while RAD51, for example, is required for repair of DSBs after ionizing radiation or ICLs, SNM1 is required only for ICL DSB repair. Such a comparison illustrates that some components of ICL repair may act in a general DSB response whereas others act specifically. It appears that the DSBs occurring during normal DNA replication use many of the same components that ICL repair utilizes for genome stability and provide a substrate for HR (Ward et al., 2007).
The mammalian ICL repair pathway is more complex than E. coli or yeast. Like yeast, ICL repair operates through a DSB intermediate similar to those occurring in S-phase during replication [reviewed in (Patel and Joenje, 2007)]. While NER and HR pathways are implicated in ICL repair (De Silva et al., 2000; Zheng et al., 2003), other proteins, not found in E. coli or S. cerevisiae are involved. The Fanconi anemia (FA) pathway, as demonstrated by the extreme sensitivity of patients, model organisms, and cell lines to ICLs, is involved in mammalian ICL repair [reviewed in (Kennedy and D’Andrea, 2005; Patel and Joenje, 2007; Wang, 2007)]. Activation of the FA pathway occurs during normal S-phase, likely in response to DSBs or other events during replication, and also occurs with hydroxyurea (HU) treatment which results in stalled replication forks with no ICLs, leading to DSBs (Diffley et al., 2000; Fox, 1985). These observations suggest that DSBs caused by HU (substrate depletion) may be equivalent to those arising from ICL repair. FA patients also exhibit chromosome instability, leading to chromosomal radial formations (Figure 1), levels of which are increased as a result of ICL exposure. In fact, radials are not observed in normal cells without ICL damage at high levels. This implies an overload of the repair system in FA cells. The radial formations may occur as ‘quadriradials’ showing symmetry of four arms, or as adhesions of less than two complete sets of chromatids. The finding of radials is accompanied by apparent breaks in chromatids (Figure 1).
Figure 1.
Metaphase spread demonstrating chromosome radials.
The participation of multiple repair pathways in ICL repair raises the question of whether there is a single monolithic repair response to ICLs in mammals, or if the pathways are independent, as has been unambiguously shown for yeast (Grossmann et al., 2001; McHugh et al., 2001). While models for a unitary path involving NER, HR and post-replication have been suggested (Niedernhofer et al., 2005), results from mutant cell lines and siRNA depletion indicate there are multiple independent pathways in mammals (Hanlon Newell et al., 2008; Hemphill et al., 2008; Wang, 2007) (Jakobs, unpublished). In addition, it appears several bypass DNA polymerases, including Pol κ, may be involved (Minko et al., 2008), not just Pol ζ, further complicating interpretaion.
Stages of ICL repair: Recognition and Incision
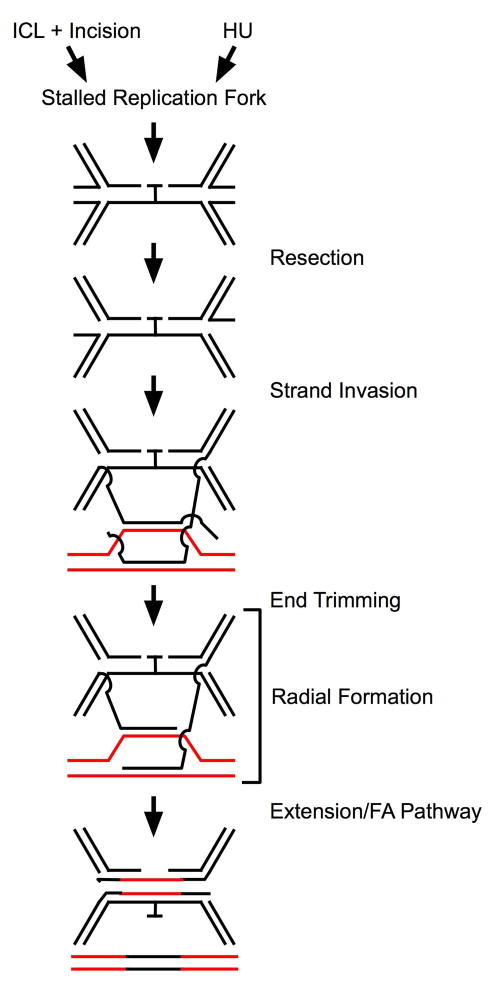
The first step of ICL repair involves recognition of the damage and incision on DNA near the crosslink (Kumaresan et al., 2007) (Figure 2). Incision at the ICL could occur, before or after bypass, leaving a DSB subject to HR or non-homologous end joining (NHEJ) (Dronkert and Kanaar, 2001). Unlike yeast, where loss of any of several NER proteins leads to ICL sensitivity, only loss of either ERCC1 or XPF, which form a heterodimer, leads to exquisite ICL sensitivity in mammalian cells (Collins, 1993; De Silva et al., 2000). The ERCC1/XPF heterodimer can incise