Abstract
The vast majority of clinical tissue samples are formalin-fixed and paraffin-preserved. This type of preservation has been considered an obstacle to protein extraction from these tissues. However, these are the very tissue samples that have associated patient histories, diagnoses and outcomes — ideal samples in the quest to translate bench research into clinical applications. Thus, until recently, these valuable specimens have been unavailable for proteomic analysis.
Over the last decade, researchers have been exploring efficient methods to undo protein cross-linking caused by standard tissue fixatives and extract proteins from archived tissue specimens. These methods have been applied in different clinical proteomic studies. In this report, we attempt to review the development of these techniques, summarize the proteomic findings, and discuss the impact on future clinical proteomics.
Keywords: proteomics, protein extraction, fixed tissue, paraffin, formalin, Hollandes, laser capture microdissection
INTRODUCTION
Proteins are known to play key roles in biologic disease processes. The medical community is interested in proteins both as biomarkers and as therapeutic targets for disease. The field of proteomics has blossomed by characterizing and quantifying proteins from biologic tissue samples. Initially, proteomic assessment could only be performed on fresh frozen tissue samples, since standard histologic fixatives, like formalin, were known to crosslink proteins. Frozen tissue, however, is difficult and costly to preserve and store; therefore, the vast majority of clinical tissue samples are formalin-fixed and paraffin-embedded (FFPE). This type of preservation is the gold standard for hospital pathology departments and clinical labs. These stored, clinical FFPE tissues are those with associated data regarding patient characteristics, disease characteristics, prognosis, and outcome — incredibly valuable from the standpoint of translating tissue protein expression into clinical applications.
Until recently, it was believed that the formalin-induced protein cross-links would obstruct methods of protein evaluation, with immunohistochemistry (IHC) being the exception. Recently, a growing number of research groups have developed and described proteomic evaluation of formalin-fixed, paraffin-embedded tissues. This has been heralded as an important step in providing the starting material that enables us to enrich our understanding of human disease. This review examines the main methods of protein assessment and the recent developments in the evaluation of fixed, paraffinized tissues using these methods.
METHODS OF PROTEIN ANALYSIS
A variety of methods have been used to characterize proteins after their extraction from fixed tissue. The procedures and materials used for protein extraction are inherently molded by the method of subsequent analysis.
Immmunohistochemistry (IHC), a technique first described in the 1940s, is used to ascertain the location and distribution of a specific protein in tissue. After cell membranes are disrupted via detergent treatment, an antibody to a specific protein is applied to the tissue. Either that antibody, or a secondary antibody, is labeled or stained to enable protein visualization (Coons et al., 1941).
Western Blotting, also known as immunoblotting, involves the separation of the protein(s) via sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and subsequent immobilization of those proteins on a nitrocellulose membrane. Protein detection on the membrane is identified via antibody binding (Laemmli 1970, Towbin et al., 1979, Burnette 1981).
Mass Spectrometry (MS) — based proteomics enables the measurement of complex protein mixtures. In general, the protein mixture is first digested into peptides, followed by chromatographic separation and mass spectrometry analysis. Computational analyses of the obtained spectra are then performed and result in identification of peptides and their associated proteins. To pursue maximal peptide identification in samples with typically small (1-10ug) amounts of protein, a sample can be separated by multi-dimensional chromatography before MS analysis. While the technology involved in liquid chromatography-tandem mass spectrometry (LC-MS/MS) is much more complex than IHC or Western Blotting, no a priori knowledge of a sample’s proteome is required. Also, every protein in the sample can be identified at once. This makes LC-MS/MS ideal for proteome characterization of biologically interesting lesions. To date, ESI (Electrospray Ionization) LC-MS/MS has become the predominant method for protein analysis from FFPE tissues. Variants of the LC technique that increase sample separation (reverse-phase liquid chromatography, RPLC) and use smaller quantities of starting material (nano-) have also been developed. In addition to ESI, MALDI (matrix assisted laser desorption/ionization) has also recently been explored in FFPE tissues (Lemaire 2007, Ronci et al., 2008). One group has met success with O18 labeling (Zang 2004).
