Abstract
Data analysis is an essential tenet of analytical chemistry, extending the possible information obtained from the measurement of chemical phenomena. Chemometric methods have grown considerably in recent years, but their wide use is hindered because some still consider them too complicated. The purpose of this review is to describe a multivariate chemometric method, principal component regression, in a simple manner from the point of view of an analytical chemist, to demonstrate the need for proper quality-control (QC) measures in multivariate analysis and to advocate the use of residuals as a proper QC method.
Keywords: Chemometrics, Concentration, Data analysis, Determination, Multivariate data analysis, Partial least squares (PLS), Principal component analysis (PCA), Principal component regression (PCR), Quality control, Residual analysis
1. Introduction
Advances in electronics and computing over the past 30 years have revolutionized the analytical laboratory. Technological developments have allowed instruments to become smaller, faster and cheaper, while continuing to increase accuracy, precision and availability. Data-analysis methods have also benefitted from advances in technical computing; commercially-available mathematical programming packages allow scientists to perform complex calculations with a few simple keystrokes. Furthermore, software sold with many commercial instruments contains automatic data-processing algorithms (e.g., Fourier transform analysis, data filtering and peak recognition). The advances in computing allow researchers to obtain increasing amounts of chemically-relevant information from their data; however, this is not always achieved using simple data-processing techniques.
Svante Wold first coined the term “kemometri” (“chemometrics” in English) in 1972 by combining the words kemo for chemistry and metri for measure [1]. Presently, the journal Chemometrics and Intelligent Laboratory Systems defines chemometrics as: “the chemical discipline that uses mathematical and statistical methods to design or select optimal procedures and experiments, and to provide maximum chemical information by analyzing chemical data” [2]. The field of chemometrics has also benefitted from technological advances in the past 30 years, causing the number of researchers using chemometric methods to grow [3–5]. Unfortunately, however, chemometrics has not been as rapidly integrated into the analytical laboratory as other advances.
The slow adoption of these methods may be attributed to several factors. Technical articles on the subject are often written by chemometricians for chemometricians; it can be difficult for the general scientist to approach this field and comprehend the material presented. Even introductory texts and review articles often require working knowledge of linear algebra and matrix manipulations. Chemometric methods have developed so that they are readily available to any scientist and, in this article, we hope to show the importance of chemometrics to the bench-top analytical chemist in concentration determination using a technique known as principal component regression (PCR).
2. Multivariate analysis in analytical chemistry
Traditional concentration determinations are usually univariate, isolating one variable (e.g., peak current at one potential in an electrochemical measurement or the wavelength of maximum absorbance in a spectroscopic measurement). While intuitive and simple, this approach to data analysis is limited and wasteful. As an example, consider a UV-VIS spectrum of a particular analyte containing 500 data points. With only one data point being used for concentration determination (absorbance at one wavelength), after identification, 99.8% of the data will be discarded. Data collection can limit the throughput of an analytical methodology; it is not efficient to collect data that will not be used. In addition, a univariate measurement is extremely sensitive to interferents. It is often impossible to differentiate an analyte-specific signal from an interferent when looking only at one point of a data spectrum.
Multivariate calibration methods involve the use of the multiple variables (e.g., the response at a range of potentials or wavelengths, or even over the entire range collected to calculate concentrations). This offers several advantages, often reducing noise and removing interferents [5]. It can be easier to identify and to remove noise when looking at the entire data set, rather than one point. In addition, interferents can be taken into account, provided their measurement profile differs sufficiently from the analyte of interest [6]. Multivariate methods are generally better than univariate methods. They increase the amount of possible information that can be obtained without loss; multivariate models can always be simplified to a univariate model [5]. The advantages of multivariate methods come at a cost of computational power and complexity, but these drawbacks are easily handled with common mathematical software packages (e.g., Matlab).
Analytical techniques are often misused because their limitations are not always clearly understood. Multivariate analysis methods are no different and have the potential to be misused more than instrumental techniques because all the computations are performed behind the computer screen. Chemometricians have derived a series of rules, statistical tests and other criteria for users to judge and to validate the accuracy of the information obtained with multivariate methods [7]. It is important for any new user of multivariate methods to remember that the computer will always give an output but it is up to the scientist to make sure that both precautions are taken and the answers obtained make chemical sense.
2.1. Principal component regression
PCR is a basic, but very powerful, multivariate calibration method. In this article, we present a brief overview of PCR, but, for a more detailed explanation, readers are referred elsewhere [8]. In addition, Kramer offers an excellent review of the topic in a manner that the bench-top analytical chemist can understand and use, and we highly recommend it to anyone interested in using the technique [9]. PCR is a combination of principal component analysis (PCA) and least-squares regression.
When discussing multivariate analysis techniques, including PCR, three terms are often used: variance, vector, and projection. Variance is another word for information of a data set. Sources of variance within a data set include:
changes in the chemical make-up of analyzed samples (concentrations and/or composition)
changes in environmental parameters (e.g., temperature and pressure); and
changes in instrument performance such (e.g., a drifting baseline).
The term “vector” is used to describe a line segment in a coordinate system with a specific direction, and the term “projection” is used to describe the distance of a point along a vector. To illustrate the point more clearly, consider the simple three-dimensional Cartesian coordinate system. The x, y, and z axes of this coordinate system are defined by the vectors i, j, and k, respectively. The point (3,4,5) has a projection of 3 onto the vector i, a projection of 4 onto the vector j, and a projection of 5 onto the vector k.
We will illustrate how PCR is performed using simulated data taken from a hypothetical UV-VIS experiment. This example is an oversimplification, but explains the technique of PCR in a manner that can be easily understood without overbearing mathematical descriptions.