near a crosslink in vitro and in vivo (Fisher et al., 2008; Kumaresan et al., 2002). ERCC1/XPF also appears to incise crosslinked DNA as measured by the comet mobility assay (Rothfuss and Grompe, 2004). However, other work demonstrates that proteins other than ERCC1 and XPF may play a role in the initial recognition and incision of ICLs (Ahn et al., 2004; Hanada et al., 2006; Thoma et al., 2005). It appears that the Mus81-Eme1 complex is capable of incision at ICLs and cells deficient in the proteins do not create DSBs at stalled replication forks (Hanada et al., 2006). The question arises, if this is the case, why cell lines lacking ERCC1/XPF are so sensitive to ICLs. Possible explanations might rest on the action of ERCC1/XPF in incision in several independent pathways, or alternatively, that ERCC1/XPF has a post-incision function required for several pathways. That is, it is possible ERCC1/XPF may act in incision, but is not strictly required.
Figure 2. Mechanistic model for ICL repair.
The model shows the ICL residue throughout the path, not specifying the point of final removal, and not affecting DSB formation or repair. Neither the ICL nor the accompanying incisions would be present after HU treatment.
Stages of ICL Repair: DSB Formation, Resection, and Strand Invasion
Following recognition and incision, several possibilities exist for ICL repair, based on the pathways involved. The resulting DSBs, from the HR process, stalled replication forks, or activity of Mus81-Eme1 incision, would require resection to allow strand invasion. In addition to function in excision of bulky adducts in NER, ERCC1/XPF has also been shown to trim overhanging non-homologous single-stranded DNA (ssDNA) from HR intermediates to facilitate extension of the duplex DNA recombination intermediate, indicating a post-incision role for ERCC1 (Adair et al., 2000; Niedernhofer et al., 2001; Niedernhofer et al., 2004). Recent work from our group also supports a post-incision function for ERCC1, needed for normal activation of the FA pathway. Interestingly, ERCC1 is required in response to ICL or HU, a potent inducer of stalled replication forks, but with no ICLs (McCabe et al., 2008). This action occurs after γH2AX formation, an early indicator of DSB. These findings suggest a common DSB repair intermediate occurs in ICL repair and HU-caused stalled replication forks, both requiring the subsequent action of ERCC1/XPF for resolution (Figure 2).
The suggestion of a common intermediate arising from ICL or HU-induced stalled replication forks reinforces the view that, though the ICL has been incised, the intermediate remains impassible to replication forks, The model we present (Figure 2) requires bidirectional replication in order to lead to structures that could be intermediates in ICL repair. When a replication fork reaches the incised ICL, replication stalls and the replication fork collapses (Heller and Marians, 2006; Shrivastav et al., 2008), leading to a “chicken foot” formation, which is functionally a DSB. In that case, stalled replication forks would represent the common repair intermediate between ICL and HU damage. One strand of the collapsed fork could be resected, creating a substrate for strand invasion (Figure 2). This is a potential post-incision site for ERCC1 action. It is possible that resection of the arrested fork by ERCC1/XPF would alter mobility in the gel-based comet assay and therefore appear as ‘incision’ reconciling the action with reported observations.