FORMALIN FIXATION
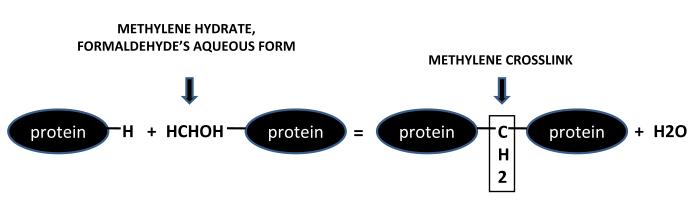
It is believed that when tissue is fixed, the formalin adds methylene hydrate groups to the side chains of the amino acids and this, in turn, leads to methylene bridge formation -- the basis for protein cross-linking (Hopwood et al., 1988, Rait et al., 2004) (see Figure (1)).
Figure 1.
Formalin-bound protein reacts with another protein molecule to form methylene bridges.
The cross-links can lead to polymer formation and masking of the native proteins from identification by standard methods. For this reason, many scientific methods of protein study have required the use of unfixed tissue. The cross-links induced by formalin can be cleaved through heating; however, the native conformation of a peptide/protein can also be influenced by pH, hydrophobic and ionic forces. Thus, approximation of a native non-fixed protein/peptide is influenced not only by the removal of cross-links, but also by the solutions in which they sit; salt content, denaturing agents, and pH can all influence protein chemistry and discovery. Moreover, the buffers used for reconstituting fixed-tissue may not be compatible with the conditions required for impending experiments, such as proteomics. In order for formalin fixed tissues, with their associated clinical data, to be used for protein molecular studies, these issues needed to be resolved.
The studies reviewed here set out to do just that. Table 1 summarizes their efforts.
Table 1.
Comparison of Fixed Tissue Proteomic Studies, organized chronologically
| CELL COUNT |
GRAMS PROTEIN |
BUFFER | Mass Spectrometer |
PEPTIDES | PROTEINS | PROTEINS ID’D BY MULT PEPTIDES |
|
|---|---|---|---|---|---|---|---|
| Hood ’05 - Cancer |
200,000 cells |
Not noted | Liquid Tissue |
either a linear ion trap (LTQ, Thermo Electron, San Jose, CA) or a hybrid LIT-FT- ICR MS. |
>2200 | 1156 | 282 |
| Crockett ‘05 |
10,000,000 cells |
600ug | RIPA | LCQ Deca XP ion trap (Thermo Electron, San Jose, CA, USA) |
10,043 | 324 | 324 |
| Palmer -Toy ‘05 |
Not noted |
Not noted | 2% SDS/100mM NaHCO3/ 20mM DTT |
LCQ DECA XP plus Proteome X workstation (Thermo Electron Corporation, San Jose, CA). |
412 | 123 | 42 |
| Shi ‘06 | Not noted |
1.67 ug/uL | 20 mM Tris- HCl buffer (pH 7 or 9) w/2%SDS |
LTQ (Thermo Finnigan San Jose, CA) |
4074 (avg, 2 samples) |
3259 (avg, 2 samples) |
1896 |
| Hwang ‘07 |
675,000 cells (cells in 10 spots ×6 × 2 arrays) |
82 ug | 100mM ammonium bicarbonate |
LTQ-linear ion- trap, Finnigan |
12,631 | 428 | 361 |
| Guo ‘07 |
100,000 cells |
Not noted | 20mM Tris, 2% SDS |
LTQ linear ion trap (LTQ, Thermo- Finnigan; San Jose, CA) |
11,398 | 2265 | Not noted |
| Jiang ‘07 |
Not noted |
10.9 ug/uL | 40mM Tris, 6 M guanidine- HCl, and 65 mM DTT, pH 8.2 |
LTQ linear ion trap (Thermo, San Jose, CA) |
3005 | 827 | 470 |
| Patel ‘08 — WD HNSCC |
80,000 cells per lesion type |
Not noted | Liquid Tissue |
LTQ linear ion trap (Thermo Electron, San Jose, CA) |
1400-1800 | 323-376 | 323-376 |
USE OF FIXED TISSUE FOR SIMPLE PROTEIN ASSESSMENTS
In 1998, Ikeda et al. described the extraction of proteins from FFPE tissue (Ikeda et al, 1998). A fine needle was used to macrodissect 5mm2 areas of human colorectal cancer, adenoma, and normal tissue from 50um thick tissue sections in 6 patients. Results were compared with fresh frozen tissue using Western Blotting. A Radio-Immuno Precipitation Assay (RIPA) extraction buffer containing 2% sodium dodecyl sulfate (SDS) was utilized, and lysates were heated at 100C ×20min, followed by 60C × 1hr. The average amount of protein extracted was 121.5ug. Western Blotting identified several types of tissue proteins, and noted common proteins between fresh frozen and FFPE tissue.
