Abstract
BACKGROUND
The detection of circulating tumor cells (CTCs) in the peripheral blood of melanoma patients by quantitative real-time reverse-transcription PCR (qRT-PCR) analysis correlates with a poor prognosis. The assessment of CTCs from blood has been difficult because of a lack of a good monoclonal antibody (mAb) directed against surface cell antigens to capture melanoma cells.
METHODS
Blood was collected prospectively from 57 melanoma patients (43 test and 14 test-development cases) and 5 healthy donors. High molecular weight melanoma-associated antigen (HMW-MAA)-specific mAbs bound to immunomagnetic beads were used to isolate CTCs. mRNA and/or DNA were extracted from CTCs. Testing for the expression of a melanoma-associated gene panel (MLANA, MAGEA3, and MITF) with qRT-PCR and for the presence of BRAFmt (a BRAF gene variant encoding the V600E mutant protein) verified the beads-isolated CTCs to be melanoma cells. A peptide nucleic acid– clamping PCR assay was used for BRAFmt analysis.
RESULTS
Spiking of peripheral blood cells (PBCs) with melanoma cells showed that the beads-based detection assay can detect approximately 1 melanoma cell in 5 × 106 PBCs. qRT-PCR analysis detected MLANA, MAGEA3, and MITF expression in 19 (44%), 29 (67%), and 19 (44%) of the patients, respectively. At least one biomarker of the panel was positive in 40 (93%) of the 43 melanoma patients. BRAFmt was detected in 17 (81%) of the 21 assessed stage IV melanoma patients.
CONCLUSION
The assay of bead capture coupled with the PCR has utility for assessing CTCs in melanoma patients, which can then be characterized for both genomic and transcriptome expression.
The metastasis of melanoma cells to distant sites often portends a poor prognosis (1, 2 ), and detection of melanoma cells in the circulation is useful to assess disease progression (3). Previous studies have shown that the detection of melanoma cells in blood by direct isolation of mRNA without isolation of tumor cells is associated with a poor disease outcome (3–9) and is a predictor of subclinical disease. Furthermore, we and others have reported that serial monitoring of circulating tumor cells (CTCs)5 may be useful for predicting disease outcome for melanoma patients receiving adjuvant therapy (8–10).
To date, investigators have identified a limited number of biomarkers that are expressed by melanoma cells but are not detectable in healthy cells. We have developed a multimarker quantitative real-time reverse- transcription PCR (qRT-PCR) assay that uses specific prognostic biomarkers to detect melanoma cells in the blood and identify lymph node metastasis (9, 11, 12). This panel of genes includes MLANA6 (melan-A; also known as MART-1, melanoma antigen recognized by T cells 1), MAGEA3 (melanoma antigen family A, 3) (12, 13), and MITF (microphthalmia-associated transcription factor) (9). MLANA is a melanocyte-differentiation antigen that is frequently synthesized by melanoma cells (12, 14). MAGEA3 is commonly synthesized by malignant cells of different embryologic origin and is not detectable in healthy tissues except male germline cells and placenta (12, 13). MITF plays an important role in melanocyte development and melanoma growth (9, 15). These qRT-PCR markers are able to detect both occult metastatic melanoma cells in sentinel lymph nodes (12) and CTCs in the blood, demonstrating their prognostic utility for melanoma patients (3, 9, 10).
A mutant form of the BRAF (v-raf murine sarcoma viral oncogene homolog B1) gene (BRAFmt) encoding the V600E variant protein occurs at high frequency in melanomas (16). BRAF encodes a serine/threonine kinase downstream in the RAS-MAPK pathway that transduces regulatory signals from RAS through MAPKs (mitogen-activated protein kinases) (17, 18). BRAFmt has been detected in 50%–80% of metastatic lesions (16, 19). In addition, we have described the presence of circulating BRAFmt DNA in the serum of melanoma patients and demonstrated its clinical utility (19).
Direct RT-PCR analysis of blood permits transcriptome and phenotypic characterization of CTCs. To date, characterization of CTCs for abnormalities in genotypic and mRNA expression has not been well studied in melanoma patients. This situation is due in part to the difficulty of isolating CTCs with monoclonal antibodies (mAbs) capable of recognizing cell surface markers.
Characterizing the genotypic and phenotypic defects in CTCs would assist researchers in understanding the mechanisms that CTCs use to escape immune recognition and may suggest strategies for counteracting them. Therefore, in the present study we used high molecular weight melanoma-associated antigen (HMW-MAA) as a marker to develop a method for isolating CTCs from peripheral blood cells (PBCs). This cell surface antigen is highly expressed on melanoma cells in at least 85% of melanoma patients and is not detectable on healthy PBCs. We were able to obtain high-affinity mAbs that recognize distinct and spatially distant antigenic determinants of HMW-MAA and used a mixture of these HMW-MAA mAbs to capture CTCs.
Materials and Methods
MELANOMA CELL LINES
The human melanoma cell lines MA, MB, MC, MD, ME, MF, MG, MH, MI, MJ, MK, ML, and MM established and characterized at the John Wayne Cancer Institute were used for in vitro studies. Cells were grown in RPMI 1640 medium containing 100 mL/L heat-inactivated fetal calf serum and 10 mL/L penicillin–streptomycin (Gibco/Invitrogen) in a T75 (75-cm2) culture flask. Cells were harvested for mRNA analysis when they reached 70%–80% confluence, as previously described (20).