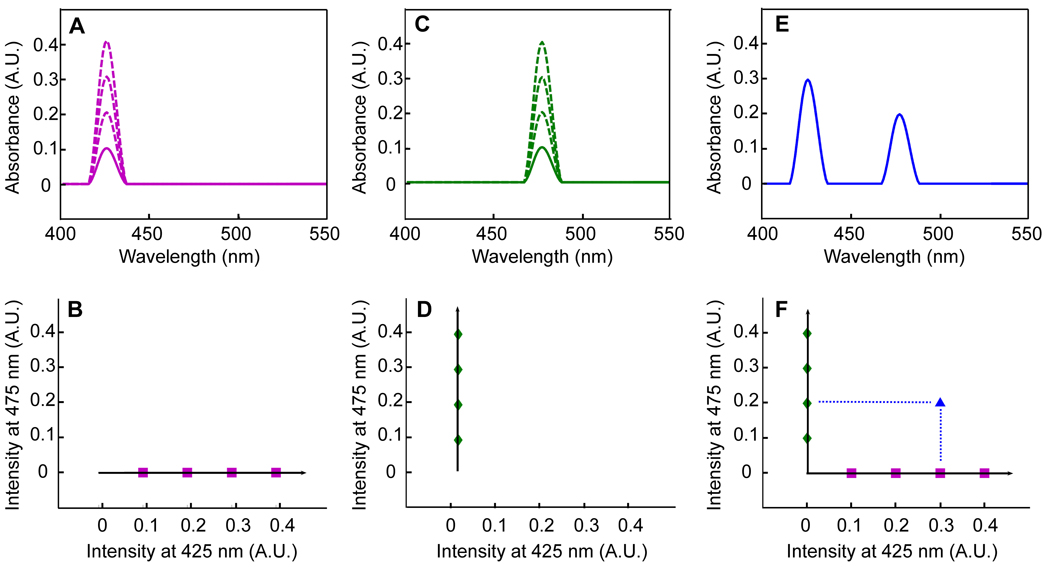
The solid line in Fig. 1A shows an example of a UV-VIS absorption trace of component X at a specific concentration, [X]. The information in the absorption spectrum of component X can be plotted in a different manner (Fig. 1B), which shows a plot of the intensities at 425 nm and 475 nm. Component X has intensities of 0.1 AU and 0 AU at 425 nm and 475 nm, respectively, and can be represented as the point (0.1, 0) in the two-dimensional coordinate system shown in Fig. 1B. According to Beer’s law, if analyte X is doubled ([2X]), tripled ([3X]), and quadrupled ([4X]), the absorbance spectrum will increase by 2-fold, 3-fold and 4-fold, respectively, as shown in the dashed lines in Fig. 1A. These absorption spectra can also be plotted the same way as the first spectrum in a two-dimensional manner as shown in Fig. 1B (purple squares). Similarly, component Y, which has a different absorption spectrum (Fig. 1C) and at concentrations [Y], [2Y], [3Y] and [4Y] can be plotted in a two-dimensional manner as shown in Fig. 1D (green diamonds) as multiples of the point (0, 0.1).
Figure 1.
Representation of UV-VIS data in an intensity space. A) UV-VIS spectra of component X in concentrations [X] (solid line) to [2X], [3X], and [4X] (dashed lines). B) Spectra in A) plotted in an intensity coordinate system with intensity at 425 nm on the x-axis, intensity at 475 nm on the y-axis, and intensity at 525 nm on the z-axis (going in and out of the page, omitted for clarity). C) UV-VIS spectra of component Y in concentrations [Y] (solid line) to [2Y], [3Y], and [4Y] (dotted lines). D) Spectra in C) plotted in an intensity coordinate system. E) UV-VIS spectra of an unknown mixture of X and Y. F) Spectrum in E) plotted in an intensity coordinate system with the dotted lines representing projections onto each principal component.
As shown in Fig. 1B and Fig. 1D, lines can be drawn through the two-dimensional representations of the absorption spectra of components X and Y. Each of these lines describes important information about the measured absorption spectra. The horizontal line in Fig. 1B describes how intensities change based on [X] and the vertical line in Fig. 1D describes how intensities change based on [Y]. In this simplified case, moving in a horizontal direction in these graphs describes only how [X] is changing and says nothing about how [Y] is changing. Conversely, moving in a vertical direction in these graphs describes only how [Y] is changing and says nothing about how [X] is changing. Mathematically speaking these lines are orthogonal, meaning that each describes information that the other does not. These lines, which each describe different information about the original data drawn in an alternative coordinate system, can be thought of as principal components (PCs). Stated another way, PCs can be thought of as vectors in an abstract coordinate system that describe sources of variance of a data set. Chemometricians and mathematicians advocate the use of a slightly different definition of a PC, but our definition is common and is used in many introductory texts [9–11].
The projection of the points onto the PCs shown in Fig. 1B and Fig. 1D is related to concentration, just like a traditional univariate calibration curve. Fig. 1E shows an example of an absorption spectrum from an unknown mixture of components X and Y. It can be represented as the point (0.3, 0.2) in the two-dimensional space depicted in Fig. 1F. This unknown sample has a projection along the horizontal PC of 0.3 and a projection along the vertical PC of 0.2, correspond to concentrations of [3X] and [2Y]. Comparing the unknown spectrum in Fig. 1E with the standards in Fig. 1B and Fig. 1D confirm this result. Mathematically, the projection onto a PC is related to concentration by performing a simple least-squares regression.