Ahram et al. followed in 2003 with MS analysis of ethanol-fixed, paraffin-embedded tissues, and compared this with FFPE and frozen tissue (Ahram et al., 2003). Laser capture microdissection (LCM) was used to separate 50,000-70,000 prostate epithelial cells from both normal and adenocarcinoma specimens. Proteins were extracted using a 40mM Tris-HCl buffer, and separated using 2-dimensional gel electrophoresis. Trypsin was added to protein spots for digestion, and MS/MS analysis was performed on the resultant peptides. While frozen tissue yielded a greater quantity of protein (40-80ug protein: 1-1.8ug/mm2 tissue; 550 protein spots), they were able to identify protein in the ethanol-fixed paraffin-embedded tissue (20-40ug protein: 0.7-1.4ug/ mm2 tissue; protein spots). Unfortunately, they found almost no protein - almost no visible 2DE protein spots to digest and use for MS analysis - in the FFPE tissue. So while this showed that ethanol-fixed tissue could be used for MS analysis, it was unable to solve the riddle of formalin fixation and MS analysis.
Previous work in antigen retrieval assays had suggested that high temperatures and/or denaturing reagents, such as urea or SDS, could help break the cross-link bridges, and produced almost the same electrophoretograms as the native non-fixed proteins. In efforts to understand the retrieval mechanisms for proteins that had been formalin fixed, Yamashita and Okada expanded work initiated in the field of antigen retrieval by demonstrating that high heat treatment cleaves the formalin-induced intra- and intermolecular cross-links of proteins, including the methylene bridges that are believed to be the main structural element of cross-links (Yamashita and Okada, 2005). The investigators selected five proteins that were formalin-fixed, and then disrupted protein cross-linking using different forms of heat — boiling, autoclaving, microwaving. Two-dimensional electrophoresis with SDS/Page gels demonstrated that cross-links had been broken, and that the proteins were similar in electrophoretic characteristics to the native unfixed proteins. The investigators trialed a variety of salt and pH concentrations in the buffers used for the reconstitution of formalin-fixed proteins. The pH level appeared to be quite important in protein behavior. The outcomes of pH variation and heat demonstrated by electrophoresis on the formalin-fixed proteins were confirmed by Western Blot experiments on deparaffinized, heated mouse uterine sections. This work demonstrated the mechanisms of abnormal protein behavior caused by formalin cross-linking and the methods to restore fixed proteins for use in protein analysis.
USE OF FIXED TISSUE FOR COMPLEX PROTEIN ASSESSMENTS
A key step in transforming FFPE tissues into a dependable starting material for complex proteomic analyses was the development of specialized kits designed for this purpose. Such kits include PicoPure (Arcturus Engineering, Mountain View, CA) and Liquid Tissue (Expression Pathology, Inc., Gaithersburg, MD), and provide buffers for protein extraction, reduction, blocking, and enzymatic digestion. Additionally, these kits offer a standardized protocol for FFPE protein preparation. Prieto et al validated the utility of the Liquid Tissue kit in a 2005 proteomics study (Prieto et al., 2005). The efficacy of these commercially prepared kits was further highlighted by a seminal study performed by Hood et al., where investigators performed a direct comparison of protein evaluation in frozen mouse liver versus FFPE mouse liver. LCM was performed to obtain 30,000 cells, Liquid Tissue was used for extraction, and nanoRPLC-MS/MS performed. Frozen mouse liver yielded 2001 unique peptides and 776 unique proteins. FFPE mouse liver yielded similar numbers with 1710 unique peptides and 684 unique proteins. Gene ontology classifications and MS spectra were similar between the fixed and non-fixed tissue (Hood et al., 2005).