If extensive homology, perhaps in the form of a homologous chromosome or orthologous gene, is available as a recombination substrate, it may be that extension and bypass of the lesion occurs rapidly, utilizing HR repair mechanisms. If the homologue is damaged or not available, repair might result from strand invasion utilizing microhomology on non-homologous chromosomes (Figure 2). The notion of interaction between non-homologous chromosomes is supported by observations that radial formations after ICL damage are only observed between non-homologous chromosomes (Hanlon Newell et al., 2008). Non-allelic homologous recombination (NAHR) is a well-recognized mechanism utilizing short regions of 90% or greater homology for recombination in the mammalian genome (Gu et al., 2008; Lupski and Stankiewicz, 2005).
Bringing together the idea of fork stalling and the non-homologous chromosome radial formation, the recently described fork stalling, template-switching (FoSTeS) model (Gu et al., 2008; Lee et al., 2007) suggests a mechanism by which stalled replication fork restart might utilize non-homologous chromosomes, utilizing microhomology {Carvalho, 2009#194}. The concept of the FoSTeS mechanism acting in ICL and HU-induced stalled replication fork repair is attractive, but as yet has no supporting evidence for radial formation. RAD18 and RAD5 are known to be required for FoSTeS in yeast (Pages et al., 2008; Zhuang et al., 2008), so testing this model is feasible.
Stages of ICL Repair: End Trimming and Action of the FA Pathway
If the NAHR does not extend to the end of the strand, the resulting unpaired tail could prevent extension. The action of ERCC1/XPF in the trimming of overhanging non-homologous ssDNA from homologous recombination intermediates to facilitate resolution of the recombination intermediate might be required to continue the ICL repair process at this stage (Figure 2) (Adair et al., 2000; Niedernhofer et al., 2001). Such a role for ERCC1/XPF, well down-stream of the incision step, could also represent a basis in addition to resection, for sensitivity of ERCC1 and XPF mutants to ICL damage. Thus end trimming by ERCC1/XPF represents a second potential post-incision function for the ERCC1 protein. In addition, the observation of reduced radials with ERCC1 depletion (McCabe et al., 2008) might be explained by a failure to extend from the limited homology between these interacting non-homologous chromosomes; extension might stabilize this interaction between non-homologous chromosomes.
The inherited disease Fanconi anemia is characterized by several congenital abnormalities. While the phenotype is somewhat variable, patients typically exhibit short stature and other skeletal abnormalities, skin pigmentation abnormalities, bone marrow failure leading to anemia and leukemia, increased risk of solid tumors, and cellular sensitivity to ICL-inducing agents (Bagby and Alter, 2006). There are thirteen identified Fanconi genes: A, B, C, D1/BRCA2, D2, E, F, G, I, J, L, M and N [reviewed in (Patel and Joenje, 2007; Wang, 2007)].
Central to the FA pathway are FANCD2 and its paralog, FANCI, the ID complex (Smogorzewska et al., 2007). FANCD2 monoubiquitination is the marker of activation of the FA pathway and is required for nuclear focus formation, a necessity for normal ICL repair. FANCA, B, C, E, F, G, L and M form a core complex, which is required for monoubiquitination of FANCD2 at lysine 561 by the E3 ligase FANCL, in concert with the E2 subunit UBE2T (Machida et al., 2006). FANCI is monoubiquitinated, like FANCD2, in a core complex-dependent manner on lysine 523 in response to the cell cycle and DNA damage.