PATIENTS
All melanoma patients enrolled in this study signed consent forms for the use of blood samples and physical and medical histories. The study was carried out according to the guidelines set forth by the Saint John’s Health Center/John Wayne Cancer Institute Institutional Review Board. Disease stage was determined according to the American Joint Committee on Cancer (AJCC) and recorded at the time of blood draw. Blood was obtained from 14 stage IV melanoma patients for pilot experiments to optimize assays, and blood samples from 43 stage IV melanoma patients were used to validate assays. The study was conducted in a double-blind fashion. The investigators who performed the PCR assays did not know the patients’ disease status.
MONOCLONAL AND POLYCLONAL ANTIBODIES
Mouse mAbs 225.28, 763.74, TP61.5, VF1-TP41.2, and VF4-TP108, which recognize distinct and spatially distant determinants of HMW-MAA [as described previously (21, 22)], were purified from ascitic fluid by sequential precipitation with ammonium sulfate and caprylic acid (23). The purity of mAb preparations was assessed by SDS-PAGE analysis, and the activity of mAb preparations was monitored by testing with HMW-MAA–bearing melanoma cells in a binding assay. R-phycoerythrin–labeled F(ab′)2 fragments of goat antimouse IgG antibodies were purchased from Santa Cruz Biotechnology.
FLOW CYTOMETRIC ANALYSIS
Cells were incubated at 4 °C for 1 h with each HMW-MAA–specific mAb (1µg) or with an isotype-matched control mAb. Cells were washed twice with PBS containing 5 g/L BSA, incubated at 4 °C for an additional 30 min with an optimal amount of R-phycoerythrin–labeled F(ab′)2 fragments of goat antimouse IgG Abs, washed twice again, fixed in 40 g/L paraformaldehyde, and analyzed with a flow cytometer (FACSCalibur; BD Biosciences). Melanoma cells (104) were acquired for each sample. Debris, cell clusters, and dead cells were gated out by light-scatter assessment before single-parameter histograms were drawn. Data were analyzed with the aid of CellQuest software (BD Biosciences).
ISOLATION OF MELANOMA CELLS FROM PBCs WITH IMMUNOMAGNETIC BEADS
Blood samples (1 mL) were collected into sodium citrate–containing tubes, and the first several milliliters were discarded (10). Total cells in blood were collected with Purescript PBC lysis solution (Gentra Systems) according to the manufacturer’s instructions. CTCs were isolated from peripheral blood samples with immunomagnetic beads (CELLection™ Pan Mouse IgG Kit; Invitrogen) according to the manufacturer’s recommendations, with minor modifications. In brief, HMW-MAA–specific mouse mAbs 225.28, 763.74, TP61.5, VF1-TP41.2, and VF4-TP108 (3µg each) were added separately or in combination to cells purified from PBCs and resuspended in 4.5 mL PBS containing 1g/L BSA. Samples were incubated at 4 °C overnight with rotation on an RKDynal rotor (Dynal Biotech/Invitrogen). After 2 washes with this PBS/BSA solution, immunomagnetic beads (antimouse IgG–coated) were added to the samples, and incubation was continued at 4 °C for an additional 40 min with rotation on the RKDynal rotor. Samples were then placed in a Dynal MPC-6 magnetic device (Invitrogen) at 4 °C for 2 min. The fluid was removed by careful pipetting, and the beads were resuspended in the PBS/BSA solution. The beads were washed twice with this washing procedure and then resuspended in RPMI 1640 medium containing 10 mL/L fetal bovine serum. Cells were released from the beads by the addition of Releasing Buffer (Invitrogen) and a 20-min incubation on a mixing device. The mixture was then vigorously pipetted and placed in the Dynal MPC-6 magnet for 2 min. The medium containing the released CTCs was collected with a pipet.
RNA ASSAYS
Total RNA was extracted with Tri Reagent (Molecular Research Center) as described previously (10). RNA was quantified and assessed for purity by ultraviolet spectrophotometry and the RiboGreen assay (Molecular Probes/Invitrogen). Blood processing, RNA extraction, qRT-PCR assay setup, and post-PCR product analysis were carried out in separate designated rooms to prevent cross-contamination. Reverse-transcription reactions were performed with Moloney murine leukemia virus reverse transcriptase (Promega) with oligo( dT) primer. qRT-PCR assays were performed with the ABI 7900HT instrument (Applied Biosystems). Primer and probe sequences designed for qRT-PCR analyses have previously been described (3, 9, 11, 12). The fluorescence resonance energy transfer (FRET) probe sequences were 5′–FAM-CAGAACGTCACCACCACCT TATT-BHQ-1–3′ for MLANA, 5′–FAM-AGCTCCT GCCCACACTCCCGCCTGT-BHQ-1–3′ for MAGEA3, 5′–FAM-AGAGCACTGGCCAAAGAGAGGCA-BHQ-1–3′ for MITF, and 5′–FAM-CAGCAATGCCTCCTG CACCACCAA-BHQ-1–3′ for GAPDH (glyceraldehyde-3-phosphate dehydrogenase). Five microliters of cDNA synthesized from 250 ng total RNA were transferred to a well of a 96-well PCR plate (Fisher Scientific), along with each primer, probe, and custom iTaq Supermix (Bio-Rad Laboratories). After a precycling hold at 95 °C for 10 min, samples were amplified with specific numbers of PCR cycles for each marker: denaturation at 95 °C for 1 min; annealing for 1 min at 55 °C for GAPDH, at 58 °C for MAGEA3 and MITF, and at 59 °C for MLANA; and extension at 72 °C for 1 min. We generated a calibration curve with threshold cycle values for 9 serial dilutions of plasmid templates (100 to 108 copies) (3). The threshold cycle of each sample was interpolated from the calibration curve, and the number of mRNA copies was calculated by the iCycler iQ Real-Time PCR Detection System software (Bio-Rad). Thirteen melanoma lines and blood samples from 49 healthy donors were used to optimize the assay, as has previously been described (3, 9, 10). We performed each assay at least twice, and each assay included positive controls (melanoma lines), negative mRNA controls (PBCs), and PCR reagent controls without template. GAPDH, a so-called housekeeping gene, was used as an internal control to verify the integrity of the RNA and reverse-transcription efficiency. Any sample with an inadequate quantity of GAPDH mRNA was excluded from the study. The mean mRNA copy number calculated was used for analysis (10).