Since the comparison of fixed and unfixed tissue showed no discernible loss of information in using formalin-fixed tissue for protein analysis, the authors then performed LCM on 10um thick sections of FFPE human prostate tissue, dissecting out benign prostatic hypertrophy (BPH) lesions and prostate cancer (200,000 cells for each lesion). They dissected 100,000 cells of stroma as a control. Using the Liquid Tissue buffers for preparation, tissue proteins were evaluated by nanoRPLC-MS/MS; 1300 unique peptides and 702 proteins were identified from 200,000 cells of BPH tissue. Cancer tissue (200,000 cells) yielded 2200 unique peptides and 1156 proteins. Approximately 25% of the unique proteins in both types of tissue were identified by 2 or more unique peptides. The remaining 75% were identified by one peptide. Gene Ontology classification of the proteins demonstrated a wide array of cellular locations and functions (Hood et al., 2005).
COMPLEX PROTEIN ANALYSIS COMPARING FIXED AND FROZEN TISSUES
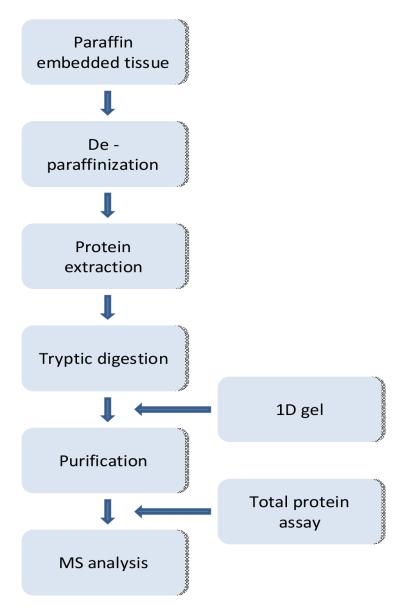
A host of papers followed the study by Hood et al to substantiate the data favorably comparing FFPE tissue to frozen. Many of these investigators forewent commercially available kits and developed or adapted existing methods for complex protein analysis of fixed tissue. “Figure (2)” demonstrates a generalized workflow for their techniques of protein extraction from fixed tissues.
Figure 2.
Schematic illustration of proteomic flow for fixed, paraffin-embedded tissue
Crockett et al. demonstrated decreased but respectable protein yield in FFPE human cells, in comparison to matched frozen human cells (Crockett et al., 2005). The cells used were from a transformed follicular lymphoma-derived cell line. Frozen and FFPE cell pellets were re-suspended; lysates were extracted using a Radio-Immuno Precipitation Assay (RIPA) buffer containing SDS; lysates were digested using trypsin, and nanoRPLC —MS/MS was performed. Fresh frozen cells yielded 514 proteins and 10,043 peptides, while FFPE cells yielded 324 proteins and 4876 peptides. Here, all proteins were identified by 2 or more unique peptides. Identifying proteins by 2 or more unique peptides has since become the standard for protein identification. Additionally, these findings were supported with Western Blotting, immunofluorescence microscopy, and a functional protein study.
Additional work by Palmer-Toy et al. confirmed protein yield in fixed tissue was comparable to, if not better than, frozen tissue (Palmer-Toy et al., 2005). They used 8um ear canal tissue sections; four 8um sections were placed in a 100mM ammonium bicarbonate/2% SDS/20mM Dithiothreitol (DTT) buffer for extraction — no microdissection of specific cell populations was performed on the ear canal sections. A FFPE cochlear ligament sample from a second patient was manually dissected from its surrounding tissue to undergo similar protein extraction. After trypsin digestion, LC-MS/MS was performed, with five subsequent MS analyses of the most abundant peptides. The first patient sample demonstrated more identified proteins and peptides in FFPE tissue (123 proteins, 412 peptides) than frozen tissue (94 proteins, 266 peptides). When using 2 or more peptides for protein identification, FFPE again yielded more than frozen (42 proteins FFPE, 31 proteins frozen). The FFPE cochlear ligament sample identified 125 proteins; it also identified peptides which encompassed known areas of mutation in that patient’s disease. Despite the use of SDS in the extraction buffer for improved peptide extraction, it was not noted how SDS was removed down line for MS.