The FA pathway acts in maintenance of genome stability after ICL damage. In accord, the FA proteins interact with other DSB repair proteins. FANCD2 co-localizes with BRCA1 in response to DNA damage and at synaptonemal complexes (Garcia-Higuera et al., 2001). The identification of FANCD1 as BRCA2 directly linked the FA pathway and HR pathway (Hirsch et al., 2004). FANCD2 also has been shown to interact in a constitutive manner with FANCD1 (BRCA2) and co-localizes with RAD51 in nuclear foci (Hussain et al., 2004). Another link between DSB repair and FA was uncovered with the identification of FANCN as the partner and localizer of BRCA2 (PALB2) (Reid et al., 2007).
FANCD2 also directly interacts directly with the histone acetylase (HAT) Tip60 (Hejna et al., 2008). Tip60 is known to act in DNA DSB repair and to acetylate ATM prior to auto-phosphorylation of that protein (Ikura et al., 2000; Squatrito et al., 2006; Sun et al., 2005; Wong et al., 2006). Depletion of Tip60 makes cells sensitive to ICLs, as it does for ionizing radiation. Thus the function of FA in DSB repair may involve the remodeling protein, Tip60, acting as a protein acetylase in the FA pathway.
Formation and Suppression of Radials
Chromosome radials are routinely used in the diagnosis of FA, and deficiencies or depletions in FA genes lead to increased radials (Hanlon Newell et al., 2008). Radials are also observed after ICL formation in Bloom syndrome (BS) cells, so the structure is non-specific with regard to the FA pathway. The basis for formation of radials is not well understood although it does appear DSB are required (Hanlon Newell et al., 2008). Deficiencies or depletions for several genes acting in DNA repair, including BRCA1, BRCA2, and RAD51, as well as the NHEJ proteins Ku70, Lig4 and XRCC4, have also been shown to increase radial formation (2004, Bruun et al., 2003, Hanlon Newell et al., 2008). As Rad51 is required for HR, the finding that RAD51 depletion/loss, as well as RAD52 depletion, led to increased radial formation (Hanlon Newell et al., 2008) appears to exclude HR from the basis for radial formation.
Loss of ERCC1 has been shown to be associated with formation of radials (Niedernhofer et al., 2004) in mouse fibroblasts. Work from our group, however, indicates ERCC1 depletion reduces radial formation in normal human fibroblasts (McCabe et al., 2008), as well as ERCC1 co-depletion with FANCA or in FA-A cells, placing ERCC1 upstream of radial formation and FA action (Figure 2), leading to the suggestion that the action of ERCC1 may stabilize an intermediate of ICL repair, prior to the actions of the FA core complex, which would form radials (Figure 2) in the absence of FA function. In the scheme of ICL repair, it appears the FA pathway is a later actor than ERCC1, and deficiencies in FA proteins lead to radials, so the point of formation of radials is placed later than ERCC1 function, but depends on ERCC1 for normal levels (Figure 2).
The model presented indicates that DNA replication would be required for radial formation. This would be in agreement with observations that HU treatment, reducing replication, produces DSBs, but reduced radials (Johnstone et al., 1997). A tantalizing, but unexplained, finding regarding radials formed after ICL damage is that only autosomes are involved in radial formation, and radials do not form between homologous chromosomes (Newell et al., 2004); indeed radials can form between human and mouse chromatids {Hanlon Newell, 2008 #154}. This observation reflects a basic mechanism of radial formation that is not understood.
Conclusions
On the basis of the selected observations noted in this brief review we conclude that: (A), ERCC1 plays a significant post-incision role in ICL repair. This is supported by observations that a normal response to HU, which introduces DSBs, but no crosslinks, requires ERCC1. Given the structure-specific nature of the nuclease function of the ERCC1/XPF heterodimer, post-incision action might be at the stage of resection or trimming of overhanging DNA ends to allow propagation of HR strand invasion in NAHR; (B), ERCC1 action is prior to the point of radial formation in the ICL repair pathway, and therefore, prior to action of the FA pathway; (C), the molecular mechanisms of radial formation remain a mystery; however, notable specifics regarding radials do emerge: radials result from failure of steps in ICL repair at the point of action of the FA pathway, radials form between non-homologues, and radials are not seen following other types of DNA damage such as UV, but are seen following IR or HU, indicating DSBs are a basis for radial formation.