In a univariate calibration, known concentrations of standards are assembled. Peak responses are plotted as a function of concentration and a regression is performed relating a measured value to concentration. Finally, the measured response is projected back onto the calibration line in order to determine a concentration. PCR is a multivariate calibration method that works in a similar manner using up to all the data points in a spectrum instead of just one. First, a series of known spectra and concentrations known as a training set is assembled. Second, PCs are calculated and describe relevant portions of the assembled calibration spectra using PCA. Third, a regression is performed that relates concentrations to distances along PCs. Finally, concentrations are predicted by projecting an unknown sample onto the PC and relating its distance back to concentration [12].
The number of PCs calculated equals the number of spectra in the training set that are input into the algorithm, but PCs themselves are not always directly interpretable. The above example showed that one PC described only component X and one PC described only component Y, but PCs are abstract and should not be thought of as belonging solely to one component or as pure analyte spectra [13]. Sometimes, however, mathematical manipulations can be performed on the PCs in order to give the user something that relates back to a specific source of variance in the experiment [14].
PCR offers an analytical chemist several advantages. First, one can separate and retain PCs that describe relevant information and discard PCs that contain noise, thereby eliminating sources of random error. PCs that describe relevant information should have larger projections because they describe more of the collected dataset than those that describe noise, which should be a small percentage of the overall measured signal. There are numerous ways to decide how many PCs to keep, but all rely on the same basic assumption that PCs that describe relevant information will describe more of the collected data than PCs that describe only noise [15–17]. Second, the size of a data matrix is drastically reduced [6]. An entire spectrum can be replaced by its distance (or projection) along a few PCs. For example, a data set comprising a 1000 data-point cyclic voltammogram measured at 10 Hz for 60 s contains 600,000 data points. If only three PCs are needed to fully describe all the relevant information of the collected data set, the number of data points can be reduced from 600,000 (1000 points × 10 Hz × 60 s) to 1800 (3 × 10 Hz × 60 s), or 0.3% of the size of the original data set. This example illustrates how PCA can reduce the dimensionality, or size, of a data set by orders of magnitude and still keep the relevant information.
Samples used in multivariate training sets must meet several requirements [7,18]. First, training-set samples must contain all expected components because concentrations obtained may not be accurate if the unknown sample contains spectral information not present in the training set. Second, training-set samples must uniformly span the expected concentrations of each of the components to ensure that unknown concentrations fall within the calibration range. Third, training-set samples must span the conditions of interest in order to account properly for environmental parameters and sample matrix. Fourth, training-set samples must be mutually independent. Samples created by serial dilutions are examples of samples that are not mutually independent because relative concentrations of the different components and relative errors in the concentration values are do not vary. Finally, there needs to be sufficient training-set samples to build an accurate model. For infrared data, ASTM International recommends at least 24 samples for a model that contains up to 3 relevant PCs and 6 samples per relevant PC for a model with more than 3 relevant PCs. Unfortunately, this means that a user will only know if there are enough training-set samples after a model is constructed.
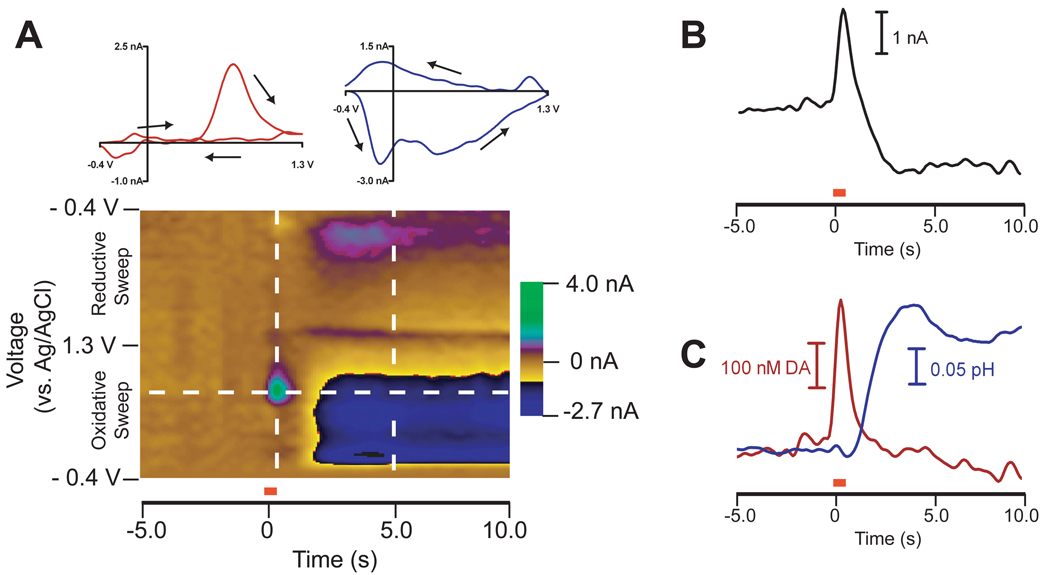
PCR has been used in order to predict concentrations of in vivo electroactive species using fast-scan cyclic voltammetry [6,19,20]. Fig. 2 shows how PCR can be used to separate neuromodulators dopamine and pH during stimulated release. A carbon-fiber microelectrode is placed in a region of the brain containing dopaminergic neuron terminals while a stimulating electrode is placed in a region containing dopaminergic cell bodies. Fig. 2A displays in vivo cyclic voltammograms in the form of a color plot, with each vertical slice a cyclic voltammogram at a specific time point, each horizontal slice a current versus time trace at a specific potential, and current in false color. The cyclic voltammograms taken around 0 s are characteristic of the neurotransmitter dopamine while those taken between 2 s and 10 s are characteristic of a pH change (Fig. 2A inset). The increase in dopamine concentration occurs due to local stimulation given to the cell bodies of dopaminergic neurons that causes release in the terminal region. The observed pH response is due to changes in blood flow and metabolism accompanying terminal activity, which cause a decrease in carbon dioxide, a component of the extracellular buffering system of the brain [21]. The current versus time trace taken at the oxidation potential of dopamine in Fig. 2B shows a convoluted response between dopamine and pH so a univariate calibration would be insufficient to determine dopamine concentration as a function of time. Using a training set of in vivo cyclic voltammograms of dopamine and pH at varying intensities, PCR can separate these two components and generate concentration traces for each analyte, as shown in Fig. 2C.