Shi et al also published work demonstrating greater protein extraction in FFPE tissue than frozen (Shi et al., 2006). This work used one case of human renal carcinoma for its final LC-MS/MS analysis, after vetting multiple protocols on human and mouse tissue, and confirming all results with SDS-Page and Western Blot. Five 10 um tissue sections of FFPE renal carcinoma were used, but the lesion was not microdissected. Analysis was performed by capillary isoelectric focus(CIEF)-nanoRPLC-MS/MS, with 5 subsequent MS analyses of the most abundant peptides. Two FFPE samples demonstrated an average yield of 3259 proteins (frozen 2404) and 4074 peptides (frozen 3305). This non-microdissected FFPE tissue showed excellent yield, due in part to further sample separation with both the capillary isoelectric focusing and the nano RPLC. The extraction buffer contained SDS, commonly thought to be necessary for extraction. As SDS is not compatible with MS, overnight dialysis of the samples was required.
Jiang et al went on to compare 5 extraction buffers and protocols (Jiang et al., 2007). While using nonspecific sections of mouse liver tissue, the group was able to demonstrate that heating the homogenized, sonicated tissue in a 40 mM Tris, 6 M guanidine-HCl, 65 mM DTT buffer at 100C ×30 min gave the greatest protein yield. The protocol using a SDS buffer + heating gave the next greatest yield. The guanidine-HCL protocol produced 11 ug/uL protein, and its LC-MS/MS analysis yielded 470 proteins identified by multiple peptides and 3005 peptides. Of note, both the guanidine-HCL and the SDS buffers required 6-20 fold dilution before trypsin digestion and subsequent MS.
The above proteomic studies compared frozen to fixed tissue using mouse liver, a human lymphoma cell line, human ear, and human renal cell carcinoma. Our group sought to evaluate an extraction protocol in a variety of organs from the human gastrointestinal system (pancreas, colon, and liver), using two different tissue fixatives commonly used in clinical settings (formalin and Hollandes) (ref-supplemental data). Single or multiple 5um tissue sections were macrodissected to total 100mm2 of tissue for each organ. Proteins were extracted in a 100mM ammonium bicarbonate/30% acetonitrile buffer heated to 94°C × 30 minutes, followed by 60°C × 3 hours. After trypsin digestion, LC-MS/MS was performed, with 5 subsequent MS analyses of the most abundant peptides. Results were comparable between FFPE (370 proteins, 1168 peptides) and frozen (340 proteins, 990 peptides) pancreas tissue, with a 69% overlap between the tissues in regard to peptide identification. The FFPE liver tissue yielded 469 proteins, 1453 peptides, and the Hollandes-fixed paraffin-embedded colon tissue yielded 279 proteins, 757 peptides.
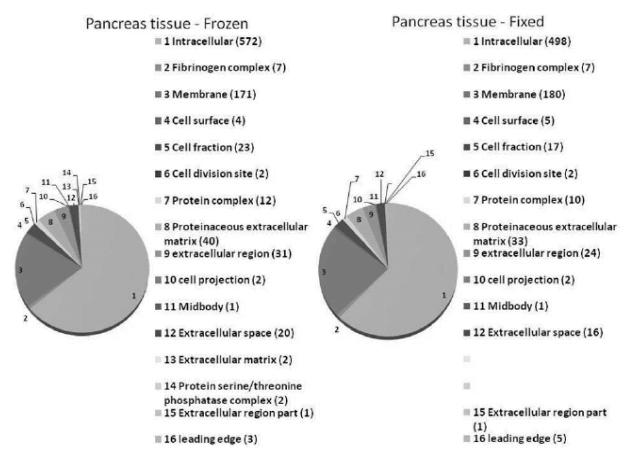
There was a similar cellular distribution of proteins in both the fixed and frozen tissue samples. The vast majority of proteins in either sample were intracellular proteins, followed by cell membrane proteins. “Figure (3)” demonstrates Gene Ontology (GO) — derived cellular locations of the identified proteins.
Figure 3.
After extraction and nanoRPLC-MS/MS, identified protein from frozen and FFPE pancreas tissue showed similar cellular distribution, as per Gene Ontology results.
LASER-CAPTURE MICRODISSECTION ISOLATION OF SPECIFIC LESIONS FROM FIXED TISSUE
While great strides were being made in the numbers of identified proteins and protocol optimization, there had not been isolation and subsequent extraction of specific cell populations since Hood et al.’s laser-capture microdissection of prostate lesions. Two groups sought to change this, one using a tissue array of needle biopsies, and one by performing manual microdissection. Protocols that can use microdissected tissue are important because they define and test the lowest limits possible for the quantity of starting material used for these assays.