DNA EXTRACTION AND BRAF DNA MUTATION
We collected blood samples for the BRAFmt study from a subset of 21 melanoma patients (AJCC stage IV) paired with qRT-PCR and isolated CTCs as described above. Genomic DNA was extracted from CTCs (24), and BRAFmt was assessed as previously described (19, 25). In brief, primers were designed to amplify exon 15 of the BRAF gene, which includes the mutation hot spot that encodes the V600E variant. The peptide nucleic acid (PNA) (Applied Biosystems) was designed to clamp the hot spot on the wild-type template and block the wild-type template from being amplified in the PCR (19, 25). A FRET dual-labeled locked nucleic acid (LNA) probe (Proligo/Sigma–Aldrich) was designed to recognize and hybridize specifically to the T-to-A mutation at the sequence encoding V600E, because this mutation is the most frequently seen for the BRAF gene at this hot spot. The design of a second FRET DNA probe (BioSource/Invitrogen) was based on using sequences adjacent to the LNA probe and avoiding the hot spot. This probe was used to amplify and quantify the total number of DNA templates, both wild-type BRAF and BRAFmt, in the PCR. Quantitative PCR (qPCR) with both the PNA clamp and the FRET LNA probe was used for mutation detection.
BRAF qPCR used the following primers and probe: forward, 5′–CCTCACAGTAAAAATAGGTG–3′; reverse, 5′–ATAGCCTCAATTCTTACCA–3′; LNA, 5′–CTACAGAGAAATCTCGAT-BHQ-1–3′; and PNA, 5′–CTACAGTGAAATCTCG–3′. The iCycler iQ Real-Time PCR Detection System (Bio-Rad) was used for the PCR assay. CTC genomic DNA (20 ng) was amplified by qPCR in a 20-µL reaction volume containing each PCR primer, LNA, PNA, deoxynucleoside triphosphates, MgCl2, PCR buffer, and AmpliTaq Gold Polymerase (Applied Biosystems). The PCR conditions were 50 cycles of 94 °C for 1 min, 72 °C for 50 s, 53 °C for 50 s, and 72 °C for 1 min. Each sample was assayed in triplicate; control reactions included PCRs with templates derived from the appropriate positive and negative cell lines and a PCR reagent control without template.
DNA from the MA cell line was established as the reference for measuring units of BRAFmt target DNA (heterozygous). The amount of target mutant DNA in 1µg/mL MA genomic DNA was arbitrarily established as 1 U of BRAFmt. qPCR results for samples were normalized to this reference to quantify the relative units of BRAFmt in all of the samples. All PCRs for mutant-sequence analysis were done in triplicate, and the median was used for data analysis.
Representative BRAFmt and wild-type BRAF genes from V600E melanoma tumors were sequenced to confirm the accuracy of the PCR assay, as has previously been described (19, 25). The BRAF PCR used primers 5′–TGTTTTCCTTTACTTACTACACCTCA–3 (forward) and 5′–AGCATCTCAGGGCCAAAAAT–3′ (reverse). The PCR products were purified with the QIAquick PCR Purification Kit (Qiagen) and then sequenced directly at 58 °C with the GenomeLab DTCS Quick Start Kit (Beckman Coulter) according to the manufacturer’s instructions. Products of dye-termination reactions were assessed by capillary array electrophoresis on a CEQ 8000XL Genetic Analysis System (Beckman Coulter).