Figure 2.
PCR deconvolution of in vivo electrochemical data. A) Color plot representation of cyclic voltammograms taken in the brain of a freely moving rat after a stimulation given at 0 s (60 Hz, 24 pulses, 300 µA depicted by the red bar). Each vertical slice represents a cyclic voltammogram collected at a specific time point and each horizontal slice represents a current versus time trace at a specific potential. The horizontal dashed line represents the oxidation potential of dopamine, 0.6 V. Insets are cyclic voltammograms of dopamine (red, taken at the dashed line at 0 s) and pH (blue, taken at the dashed line at 5 s) with arrows drawn indicating the direction of the voltammetric sweep. B) Current versus time trace at the oxidation potential of dopamine with the red bar marking the onset of the stimulation. C) Dopamine (DA) and pH concentrations predicted using PCR.
3. PCR-model validation
When fitting any calibration model to a data set, univariate or multivariate, an analytical chemist should ask two questions:
How accurate is my calibration model at predicting concentrations?
How applicable is my calibration model an unknown data set?
When using multivariate calibrations, the accuracy of a model is addressed with a process called validation. A set of test samples distinct from the calibration set with known concentrations is used to determine the accuracy of the calibration at predicting unknown concentrations. The predicted residual error sum-of-squares (PRESS) is the squared difference between the actual and the predicted concentrations for all validation samples and serves as a figure of merit for the multivariate model [22]. PRESS gives the experimenter an idea of how well the model can predict new concentrations and how much error can be expected in the concentrations obtained from the analysis of unknowns. The extra work to validate a model before running an experiment is necessary; it is better to test the accuracy of a model first rather than using it blindly on unknowns and hoping for accuracy [22].
Unfortunately, validation samples are not always available due to cost, time constraints or other experimental conditions. In these cases, the training set can be used as a test set in a process called cross validation. When using cross validation with PCR, the regression is performed using all the samples of the training set except one. The concentration of this training set sample is predicted using the regression model and a PRESS value is calculated. The excluded sample is reintroduced into the training set and another training set sample is excluded and its concentration and PRESS value is estimated and added to the previous PRESS value. The process is repeated until all of the training set samples have been estimated and a final PRESS value is calculated [22].
A PRESS value calculated in this way can also be used as a measure of the proper number of PCs of a data set to retain. As more PCs are retained, the PCR model will predict concentrations more accurately and PRESS values will decrease. However, there will come a point where increasing the number of PCs retained does not significantly improve the accuracy of the prediction and those PCs should be discarded [12,15,17].
4. Applicability of PCR model to residual analysis
The accuracy and the applicability of a model are two distinct questions (vide supra) [23]. Some users of PCR do not address applicability of their calibration model and thus assume that the calibration model is always applicable to an unknown data set. Stated another way, one assumes that the relevant PCs of a data set describe all relevant information in the unknown data set. Instrumental errors (e.g., drift), experimental system errors (e.g., pressure and temperature) and impurities or interferents can invalidate this assumption, if they contribute significantly to the measured signal [5, 24].
There are situations in which a scientist may not always know the complete composition of the unknown data set a priori and will not be able to predict if there are any unknown components that will significantly affect the measured response. As an example, in vivo electrochemists use fast-scan cyclic voltammetry to measure electroactive species in the brain of freely moving rats. Training-set cyclic voltammograms often incorporate only dopamine and pH but measure in brain regions containing many electroactive species [25]. If dopamine and pH are the only significant current contributions to the overall measurement, concentration data should be accurate. However, if other electroactive species are present in concentrations large enough to contribute a significant amount of current, the training-set cyclic voltammograms would be insufficient to model all of the collected data, and concentration data obtained from PCR would be questionable.
Jackson and Mudholkar proposed a method in order to evaluate the goodness of fit of training-set data to an unknown data set in PCR using residuals [26,27]. In general, a residual is defined as the difference between an experimental observation and a predicted value from a model. Residual analysis has several advantages, including quality-control (QC) monitoring, interferent identification and outlier detection. An advantage of working with multivariate data is that it can sometimes be possible to visualize the data spectrum of an interferent, something that is impossible with a univariate measurement.
In PCR, residuals are a measure of the unknown signal (e.g., current) that is not accounted for by the retained PCs of the training set. This includes noise and any signal arising from the response of any interfering analytes. Ideally, the training set contains all the relevant information of an unknown data set and the residuals should contain only noise. We will continue to use in vivo electrochemical data as an example throughout this section, but the principles apply to all other fields of analytical chemistry.