Guo et al performed manual dissection of FFPE human glioblastoma multiforme (brain tumor) under the microscope, using 6um sections (Guo et al., 2007). They dissected 100,000 tumor cells and extracted their proteins in a 20mMTris/2%SDS buffer heated to 100C × 20min, then 60 C × 2hrs. Like Shi et al, they employed capillary isoelectric focusing, as well as nanoRPLC to obtain excellent sample separation. MS/MS, followed by MS on the 5 most abundant peptides, yielded an average of 2265 proteins and 11,389 peptides (2 runs averaged). It was not noted how many proteins were identified by 2 or more peptides. The FFPE results were compared to frozen, and an 83% overlap in protein quantity and identification was achieved. Due to the SDS in the extraction buffer, the samples did require overnight dialysis prior to digestion and analysis.
Hwang et al took the next step forward with the development a non-SDS protein extraction buffer that was compatible with MS (no dilution or dialysis needed), and used this on specific cell populations via a biopsy tissue array (Hwang et al., 2007). They used a FFPE tissue array containing prostate biopsies from 5 normal and 25 cancer patients. The total tissue area per array was 188.4 mm2, and yielded approximately 41ug of extractable protein. Two arrays were processed for a total of 82ug of extracted protein used for thousands of MS runs. From these, 361 proteins were identified by two or more peptides (428 proteins in total), and 12, 631 peptides were identified overall.
Laser Capture Microdissection has been used recently in conjunction with a commercially available kit (Liquid Tissue) to identify proteins associated with human head and neck squamous cell carcinoma using nanoRPLC-MS/MS. The starting material for the proteomics experiments was as low as 20,000 cells per sample tested and yielded in excess of 300 proteins identified by 3 or more peptides. When 4 samples were run, unique identifications totaled over 800 proteins for the well-differentiated cancer group. A subset of the identified proteins of interest was validated by IHC on a tissue microarray study (Patel et al., 2008).
Summary
Efforts to develop methods to use formalin-fixed tissue in protein chemistry and proteomics studies have now been successful. Effective protocols have been established which cleave protein cross-links and provide buffer conditions permitting fixed material to approximate the chemical behavior of native peptides and proteins. Initial studies focused on simple measures of protein chemistry, such as Western Blotting and 2-D electrophoresis, using larger amounts of starting material. More recent efforts have allowed for the use of very small amounts of starting material (as low as 20,000 cells) for robust complex proteomic analysis. The latest protocols can be applied through commercial kits or through bench methods that have been stream-lined and do not require kits.
There are still many opportunities to expand and improve fixed tissue proteomics. It is possible that fixed tissues, especially aged fixed tissues, can induce irreversible chemical modifications on amino acids, resulting additional unidentified MS/MS spectra. While many studies, including our data have suggested that the extraction methods work reasonably well in extracting proteins from fixed tissues for a given analytical sensitivity, a systematic study to compare statistical meaningful number of fixed and corresponding frozen tissues will be highly desirable to address such concern. Also, protein-labeling techniques need to be refined for widespread use in FFPE tissues, so that truly quantitative proteomic studies can be performed, enabling detailed analysis of differential protein expression. This will permit researchers to look carefully into the upregulation and downregulation of proteins in specific disease states. Additionally, protein extraction and processing protocols can be further developed to where smaller numbers of cells are standard starting amounts. Given the widespread availability of fixed tissue specimens, researchers can work to create databases of disease and tissue-specific proteomes. The evolution of this methodology will undoubtedly open a new era for exciting translational work that identifies proteins in clinically valuable tissue that has previously been inaccessible for scientific study.
Table 2.