Results
HMW-MAA PROTEIN EXPRESSION ON MELANOMA CELLS
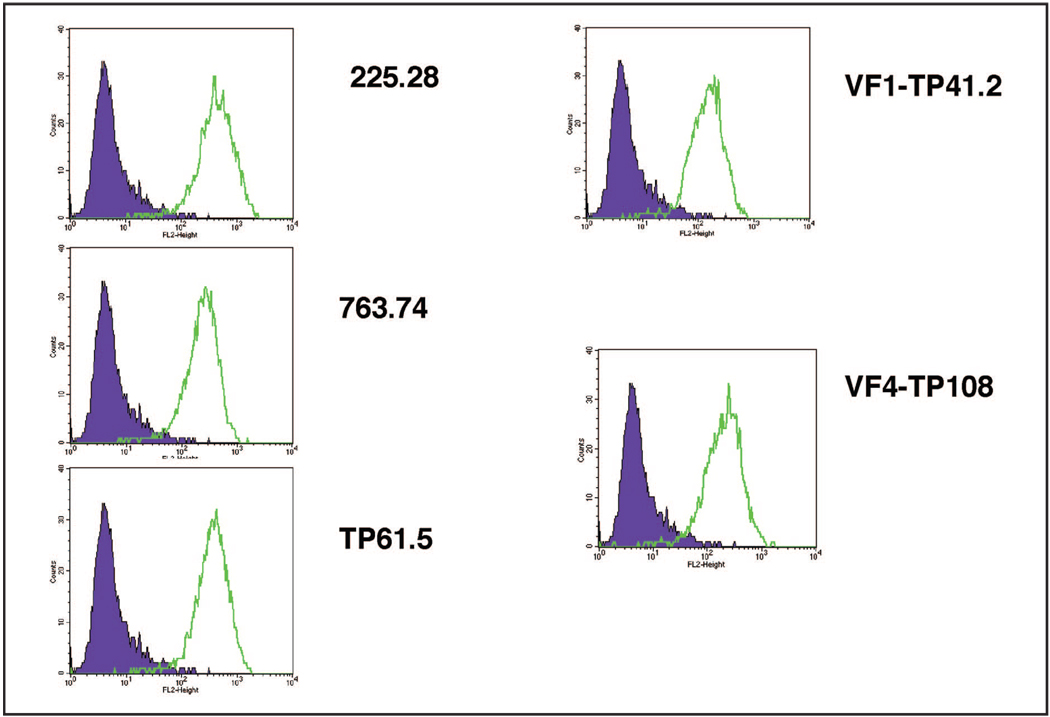
The expression of HMW-MAA protein on cells of melanoma lines was initially screened by flow cytometric analysis with HMW-MAA–specific mAbs 225.28, 763.74, TP61.5, VF1-TP41.2, and VF4-TP108 to verify the sensitivity and specificity of each mAb (Fig. 1). HMW-MAA expression was detected on all melanoma lines with all HMW-MAA–specific mAbs tested. The individual mAbs recognize different epitopes on HMW-MAA. We subsequently used a mixture of all 5 HMW-MAA–specific mAbs for this assay.
Figure 1.
HMW-MAA protein expression on melanoma cell lines by flow cytometry analysis with each of the HMW-MAA specific mAbs (225.28, 763.74, TP61.5, VF1-TP41.2, and VF4-TP108) and an isotype-matched control Ab.
ISOLATION OF MELANOMA CELLS MIXED WITH PBCs
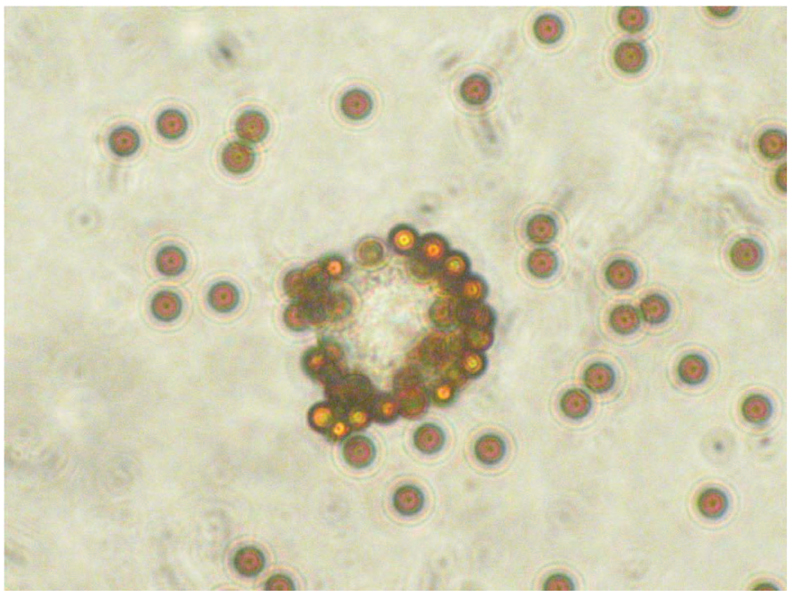
We conducted pilot experiments to optimize conditions for isolating CTCs from peripheral blood with HMW-MAA mAbs and immunomagnetic beads (Fig. 2). We used the immunomagnetic beads to assess captured CTCs via both an indirect technique and a direct technique to determine the optimal assay. In brief, the indirect technique entails first incubating melanoma cells with HMW-MAA mAbs; melanoma cells labeled with HMW-MAA mAbs are then bound and captured by the immunomagnetic beads. In the direct technique, HMW-MAA mAbs are first incubated with immunomagnetic beads; melanoma cells are then bound and captured by immunomagnetic beads with HMW-MAA mAbs. Melanoma cells were added to healthy donor PBCs at ratios of 1–103 per 5 × 106 PBCs. qRT-PCR analysis successfully detected mixtures of 1–5 melanoma cells in 5 × 106 healthy donor PBCs by the indirect technique but required at least 100 melanoma cells in 5 × 106 PBCs with the direct technique. Thus, the indirect technique was better than the direct technique for separating melanoma cells from healthy donor PBCs. A mixture of HMW-MAA–specific mAbs (225.28, 763.74, TP61.5, VF1-TP41.2, and VF4-TP108) was used to confirm the improvement in tumor cell capture. qRT-PCR analysis was always successful in detecting approximately 1 melanoma cell mixed with 5 × 106 PBCs in quadruplicate experiments that used the mAb mixture but was not always successful when a single HMW-MAA mAb was used. For in vitro studies, the mixture of HMW-MAA mAbs was better than a single HMW-MAA mAb for isolating melanoma cells from mixtures with healthy donor PBCs. In a pilot study that used samples from 14 melanoma patients, we determined that the optimal and most efficient amount of mAbs to use was 3 µg of each mAb. The optimal incubation time with CTCs and primary antibody was determined to be overnight.