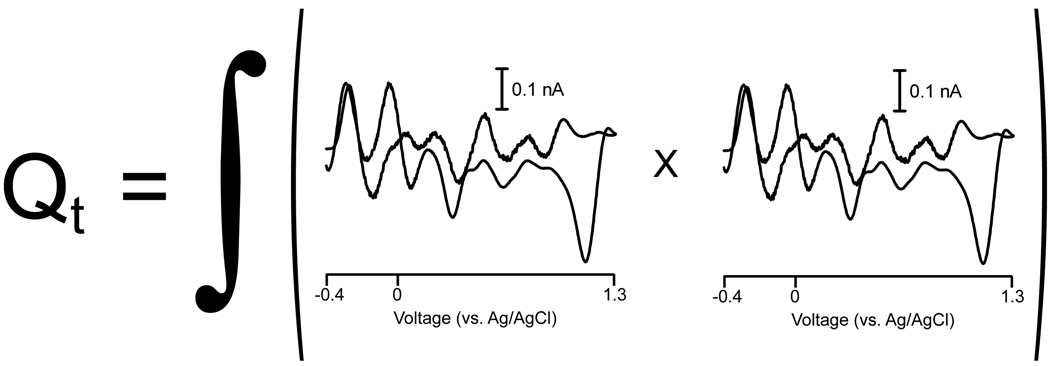
Quantity Q is defined as the sum of the squares of the residual values at each variable in each sample of the data set. Using in vivo electrochemistry as an example, one Q value is calculated for each cyclic voltammogram in the unknown data set by summing the squares of the current at each potential scanned that was not accounted for by the retained PCs of the training set used, as shown in Fig. 3. Mathematically, the Q value of a cyclic voltammogram at time t, Qt, can be represented by
| (1) |
where ix is the current at x point number of the wth point cyclic voltammogram and îx is the current predicted from the PCR model containing only the relevant PCs at x point number of the wth point cyclic voltammogram. These Qt values are tabulated for each sample and plotted consecutively for unknown data set to make a Q plot; the y-axis is in units of nA2 for this example.
Figure 3.
Calculation of a Q value at a specific time point, t. The cyclic voltammetric representation of residual current is squared and summed at time t in order to obtain a Qt value.
4.1. Qα as a measure of significance
The threshold for the sum of the squares of the residuals (Qα) is a threshold that establishes whether a satisfactory description of the experimental data by the retained PCs is achieved. The discarded PCs should contain only noise and thus provide a measure of a noise level. If the Qt values exceed Qα, then there is a measured signal that exceeds the noise anticipated by the PCs discarded. The value of Qα includes a significance level that can be set by the user for how much noise can be tolerated.
Qα is calculated using the following equations [26]:
| (2) |
| (3) |
| (4) |
where cα is the z-score that determines the (1-α)% of noise that will be tolerated, k is the number of PCs retained to describe all significant signal contributions of the training set (i.e. if the training set contained 10 cyclic voltammograms, k could vary between 1 and 10, depending on the number retained), n is the total number of PCs calculated (10 in the example described above, because the number of PCs calculated equals the number of cyclic voltammograms in the training set), and λ is the sum of the squares of the data projections from all the samples in the training set for each PC. The remaining terms (Θ1, Θ2, and Θ3, and thus h0) are simply calculated from the λ values of the discarded noise components ((k + 1) → n). From this description, the calculation of Qα is based on only two pieces of information:
a noise level threshold (cα); and
information contained in the discarded PCs of the training set (λ(k + 1) → n).
Here noise is defined as any signal that has a low probability of containing relevant information [28]. When the PCs of the training set were discarded, they were assumed to be irrelevant and thus serve as an estimated noise level.
Qα is a threshold for significance of the Qt values. Qα is an upper limit on the amount of noise or random error that will be tolerated from collected data, based on the amount of error contained in the discarded PCs of the training set. A cyclic voltammogram with a Qt value above this threshold will be considered to contain significant information not accounted for by the retained PCs and concentration values obtained with PCR would be questionable.
A chief advantage in using PCA is to help separate the significant deterministic information from non-deterministic error. Deterministic variation is a non-random change in a signal (e.g., the signature shape of the cyclic voltammogram that lets one determine its chemical identity). Non-deterministic noise or error is random and should thus follow a normal distribution. If Qt exceeds Qα, then the level of the noise is greater than expected and may contain deterministic information that is not accounted for by the retained PCs.
4.2. Interpretation of cα
The cα term in Equation 2 is the z-score corresponding to the (1-α)% of noise that will be tolerated. Qt values are the sum of differences of squares and are not normally distributed. However, Jenson and Solomon [29] have shown that the quantity (Q/Θ1)h0 can be approximated by a normal distribution with a mean (μ) and standard deviation (σ), respectively, equal to
| (5) |
| (6) |
From elementary statistics, a z-score for a normal distribution is calculated as the difference between an observed value and the mean, divided by the standard deviation. This would make the z-score for the (Q/Θ1)h0 distribution
| (7) |
Substituting cα for z and Qα for Q in Equation 7 and rearranging to solve for Qα gives Equation 2.
Approximately 95% of random, non-deterministic error will fall below a cα of 1.645 [27]. An unknown sample will be significantly different from the training set if its Qt exceeds Qα. Its signal contribution is larger than were a certain percentage of the signal contributions due solely to random error. Using our example with a cα of 1.645, a Qt value will be significant (cross Qα) only if its current contributions are larger than 95% of current contributions due to random noise. Qα is a measure of significance, not confidence. If Qt exceeds Qα, Qt has a significant value and the use of the retained PCs is insufficient to describe the experimental data. It is incorrect to say that one is (1-α)% confident that concentration data obtained from PC regression is correct if the residuals do not cross Qα. Accuracy of concentrations is addressed using validation, but if Qt crosses Qα, the validation cannot be trusted because significant interferents are present.
As cα increases, Qα increases. As an example, increasing from 95% to 99% increases cα from 1.645 to 2.326. This increase would mean that a residual (Qt) would be significant only if it has a current contribution larger than 99% of current contributions due to random noise. Qα has to increase because an extra 4% of larger random-error current contributions will have to be accounted for. Mathematically, Equation 2 shows that increasing cα increases Qα (ho is less than 1). Also, decreasing cα decreases Qα, and the smaller the Qt value will have to be in order to be deemed to contain significant information.