Acknowledgments
We thank Daniel Pauw and A. Hanlon Newell for skillful assistance. The work from the authors’ laboratories was supported by NHLBI Program Project Grant 1PO1HL48546.
References
- Adair G, Rolig R, Moore-Faver D, Zabelshansky M, Wilson J, Nairn R. Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination. EMBO J. 2000;19(20):5552–5561. doi: 10.1093/emboj/19.20.5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn B, Kang D, Kim H, Wei Q. Repair of mitomycin C cross-linked DNA in mammalian cells measured by a host cell reactivation assay. Mol Cells. 2004;18(2):249–255. [PubMed] [Google Scholar]
- Bagby G, Alter B. Fanconi anemia. Semin Hematol. 2006;43(3):147–156. doi: 10.1053/j.seminhematol.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Cole RS. Repair of DNA containing interstrand crosslinks in Escherichia coli: sequential excision and recombination. Proc Natl Acad Sci U S A. 1973;70(4):1064–1068. doi: 10.1073/pnas.70.4.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A. Mutant rodent cell lines sensitive to ultraviolet light, ionizing radiation and cross-linking agents: a comprehensive survey of genetic and biochemical characteristics. Mutat Res. 1993;293(2):99–118. doi: 10.1016/0921-8777(93)90062-l. [DOI] [PubMed] [Google Scholar]
- De Silva I, McHugh P, Clingen P, Hartley J. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol. 2000;20(21):7980–7990. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF, Bousset K, Labib K, Noton EA, Santocanale C, Tercero JA. Coping with and recovering from hydroxyurea-induced replication fork arrest in budding yeast. Cold Spring Harb Symp Quant Biol. 2000;65:333–342. doi: 10.1101/sqb.2000.65.333. [DOI] [PubMed] [Google Scholar]
- Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat Res. 2001;486(4):217–247. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- Fisher LA, Bessho M, Bessho T. Processing of a psoralen DNA interstrand cross-link by XPF-ERCC1 complex in vitro. J Biol Chem. 2008;283(3):1275–1281. doi: 10.1074/jbc.M708072200. [DOI] [PubMed] [Google Scholar]
- Fox RM. Changes in deoxynucleoside triphosphate pools induced by inhibitors and modulators of ribonucleotide reductase. Pharmacol Ther. 1985;30(1):31–42. doi: 10.1016/0163-7258(85)90046-4. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn M, Timmers C, Hejna J, Grompe M, D’Andrea A. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7(2):249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Grossmann K, Ward A, Matkovic M, Folias A, Moses R. S. cerevisiae has three pathways for DNA interstrand crosslink repair. Mutat Res. 2001;487(3–4):73–83. doi: 10.1016/s0921-8777(01)00106-9. [DOI] [PubMed] [Google Scholar]
- Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. Pathogenetics. 2008;1(1):4. doi: 10.1186/1755-8417-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. Embo J. 2006;25(20):4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon Newell AE, Hemphill A, Akkari YM, Hejna J, Moses RE, Olson SB. Loss of homologous recombination or non-homologous end-joining leads to radial formation following DNA interstrand crosslink damage. Cytogenet Genome Res. 2008;121(3–4):174–180. doi: 10.1159/000138882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejna J, Holtorf M, Hines J, Mathewson L, Hemphill A, Al-Dhalimy M, Olson SB, Moses RE. Tip60 is required for DNA interstrand cross-link repair in the Fanconi anemia pathway. J Biol Chem. 2008;283(15):9844–9851. doi: 10.1074/jbc.M709076200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol. 2006;7(12):932–943. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- Hemphill AW, Bruun D, Thrun L, Akkari Y, Torimaru Y, Hejna K, Jakobs PM, Hejna J, Jones S, Olson SB, Moses RE. Mammalian SNM1 is required for genome stability. Mol Genet Metab. 2008;94(1):38–45. doi: 10.1016/j.ymgme.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques J, Brozmanova J, Brendel M. Role of PSO genes in the repair of photoinduced interstrand cross-links and photooxidative damage in the DNA of the yeast Saccharomyces cerevisiae. J Photochem Photobiol B. 1997;39(3):185–196. doi: 10.1016/s1011-1344(97)00020-1. [DOI] [PubMed] [Google Scholar]