Comparison of Fixed Tissue Proteomic Studies, organized chronologically
| Cell count | Grams PROTEIN |
BUFFER | PEPTIDES | PROTEINS | PROTEINS ID’D BY MULT PEPTIDES |
|
|---|---|---|---|---|---|---|
| Hood ’05 - Cancer |
200,000 cells |
Not noted | Liquid Tissue | >2200 | 1156 | 282 |
| Crockett ‘05 |
10,000,000 cells |
600ug | RIPA | 10,043 | 324 | 324 |
| Palmer- Toy ‘05 |
Not noted | Not noted | 2% SDS/100mM NaHCO3/20mM DTT |
412 | 123 | 42 |
| Shi ‘06 | Not noted | 1.67 ug/uL |
20 mM Tris-HCl buffer (pH 7 or 9) w/2%SDS |
4074 (avg, 2 samples) |
3259 (avg, 2 samples) |
1896 |
| Hwang ‘07 |
675,000 cells (cells in 10 spots ×6 × 2 arrays) |
82 ug | 100mM ammonium bicarbonate |
12,631 | 428 | 361 |
| Guo ‘07 | 100,000 cells |
Not noted | 20mM Tris, 2% SDS |
11,398 | 2265 | Not noted |
| Jiang ‘07 |
Not noted | 10.9 ug/uL |
40mM Tris, 6 M guanidine-HCl, and 65 mM DTT, pH 8.2 |
3005 | 827 | 470 |
| Patel ‘08 — WD HNSCC |
80,000 cells per lesion type |
Not noted | Liquid Tissue | 1400-1800 | 323-376 | 323-376 |
Conflicts of Interest / Acknowledgements
No conflicts of interest exist. This work was supported in part by grants from National Institutes of Health (CA10720901 and CA116296), Crohn’s & Colitis Foundation of America, Gene and Mary Ann Walters Pancreatic Cancer Foundation, UW NIEHS sponsored Center for Ecogenetics and Environmental Health (NIEHS P30ES07033), AACR-PanCAN Career Development Award for Pancreatic Cancer Research, and a generous gift from the Canary Foundation.
Abbreviation List
- BPH
benign prostatic hypertrophy
- DTT
Dithiothreitol
- ESI
Electrospray Ionization
- FFPE
formalin-fixed paraffin-embedded
- IHC
immunohistochemistry
- LC
liquid chromatography LC
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- LCM
Laser capture microdissection
- MALDI
matrix assisted laser desorption/ionization
- MS
Mass Spectrometry
- RIPA
Radio-Immuno Precipitation Assay
- RPLC
reverse-phase liquid chromatography
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
REFERENCES
- Ahram M, Flaig MJ, Gillespie JW, Duray PH, Linehan WM, Ornstein DK, Niu S, Zhao Y, Petricoin EF, 3rd, Emmert-Buck MR. Evaluation of ethanol-fixed, paraffin-embedded tissues for proteomic applications. Proteomics. 2003;3(4):413–21. doi: 10.1002/pmic.200390056. [DOI] [PubMed] [Google Scholar]
- Burnette WN. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Crockett DK, Lin Z, Vaughn CP, Lim MS, Elenitoba-Johnson KS. Identification of proteins from formalin-fixed paraffin-embedded cells by LC-MS/MS. Lab Invest. 2005;85(11):1405–15. doi: 10.1038/labinvest.3700343. [DOI] [PubMed] [Google Scholar]
- Coons AH, Creech HJ, Jones RN. Immunological properties of an antibody containing a fluorescent group. Proc Soc Exp Biol Med. 1941;(47):200–202. [Google Scholar]
- Guo T, Wang W, Rudnick PA, Song T, Li J, Zhuang Z, Weil RJ, DeVoe DL, Lee CS, Balgley BM. Proteome analysis of microdissected formalin-fixed and paraffin-embedded tissue specimens. J Histochem Cytochem. 2007;55(7):763–72. doi: 10.1369/jhc.7A7177.2007. Epub 2007 Apr 4. [DOI] [PubMed] [Google Scholar]
- Hood BL, Darfler MM, Guiel TG, Furusato B, Lucas DA, Ringeisen BR, Sesterhenn IA, Conrads TP, Veenstra TD, Krizman DB. Proteomic analysis of formalin-fixed prostate cancer tissue. Mol Cell Proteomics. 2005;4(11):1741–53. doi: 10.1074/mcp.M500102-MCP200. Epub 2005 Aug 9. [DOI] [PubMed] [Google Scholar]
- Hopwood D, Yeaman G, Milne G. Differentiating the effects of microwave and heat on tissue proteins and their crosslinking by formaldehyde. Histochem J. 1988;20(67):341–6. doi: 10.1007/BF01002727. [DOI] [PubMed] [Google Scholar]