Figure 2.
HMW-MAA–positive cells were isolated from suspensions of healthy donor PBCs and melanoma cells by means of an HMW-MAA mAb mixture and immunomagnetic beads.
qRT-PCR DETECTION LIMIT FOR MELANOMA CELLS MIXED WITH PBCs
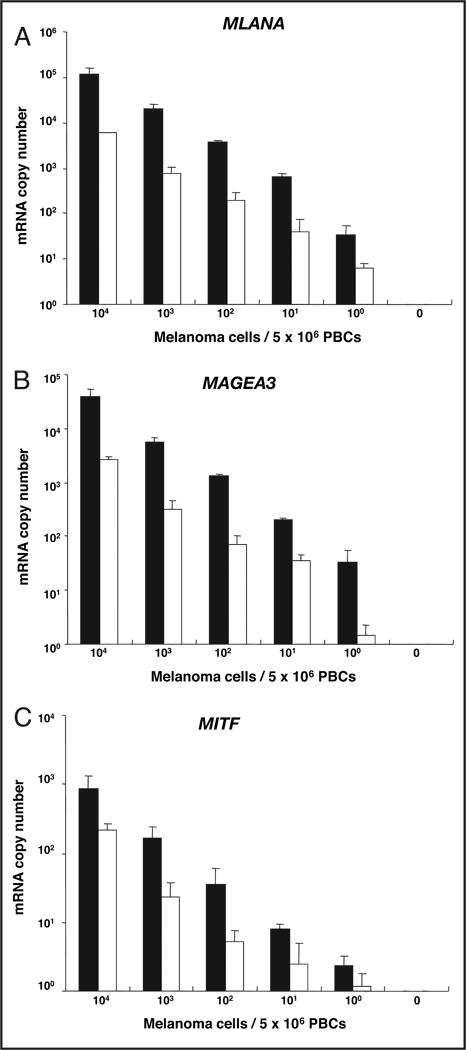
To assess the limit of detection for melanoma cells in blood with the optimized immunomagnetic bead method (i.e., indirect technique, overnight incubation with a mixture of all 5 HMW-MAA–specific mAbs), we performed immunomagnetic bead separation with a mixture of HMW-MAA mAbs and a qRT-PCR analysis of melanoma cells that we serially diluted with healthy donor PBCs. In the assays, we added serial dilutions of melanoma cells (104, 103, 102, 10, 1, and 0 cells) expressing MAGEA3, MLANA, and MITF to 5 × 106 donor-derived PBCs and assessed the qRT-PCR assay’s ability to detect each marker. This in vitro assay was performed both with and without the immunomagnetic bead technique, but always with the HMW-MAA–specific mAbs added. The assay was repeated multiple times to validate the reproducibility and robustness of the assay system. This in vitro model system was used to mimic the detection of CTCs in blood. Use of the beads permitted the detection of MAGEA3, MLANA, and MITF mRNAs from approximately 1 melanoma cell mixed with 5 × 106 PBCs (Table 1). The number of mRNA copies detected gradually decreased with serial dilution of the melanoma cells (Fig. 3). The number of mRNA copies for individual markers varied with the cell line, as expected. This heterogeneity in mRNA production is also to be expected in blood samples from patients. All biomarkers were positive for detecting 10 melanoma cells mixed with 5 × 106 PBCs in all 4 experiments (Table 1). Detection frequencies for MAGEA3, MLANA, and MITF transcripts from approximately 1 melanoma cell mixed with 5 × 106 PBCs were 100%, 100%, and 75%, respectively. We performed the same experiments without immunomagnetic beads to assess the detection of melanoma cells in blood via direct isolation of mRNA from blood (Fig. 3). The numbers of mRNA copies obtained for each marker in samples processed with immunomagnetic beads were higher for all serial-dilution experiments than for samples processed without beads. The numbers of copies of MAGEA3 and MLANA transcripts obtained for bead-processed samples were the same as those obtained for samples without the bead-capture method when approximately 100 melanoma cells were mixed with 5 × 106 PBCs.
Table 1.
qRT-PCR detection of melanoma markers in serially diluted melanoma cell lines (n = 4).a
| Cell number | ||||||
|---|---|---|---|---|---|---|
| Marker | 104 | 103 | 102 | 101 | 100 | 0 |
| GAPDH | 4 | 4 | 4 | 4 | 4 | 0 |
| MLANA | 4 | 4 | 4 | 4 | 4 | 0 |
| MAGEA3 | 4 | 4 | 4 | 4 | 4 | 0 |
| MITF | 4 | 4 | 4 | 4 | 3 | 0 |
Data represent the number of times the indicated marker was detected (of 4 experiments) at the indicated numbers of melanoma cells mixed with 5 × 106 PBCs. An HMW-MAA mAb mixture was used in an indirect assay with immunomagnetic beads. GAPDH is a housekeeping gene used as an internal control.