4.3. Qt crossing Qα
One of three possibilities occurs if Qt crosses Qα:
First, there is α% chance that random noise would cross Qα, but, since α is small, this occurrence is not very probable.
Second, too many PCs are kept and tolerance for noise is essentially zero. Each consecutive PC is calculated by determining the maximum amount of variance present not accounted for by previous PCs. The first PC describes the largest source of variance in the training set; the second PC describes the largest source of variance not described by the first PC, etc. Increasing the number of retained PCs deems more and more of a data set significant, leaving less to be counted as noise. Thus, if the amount of noise decreases, the threshold for what is significant must also decrease. Mathematically speaking, Equation 3 decreases as k increases. This possibility is also not likely if the proper number of PCs is retained.
The third, and most important, reason that Qt crosses Qα is because significant deterministic variation is present in the residual. If Qt crosses Qα, significant information is present in the residual because the PCs retained in the training set do not accurately model all of the significant current contributions in experimental data set.
Qα is a trigger of significance and is not related to the accuracy of the predicted concentrations. Qα is a threshold to determine if significant information is present in the residual. If Qt does not cross Qα, it means that all significant signals in the collected data set have been accounted for, where significance is defined as having a Qt value larger than (1-α)% of Qt values that would be calculated by chance from random noise.
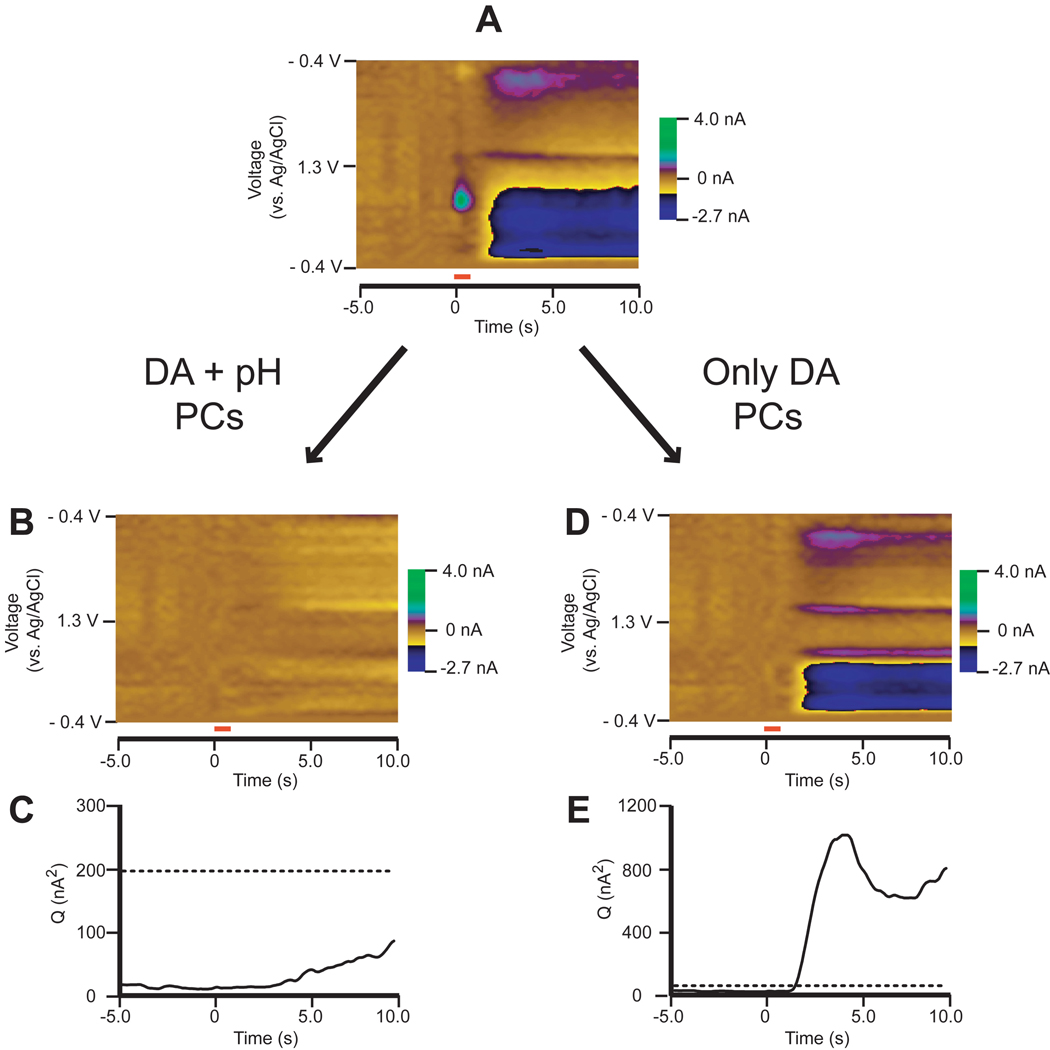
Fig. 4 shows how residuals and Qt values can be visualized for the in vivo electrochemical data set used previously. Fig. 4B shows a color-plot representation of the residual currents when both dopamine and pH are included in the training set. There are no features in the color plot, suggesting that the training set accurately describes all relevant sources of information present in the unknown data set. Furthermore, the Qplot in Fig. 4C also shows no significant current contributions at the 95%-significance level. If the training set used contains both dopamine and pH, its PCs should describe all the relevant information in the measured color plot leaving only noise.
Figure 4.