- Hwang SI, Thumar J, Lundgren DH, Reszaul K, Mayya V, Wu L, Eng J, Wright ME, Han DK. Direct cancer tissue proteomics: a method to identify candidate cancer biomarkers from formalin-fixed paraffin-embedded archival tissues. Oncogene. 2007;26:65–76. doi: 10.1038/sj.onc.1209755. Epub 2006 Jun 26. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Monden T, Kanoh T, Tsujie M, Izawa H, Haba A, Ohnishi T, Sekimoto M, Tomita N, Shiozaki H, Monden M. Extraction and analysis of diagnostically useful proteins from formalin-fixed, paraffin-embedded tissue sections. J Histochem Cytochem. 1998;46(3):397–403. doi: 10.1177/002215549804600314. [DOI] [PubMed] [Google Scholar]
- Jiang X, Feng S, Tian R, Ye M, Zou H. Development of efficient protein extraction methods for shotgun proteome analysis of formalin-fixed tissues. J Proteome Res. 2007;6(3):1038–47. doi: 10.1021/pr0605318. Epub 2007 Feb 1. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lemaire R, Desmons A, Tabet JC, Day R, Salzet M, Fournier I. Direct analysis and MALDI imaging of formalin-fixed, paraffin-embedded tissue sections. J Proteome Res. 2007;6(4):1295–305. doi: 10.1021/pr060549i. Epub 2007 Feb 10. [DOI] [PubMed] [Google Scholar]
- Palmer-Toy DE, Krastins B, Sarracino DA, Nadol JB, Jr, Merchant SN. Efficient method for the proteomic analysis of fixed and embedded tissues. J Proteome Res. 2005;4(6):2404–11. doi: 10.1021/pr050208p. [DOI] [PubMed] [Google Scholar]
- Patel V, Hood BL, Molinolo AA, Lee NH, Conrads TP, Braisted JC, Krizman DB, Veenstra TD, Gutkind JS. Proteomic analysis of laser-captured paraffin-embedded tissues: a molecular portrait of head and neck cancer progression. Clin Cancer Res. 2008;14(4):1002–14. doi: 10.1158/1078-0432.CCR-07-1497. [DOI] [PubMed] [Google Scholar]
- Prieto DA, Hood BL, Darfler MM, Guiel TG, Lucas DA, Conrads TP, Veenstra TD, Krizman DB. Liquid Tissue: proteomic profiling of formalin-fixed tissues. Biotechniques. 2005 Jun;(Suppl):32–5. doi: 10.2144/05386su06. [DOI] [PubMed] [Google Scholar]
- Rait VK, Xu L, O’Leary TJ, Mason JT. Modeling formalin fixation and antigen retrieval with bovine pancreatic RNase A II. Interrelationship of cross-linking, immunoreactivity, and heat treatment. Lab Invest. 2004;84(3):300–6. doi: 10.1038/labinvest.3700041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronci M, Bonanno E, Colantoni A, Pieroni L, Di Ilio C, Spagnoli LG, Federici G, Urbani A. Protein unlocking procedures of formalin-fixed paraffin-embedded tissues: Application to MALDI-TOF Imaging MS investigations. Proteomics. 2008;8(18):3702–3714. doi: 10.1002/pmic.200701143. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Shi SR, Liu C, Balgley BM, Lee C, Taylor CR. Protein extraction from formalin-fixed, paraffin-embedded tissue sections: quality evaluation by mass spectrometry. J Histochem Cytochem. 2006;54(6):739–43. doi: 10.1369/jhc.5B6851.2006. Epub 2006 Jan 6. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76(9):4350–4. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S, Okada Y. Mechanisms of heat-induced antigen retrieval: analyses in vitro employing SDS-PAGE and immunohistochemistry. J Histochem Cytochem. 2005;53(1):13–21. doi: 10.1177/002215540505300103. [DOI] [PubMed] [Google Scholar]
- Yamashita S, Okada Y. Application of heat-induced antigen retrieval to aldehyde-fixed fresh frozen sections. J Histochem Cytochem. 2005;53(11):1421–32. doi: 10.1369/jhc.4A6579.2005. Epub 2005 Jul 26. [DOI] [PubMed] [Google Scholar]
- Zang L, Toy D Palmer, Hancock WS, Sgroi DC, Karger BL. Proteomic analysis of ductal carcinoma of the breast using laser capture microdissection, LC-MS, and 16O/18O isotopic labeling. J Proteome Res. 2005;3(3):604–12. doi: 10.1021/pr034131l. [DOI] [PubMed] [Google Scholar]