Figure 3.
qRT-PCR detection limit for melanoma cells (104, 103, 102, 10, 1, and 0 cells) mixed with 5 × 106 PBCs from healthy donors.
Assay for MLANA (A), MAGEA3 (B), and MITF (C) with (filled columns) and without (open columns) the use of immunomagnetic beads with bound anti–HMW-MAA mAbs. Data are presented as the mean (SD).
ASSESSMENT OF CTCs IN PBCs
We performed immunomagnetic beads– based qRT-PCR assays with samples from healthy blood donors and obtained no positive signals for any of the 3 markers. These results demonstrated the specificity of the assay. In blood samples from 43 melanoma patients, MAGEA3, MLANA, and MITF mRNAs were detected in 67%, 44%, and 44% of the patients, respectively, when we used the assay with optimized CTC isolation (Table 2). At least one of the markers was detected in 40 (93%) of the 43 patients investigated when the panel of all 3 biomarkers was assessed; this detection level was higher than that obtained for any single biomarker. This analysis demonstrated the sensitivity and efficiency of the assay. The combination of the capture of melanoma CTCs with immunomagnetic beads containing HMW-MAA mAbs and qRT-PCR analysis constitutes a unique approach that integrates 2 different techniques to assess CTCs with unparalleled specificity.
Table 2.
Multimarker mRNA expression in blood samples from melanoma patients (n = 43).
| Positive expression, n (%) |
|
|---|---|
| Melanoma marker | |
| MLANA | 19 (44) |
| MAGEA3 | 29 (67) |
| MITF | 19 (44) |
| No. of markers detected | |
| 0 | 3 (7) |
| 1 | 19 (44) |
| 2 | 15 (35) |
| 3 | 6 (14) |
| 4 | 0 (0) |
| ≥1 | 40 (93) |
ASSESSMENT OF BRAF MUTATION IN CTCs
In an in vitro model, the assay detected BRAFmt from 1–5 MA cells in mixtures with 5 × 106 healthy donor-derived PBCs. BRAFmt was assessed in isolated CTCs from melanoma patients. Genomic DNA was extracted from 21 melanoma patients (AJCC stage IV), and BRAFmt was detected in 17 (81%) of these patients. The study demonstrated that HMW-MAA–positive melanoma cells frequently have BRAFmt. The analysis also demonstrated that BRAFmt is frequently detected in CTCs with this assay.
Discussion
To characterize CTCs in PBCs, we combined immunomagnetic-bead isolation with DNA and RNA PCR assays. We previously reported that qRT-PCR–based methods can detect CTCs and that positive signals for markers are associated with metastatic disease and poor prognosis in melanoma patients (3, 9, 10). The specificity of these qRT-PCR assays with respect to the cell type being assessed is controversial, however, primarily because it is not clear whether all melanoma-associated biomarkers actually are derived from CTCs in the blood of melanoma patients. The use of immunomagnetic-bead capture of melanoma cells in our approach exploits the selective expression of HMW-MAA on melanoma cells (22).
HMW-MAA has been shown to be heterogeneous in the expression of antigenic determinants recognized by mAbs. To minimize the occurrence of false-negative results caused by differential expression of HMW-MAA determinants on melanoma cells and to increase the detection capability of our isolation procedure, we used a mixture of mAbs that recognize distinct and spatially distant determinants of HMW-MAA to isolate CTCs from the PBCs of melanoma patients. The use of an HMW-MAA mAb mixture enabled us to maximize the exclusion of healthy blood cells and other types of circulating cells and to maximize the isolation of CTCs (i.e., HMW-MAA–positive melanoma cells). The immunomagnetic beads– based assay is capable of capturing intact HMW-MAA–positive melanoma cells. The identity of these CTCs as melanoma cells was established by subsequent analysis of 3 recognized melanoma-associated mRNA biomarkers frequently found in melanomas. The genes encoding these mRNA biomarkers do not have directly correlated functions (3, 9, 10).
Our in vitro model demonstrated that our assay can detect approximately 1 melanoma cell diluted into 5 × 106 PBCs from healthy donors. This assay provides a new opportunity not only to improve melanoma diagnosis but also to identify patients who have spreading systemic disease. The approach of coupling the capture of CTCs with the PCR permits improved characterization of the CTCs in blood both for the expression of tumor biomarkers and for genomic aberrations.
Our previous studies found that metastatic melanoma tumors were heterogeneous for the expression of melanoma-associated markers (3, 10–12). For the clinical application, we selected 3 mRNA biomarkers for detecting CTCs in the blood of melanoma patients. Use of a combination of melanoma-associated markers in the qRT-PCR assay can circumvent the problem of heterogeneity in mRNA expression of individual markers. We previously demonstrated that melanoma-associated genes are not expressed at 100% in all melanoma cells (3).
The MAGEA3 transcript had the highest overall detection rate in PBCs from melanoma patients, followed by MLANA and MITF transcripts. Differences between results obtained for cell lines and blood samples may be related to the physiology of cells in the circulation or to the phenotype of the clone shed into the blood (26). Detection rates of individual biomarkers with qRT-PCR assays after immunomagnetic beads– based capture was slightly higher for some biomarkers in paired patient blood samples than for individual biomarkers assessed by qRT-PCR without bead capture.