The use of residual analysis as a diagnostic tool for significance in in vivo electrochemical data. A) Color plot taken from Fig. 2. B) Residual color plot when both dopamine (DA) and pH were used in the PCR training set. Malinowski’s F-Test was used in order to determine the proper number of principal components (PCs) to keep (k = 2) [9,16]. C) Q trace of the residuals from B) with the dashed line marking Qα at 95% significance. All Q values are below Qα, indicating the PCs of the training set accurately describe all relevant sources of information. D) Residual color plot when only DA was used in the PCR training set. Malinowski’s F-Test was used in order to determine the proper number of PCs to keep (k = 1). E) Qt trace of the residuals from D) with the dashed line marking Qα at 95% significance. Qt values crossed Qα in the pH region of the color plot, indicating significant information present in the residual.
However, if we construct a model with a training set that includes only dopamine, its PCs should fail to describe all relevant information in the measured color plot. The residual color plot in Fig. 4D shows features in the pH region and its Q plot in Fig. 4E crosses Qα at the 95%-significance level, meaning that the dopamine PCs fail to describe all significant current contributions in the cyclic voltammograms taken between 2 s and 10 s, so the concentration values should not be trusted. The residual cyclic voltammograms do not look identical to pH, but they have some pH-like features. Residuals cannot always be directly interpreted as an interferent spectrum, as shown in Fig. 4, but the Q residual plot will inform the experimenter of any samples in the unknown data set that possibly contain an interferent [30].
It is not always true that a training set with 4 relevant PCs will have a larger Qα than a different training set with 3 relevant PCs. This is an erroneous assumption because the Qα threshold for two different training sets cannot be compared in this way. This statement is true only if one is referring to the same training set when the PCA decomposition is calculated. The PCs in the two training sets shown in Fig. 4 are calculated differently because the two training sets have different sources of variation (one training set contains dopamine and pH, and the other training set contains only dopamine). Further, the pH cyclic voltammograms used in the training set are noisier than the dopamine cyclic voltammograms, so the training set with only dopamine cyclic voltammograms contains less noise, so the noise threshold, and thus Qα, is smaller.
Any multivariate model used should fulfill four requirements [31]:
First, it should provide a “Yes” or a “No” answer as to whether the model used accurately describes all relevant measured responses of an unknown data set.
The rate of false positives, concluding the model does not accurately model all relevant measured responses of an unknown data set when it actually does, should also be specified.
Any relationships that exist between experimental variables must be taken into account.
Finally, there should be a way to identify why the model does not accurately describe an unknown data set.
All of these points, especially the last, are very interesting to the analytical chemist, and residual analysis is an excellent tool that meets all of these requirements.
5. Future outlook and conclusions
PCR is a powerful data analysis tool used in analytical chemistry [19,20,32,33], but another technique called partial least-squares (PLS) [34] has become the de facto standard in multivariate calibration in recent years due to a technical advantage and availability of commercial software programs [3,35]. PCR calculates each PC of data matrix to maximize the amount of variance described without using concentration information, so there is no guarantee that the calculated PCs are important for concentration prediction [36]. PLS calculates PCs using concentration information, enabling better prediction while sacrificing some spectral fit. For example, if some of the training-set spectra contained a substantial linear baseline shift, PCA decomposition of the data matrix would be significantly altered while PLS should disregard the baseline shift, since it has little to do with concentration of an analyte. PCR and PLS have been extensively compared theoretically and practically. However, despite their theoretical difference, both methods offer similar predictive abilities with only a slight advantage to PLS in some cases [37].
Multivariate techniques offer several advantages over univariate calibration methods. Noise is more easily removed and interferents can be identified. PCR can dramatically reduce the dimensionality of a data set, while still retaining all the pertinent information. Residual analysis assures users that the calibration data take into account all relevant components of measured data and can identify specific samples that contain significant amounts of an interfering signal.
Acknowledgements
The authors wish to thank Leslie Sombers at North Carolina State University for the use of her data. Richard Keithley is supported with a National Defense Science & Engineering Graduate Fellowship. Research in this area in the Wightman Laboratory has been supported by the National Institutes of Health (NS 15841).
Appendix: Principal Component Regression Command Lines
Below is a list of command lines that can be directly typed into Matlab for any user to begin work with PCR. These command lines do not include mean-centering and scaling, which is necessary before PCR usage. Anyone interested in performing PCR is urged to read Kramer before proceeding [9]. As a note, all matrices are defined by bold letters in the text and any command lines with an apostrophe (') indicate the transpose of a matrix.
To begin, a training set of known spectra and known concentrations must be assembled under the same experimental conditions as the unknown. For the following command lines, an (n x m) matrix containing all spectra must be assembled such that each column contains an n-point data spectrum for all m samples in the training set. Again, for infrared data ASTM International recommends at least 24 samples for a model that contains up to 3 relevant PCs and 6k samples for a model with more than 3 relevant PCs [7]. In addition, a (l × m) concentration matrix containing all known concentrations must be assembled such that each row contains concentrations of each of the l components in all of the m samples.