- Hirsch B, Shimamura A, Moreau L, Baldinger S, Hag-alshiekh M, Bostrom B, Sencer S, D’Andrea A. Association of biallelic BRCA2/FANCD1 mutations with spontaneous chromosomal instability and solid tumors of childhood. Blood. 2004;103(7):2554–2559. doi: 10.1182/blood-2003-06-1970. [DOI] [PubMed] [Google Scholar]
- Hussain S, Wilson J, Medhurst A, Hejna J, Witt E, Ananth S, Davies A, Masson J, Moses R, West S, de Winter J, Ashworth A, Jones N, Mathew C. Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways. Hum Mol Genet. 2004;13(12):1241–1248. doi: 10.1093/hmg/ddh135. [DOI] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102(4):463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Jachymczyk W, von Borstel R, Mowat M, Hastings P. Repair of interstrand cross-links in DNA of Saccharomyces cerevisiae requires two systems for DNA repair: the RAD3 system and the RAD51 system. Mol Gen Genet. 1981;182(2):196–205. doi: 10.1007/BF00269658. [DOI] [PubMed] [Google Scholar]
- Johnstone P, Reifsteck C, Kohler S, Worland P, Olson S, Moses RE. Fanconi anemia group A and D cell lines respond normally to inhibitors of cell cycle regulation. Somat Cell Mol Genet. 1997;23(6):371–377. doi: 10.1007/BF02673747. [DOI] [PubMed] [Google Scholar]
- Kennedy R, D’Andrea A. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19(24):2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- Kumaresan KR, Hwang M, Thelen MP, Lambert MW. Contribution of XPF functional domains to the 5′ and 3′ incisions produced at the site of a psoralen interstrand cross-link. Biochemistry. 2002;41(3):890–896. doi: 10.1021/bi011614z. [DOI] [PubMed] [Google Scholar]
- Kumaresan KR, Sridharan DM, McMahon LW, Lambert MW. Deficiency in incisions produced by XPF at the site of a DNA interstrand cross-link in Fanconi anemia cells. Biochemistry. 2007;46(50):14359–14368. doi: 10.1021/bi7015958. [DOI] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131(7):1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Li X, Hejna J, Moses RE. The yeast Snm1 protein is a DNA 5′-exonuclease. DNA Repair (Amst) 2005;4(2):163–170. doi: 10.1016/j.dnarep.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Li X, Moses RE. The beta-lactamase motif in Snm1 is required for repair of DNA double-strand breaks caused by interstrand crosslinks in S. cerevisiae. DNA Repair (Amst) 2003;2(1):121–129. doi: 10.1016/s1568-7864(02)00192-1. [DOI] [PubMed] [Google Scholar]
- Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1(6):e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida Y, Machida Y, Chen Y, Gurtan A, Kupfer G, D’Andrea A, Dutta A. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23(4):589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Magana-Schwencke N, Henriques J, Chanet R, Moustacchi E. The fate of 8-methoxypsoralen photoinduced crosslinks in nuclear and mitochondrial yeast DNA: comparison of wild-type and repair-deficient strains. Proc Natl Acad Sci U S A. 1982;79(6):1722–1726. doi: 10.1073/pnas.79.6.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe KM, Hemphill A, Akkari Y, Jakobs PM, Pauw D, Olson SB, Moses RE, Grompe M. ERCC1 is required for FANCD2 focus formation. Mol Genet Metab. 2008;95(1–2):66–73. doi: 10.1016/j.ymgme.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2(8):483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- Meetei A, Sechi S, Wallisch M, Yang D, Young M, Joenje H, Hoatlin M, Wang W. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol Cell Biol. 2003;23(10):3417–3426. doi: 10.1128/MCB.