We previously reported that the presence of a BRAF mutation in circulating DNA in serum may have clinical utility in predicting tumor response and disease outcome (19); however, whether the BRAF mutation in serum DNA actually derives from metastatic melanoma, tumors, or CTCs remains to be determined. Our results demonstrated a high frequency of BRAFmt in cells captured with HMW-MAA mAbs. BRAFmt analysis in the present assay may be useful as a melanoma-associated biomarker. Our description of this assay for a surrogate biomarker of CTCs in the blood of melanoma patients is the first such report. The BRAF mutation encoding the V600E variant is specific to tumor cells, particularly in melanoma, although nevi cells have been reported to possess the V600E variant (27). This mutation can be found in up to 80% of patients with metastatic cutaneous melanomas. A recent study found that circulating non-small cell lung cancer cells were captured and that the cells featured epidermal growth factor receptor mutation in 11 of 12 patients (28).
Currently, melanoma prognosis is based on the tumor and demographic and prognostic factors of the host, but ongoing dynamic factors of tumor metastasis are also important. Our findings demonstrate the potential clinical usefulness of an immunomagnetic beads– based assay for detecting CTCs in the blood of melanoma patients. Future studies involving serial blood analysis and long-term follow-up of patients may be needed for a more detailed assessment of the clinical utility of this assay.
Acknowledgments
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, or preparation or approval of manuscript.
Footnotes
Nonstandard abbreviations: CTC, circulating tumor cell; qRT-PCR, quantitative real-time reverse-transcription PCR; BRAFmt, BRAF gene variant encoding the V600E mutant protein; MAPK, mitogen-activated protein kinase; mAb, monoclonal antibody; HMW-MAA, high molecular weight melanoma-associated antigen; PBC, peripheral blood cell; AJCC, American Joint Committee on Cancer; FRET, fluorescence resonance energy transfer; PNA, peptide nucleic acid; LNA, locked nucleic acid; qPCR, quantitative PCR.
Human genes: MLANA, melan-A (also known as MART-1, melanoma antigen recognized by T cells 1); MAGEA3, melanoma antigen family A, 3; MITF, microphthalmia-associated transcription factor; BRAF, v-raf murine sarcoma viral oncogene homolog B1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Authors’ Disclosures of Potential Conflicts of Interest: Upon manuscript submission, all authors completed the Disclosures of Potential Conflict of Interest form. Potential conflicts of interest:
Employment or Leadership: None declared.
Consultant or Advisory Role: None declared.
Stock Ownership: None declared.
Honoraria: None declared.
Research Funding: D.S.B. Hoon, NIH.
Expert Testimony: None declared.
References
- 1.Balch CM, Buzaid AC, Soong SJ, Atkins MB, Cascinelli N, Coit DG, et al. Final version of the American Joint Committee on Cancer staging system for cutaneous melanoma. J Clin Oncol. 2001;19:3635–3648. doi: 10.1200/JCO.2001.19.16.3635. [DOI] [PubMed] [Google Scholar]
- 2.Greene FL, Page DL, Fleming ID, Fritz A, Balch CM, Haller DG, Morrow M. Cancer staging manual. 6th ed. New York: Springer; 2002. [Google Scholar]
- 3.Koyanagi K, Kuo C, Nakagawa T, Mori T, Ueno H, Lorico AR, Jr, et al. Multimarker quantitative real-time PCR detection of circulating melanoma cells in peripheral blood: relation to disease stage in melanoma patients. Clin Chem. 2005;51:981–988. doi: 10.1373/clinchem.2004.045096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mellado B, Gutierrez L, Castel T, Colomer D, Fontanillas M, Castro J, Estape J. Prognostic significance of the detection of circulating malignant cells by reverse transcriptase-polymerase chain reaction in long-term clinically disease-free melanoma patients. Clin Cancer Res. 1999;5:1843–1848. [PubMed] [Google Scholar]
- 5.Palmieri G, Strazzullo M, Ascierto PA, Satriano SM, Daponte A, Castello G Melanoma Cooperative Group. Polymerase chain reaction-based detection of circulating melanoma cells as an effective marker of tumor progression. J Clin Oncol. 1999;17:304–311. doi: 10.1200/JCO.1999.17.1.304. [DOI] [PubMed] [Google Scholar]
- 6.Pantel K, Cote RJ, Fodstad O. Detection and clinical importance of micrometastatic disease. J Natl Cancer Inst. 1999;91:1113–1124. doi: 10.1093/jnci/91.13.1113. [DOI] [PubMed] [Google Scholar]
- 7.Taback B, Morton DL, O’Day SJ, Nguyen DH, Nakayama T, Hoon DS. The clinical utility of multimarker RT-PCR in the detection of occult metastasis in patients with melanoma. Recent Results Cancer Res. 2001;158:78–92. doi: 10.1007/978-3-642-59537-0_8. [DOI] [PubMed] [Google Scholar]
- 8.Wascher RA, Morton DL, Kuo C, Elashoff RM, Wang HJ, Gerami M, Hoon DS. Molecular tumor markers in the blood: early prediction of disease outcome in melanoma patients treated with a melanoma vaccine. J Clin Oncol. 2003;21:2558–2563. doi: 10.1200/JCO.2003.06.110. [DOI] [PubMed] [Google Scholar]