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minko IG, Harbut MB, Kozekov ID, Kozekova A, Jakobs PM, Olson SB, Moses RE, Harris TM, Rizzo CJ, Lloyd RS. Role for DNA polymerase kappa in the processing of N2-N2-guanine interstrand cross-links. J Biol Chem. 2008;283(25):17075–17082. doi: 10.1074/jbc.M801238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell A, Akkari Y, Torimaru Y, Rosenthal A, Reifsteck C, Cox B, Grompe M, Olson S. Interstrand crosslink-induced radials form between non-homologous chromosomes, but are absent in sex chromosomes. DNA Repair (Amst) 2004;3(5):535–542. doi: 10.1016/j.dnarep.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Niedernhofer L, Essers J, Weeda G, Beverloo B, de Wit J, Muijtjens M, Odijk H, Hoeijmakers J, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J. 2001;20(22):6540–6549. doi: 10.1093/emboj/20.22.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer L, Lalai A, Hoeijmakers J. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123(7):1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Niedernhofer L, Odijk H, Budzowska M, van Drunen E, Maas A, Theil A, de Wit J, Jaspers N, Beverloo H, Hoeijmakers J, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004;24(13):5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pages V, Bresson A, Acharya N, Prakash S, Fuchs RP, Prakash L. Requirement of Rad5 for DNA polymerase zeta-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics. 2008;180(1):73–82. doi: 10.1534/genetics.108.091066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KJ, Joenje H. Fanconi anemia and DNA replication repair. DNA Repair (Amst) 2007;6(7):885–890. doi: 10.1016/j.dnarep.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, Neveling K, Kelly P, Seal S, Freund M, Wurm M, Batish S, Lach F, Yetgin S, Neitzel H, Ariffin H, Tischkowitz M, Mathew C, Auerbach A, Rahman N. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39(2):162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- Rothfuss A, Grompe M. Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRis required. Mol Cell Biol. 2004;24(13):5776–5787. doi: 10.1128/MCB.24.1.123-134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18(1):134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald Er, Hurov K, Luo J, Ballif B, Gygi S, Hofmann K, D’Andrea A, Elledge S. Identification of the FANCI Protein, a Monoubiquitinated FANCD2 Paralog Required for DNA Repair. Cell. 2007;129(2):289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16(9):433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102(37):13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma B, Wakasugi M, Christensen J, Reddy M, Vasquez K. Human XPC-hHR23B interacts with XPA-RPA in the recognition of triplex-directed psoralen DNA interstrand crosslinks. Nucleic Acids Res. 2005;33(9):2993–3001. doi: 10.1093/nar/gki610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8(10):735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- Ward JD, Barber LJ, Petalcorin MI, Yanowitz J, Boulton SJ. Replication blocking lesions present a unique substrate for homologous recombination. Embo J. 2007;26(14):3384–3396. doi: 10.1038/sj.emboj.7601766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LY, Recht J, Laurent BC. Chromatin remodeling and repair of DNA double-strand breaks. J Mol Histol. 2006;37(5–7):261–269. doi: 10.1007/s10735-006-9047-4. [DOI] [PubMed] [Google Scholar]
- Zheng H, Wang X, Warren AJ, Legerski RJ, Nairn RS, Hamilton JW, Li L. Nucleotide excision repair- and polymerase eta-mediated error-prone removal of mitomycin C interstrand cross-links. Mol Cell Biol. 2003;23(2):754–761. doi: 10.1128/MCB.23.2.754-761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z, Johnson RE, Haracska L, Prakash L, Prakash S, Benkovic SJ. Regulation of polymerase exchange between Poleta and Poldelta by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proc Natl Acad Sci U S A. 2008;105(14):5361–5366. doi: 10.1073/pnas.0801310105. [DOI] [PMC free article] [PubMed] [Google Scholar]