- 9.Koyanagi K, O’Day SJ, Gonzalez R, Lewis K, Robinson WA, Amatruda TT, et al. Microphthalmia transcription factor as a molecular marker for circulating tumor cell detection in blood of melanoma patients. Clin Cancer Res. 2006;12:1137–1143. doi: 10.1158/1078-0432.CCR-05-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koyanagi K, O’Day SJ, Gonzalez R, Lewis K, Robinson WA, Amatruda TT, et al. Serial monitoring of circulating melanoma cells during neoadjuvant biochemotherapy for stage III melanoma: outcome prediction in a multicenter trial. J Clin Oncol. 2005;23:8057–8064. doi: 10.1200/JCO.2005.02.0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeuchi H, Kuo C, Morton DL, Wang HJ, Hoon DS. Expression of differentiation melanoma-associated antigen genes is associated with favorable disease outcome in advanced-stage melanomas. Cancer Res. 2003;63:441–448. [PubMed] [Google Scholar]
- 12.Takeuchi H, Morton DL, Kuo C, Turner RR, Elashoff D, Elashoff R, et al. Prognostic significance of molecular upstaging of paraffin-embedded sentinel lymph nodes in melanoma patients. J Clin Oncol. 2004;22:2671–2680. doi: 10.1200/JCO.2004.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyashiro I, Kuo C, Huynh K, Iida A, Morton D, Bilchik A, et al. Molecular strategy for detecting metastatic cancers with use of multiple tumor-specific MAGE-A genes. Clin Chem. 2001;47:505–512. [PubMed] [Google Scholar]
- 14.Kawakami Y, Eliyahu S, Delgado CH, Robbins PF, Rivoltini L, Topalian SL, et al. Cloning of the gene coding for a shared human melanoma antigen recognized by autologous T cells infiltrating into tumor. Proc Natl Acad Sci U S A. 1994;91:3515–3519. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis IJ, Kim JJ, Ozsolak F, Widlund HR, Rozenblatt-Rosen O, Granter SR, et al. Oncogenic MITF dysregulation in clear cell sarcoma: defining the MiT family of human cancers. Cancer Cell. 2006;9:473–484. doi: 10.1016/j.ccr.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 16.Shinozaki M, Fujimoto A, Morton DL, Hoon DS. Incidence of BRAF oncogene mutation and clinical relevance for primary cutaneous melanomas. Clin Cancer Res. 2004;10:1753–1757. doi: 10.1158/1078-0432.ccr-1169-3. [DOI] [PubMed] [Google Scholar]
- 17.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 18.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 19.Shinozaki M, O’Day SJ, Kitago M, Amersi F, Kuo C, Kim J, et al. Utility of circulating B-RAF DNA mutation in serum for monitoring melanoma patients receiving biochemotherapy. Clin Cancer Res. 2007;13:2068–2074. doi: 10.1158/1078-0432.CCR-06-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takeuchi H, Fujimoto A, Tanaka M, Yamano T, Hsueh E, Hoon DS. CCL21 chemokine regulates chemokine receptor CCR7 bearing malignant melanoma cells. Clin Cancer Res. 2004;10:2351–2358. doi: 10.1158/1078-0432.ccr-03-0195. [DOI] [PubMed] [Google Scholar]
- 21.Goto Y, Ferrone S, Arigami T, Kitago M, Tanemura A, Sunami E, et al. Human high molecular weight-melanoma-associated antigen: utility for detection of metastatic melanoma in sentinel lymph nodes. Clin Cancer Res. 2008;14:3401–3408. doi: 10.1158/1078-0432.CCR-07-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campoli MR, Chang CC, Kageshita T, Wang X, McCarthy JB, Ferrone S. Human high molecular weight-melanoma-associated antigen (HMW-MAA): a melanoma cell surface chondroitin sulfate proteoglycan (MSCP) with biological and clinical significance. Crit Rev Immunol. 2004;24:267–296. doi: 10.1615/critrevimmunol.v24.i4.40. [DOI] [PubMed] [Google Scholar]
- 23.Temponi M, Kageshita T, Perosa F, Ono R, Okada H, Ferrone S. Purification of murine IgG monoclonal antibodies by precipitation with caprylic acid: comparison with other methods of purification. Hybridoma. 1989;8:85–95. doi: 10.1089/hyb.1989.8.85. [DOI] [PubMed] [Google Scholar]
- 24.Spugnardi M, Tommasi S, Dammann R, Pfeifer GP, Hoon DS. Epigenetic inactivation of RAS association domain family protein 1 (RASSF1A) in malignant cutaneous melanoma. Cancer Res. 2003;63:1639–1643. [PubMed] [Google Scholar]
- 25.Kim J, Giuliano AE, Turner RR, Gaffney RE, Umetani N, Kitago M, et al. Lymphatic mapping establishes the role of BRAF gene mutation in papillary thyroid carcinoma. Ann Surg. 2006;244:799–804. doi: 10.1097/01.sla.0000224751.80858.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoon DS, Wang Y, Dale PS, Conrad AJ, Schmid P, Garrison D, et al. Detection of occult melanoma cells in blood with a multiple-marker polymerase chain reaction assay. J Clin Oncol. 1995;13:2109–2116. doi: 10.1200/JCO.1995.13.8.2109. [DOI] [PubMed] [Google Scholar]
- 27.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 28.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]