The PCs are calculated for the (n x m) training set data spectrum matrix A using singular value decomposition (SVD) [38]:
| (A1) |
While there are many methods to decide how many PCs to keep, a simple, yet very subjective method is the use of a log Scree graph [15]:
| (A2) |
Once the number of relevant PCs is known, k, a data matrix Vc is constructed to contain all the relevant PCs. For a (n x m) training set data spectrum matrix A, the PCs are contained in the U matrix from SVD. If the training set data spectrum matrix A is (m × n), the PCs are contained in the V matrix from SVD.
| (A3) |
Next, the projections of the training set data spectra onto the relevant PCs (Aproj) are calculated:
| (A4) |
After the projections are calculated, a regression matrix relating the data projections to concentrations (F) is calculated:
| (A5) |
To calculate concentrations, an unknown data set D in the form of (n × m2) with m2 columns of n-point spectra must be assembled. Concentrations of each of the l components in each of the m2 samples, Cu, can be predicted by first calculating the projections of D onto the relevant PCs of A and then relating these projections to concentrations using F:
| (A6) |
| (A7) |
The residuals, E, can be calculated by subtracting the data accounted for by the relevant PCs from the unknown data set D.
| (A8) |
The Q values from equation (1) can be calculated using the following command
| (A9) |
Equations (2)–(4) can then be used to calculate Qα in order to test for significance. The S matrix from equation (A1) contains the square roots of λ (from Equation (3) in section 4.1.) along its diagonal.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Richard B. Keithley, The University of North Carolina, Department of Chemistry, B-5 Venable Hall CB#3290, Chapel Hill, NC 27599, USA
Michael L. Heien, The Pennsylvania State University, Department of Chemistry, 104 Chemistry Building, University Park, PA, 16802, USA
R. Mark Wightman, The University of North Carolina, Department of Chemistry, B-5 Venable Hall CB#3290, Chapel Hill, NC 27599, USA.
References
- 1.Kiralj MMCFR, Chemom J. 2006;20:247. [Google Scholar]
- 2.Guide for Authors. Chemom. Intellig. Lab. Syst. 2009 ( www.elsevier.com/locate/chemometrics).
- 3.Lavine B, Workman J. Anal. Chem. 2008;80:4519. doi: 10.1021/ac800728t. [DOI] [PubMed] [Google Scholar]
- 4.Brown SD, Bear RS. Crit. Rev. Anal. Chem. 1993;24:99. [Google Scholar]
- 5.Bro R. Anal. Chim. Acta. 2003;500:185. [Google Scholar]
- 6.Heien M, Johnson MA, Wightman RM. Anal. Chem. 2004;76:5697. doi: 10.1021/ac0491509. [DOI] [PubMed] [Google Scholar]
- 7.ASTM International. Standard Practices for Infrared Multivariate Quantitative Analysis, Doc. E 1655-00 in ASTM Annual Book of Standards, Vol. 03.06. West Conshohocken, PA, USA: ASTM International; 2000. [Google Scholar]
- 8.Jackson JE. Principal Component Analysis. New York, USA: Springer Science; 2004. [Google Scholar]
- 9.Kramer R. Chemometric Techniques for Quantitative Analysis. New York, NY, USA: Marcel Dekker, Inc.; 1998. [Google Scholar]
- 10.Jolliffe IT. Principal Component Analysis. New York, NY, USA: Springer Science; 2004. p. 6. [Google Scholar]
- 11.Ralston P, DePuy G, Graham JH. ISA Trans. 2004;43:639. doi: 10.1016/s0019-0578(07)60174-8. [DOI] [PubMed] [Google Scholar]
- 12.Kramer R. Chemometric Techniques for Quantitative Analysis. New York, NY, USA: Marcel Dekker, Inc; 1998. p. 99. [Google Scholar]
- 13.Brown CD, Green RL. Trends Anal. Chem. 2009;28:506. [Google Scholar]
- 14.Jolliffe IT. Principal Component Analysis. New York, NY, USA: Springer Science; 2004. p. 269. [Google Scholar]
- 15.Jolliffe IT. Principal Component Analysis. New York, NY, USA: Springer Science; 2004. p. 111. [Google Scholar]
- 16.Malinowski ER, Chemom J. 1990;4:102. [Google Scholar]
- 17.Jackson JE. A User's Guide To Principal Components. New York, NY, USA: John Wiley & Sons, Inc; 1991. p. 41. [Google Scholar]
- 18.Kramer R. Chemometric Techniques for Quantitative Analysis. New York, NY, USA: Marcel Dekker, Inc.; 1998. p. 13. [Google Scholar]
- 19.Heien M, Khan AS, Ariansen JL, Cheer JF, Phillips PEM, Wassum KM, Wightman RM. Proc. Natl. Acad. Sci. USA. 2005;102:10023. doi: 10.1073/pnas.0504657102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hermans A, Keithley RB, Kita JM, Sombers LA, Wightman RM. Anal. Chem. 2008;80:4040. doi: 10.1021/ac800108j. [DOI] [PubMed] [Google Scholar]
- 21.Venton BJ, Michael DJ, Wightman RM. J. Neurochem. 2003;84:373. doi: 10.1046/j.1471-4159.2003.01527.x. [DOI] [PubMed] [Google Scholar]
- 22.Kramer R. Chemometric Techniques for Quantitative Analysis. Vol. 17. New York, NY, USA: Marcel Dekker, Inc.; 1998. [Google Scholar]
- 23.Daszykowski M, Walczak B. Trends Anal. Chem. 2006;25:1081. [Google Scholar]
- 24.Nomikos P, Macgregor JF. Technometrics. 1995;37:41. [Google Scholar]
- 25.Justice JB., Jr . Voltammetry in the Neurosciences: Principles, Methods, and Applications. Clifton, NJ, USA: Humana Press; 1987. [Google Scholar]
- 26.Jackson JE, Mudholkar GS. Technometrics. 1979;21:341. [Google Scholar]
- 27.Jackson JE. A User's Guide to Principal Components. New York, NY, USA: John Wiley & Sons, Inc.; 1991. p. 34. [Google Scholar]
- 28.Bezegh A, Janata J. Anal. Chem. 1987;59:A494. doi: 10.1021/ac00137a011. [DOI] [PubMed] [Google Scholar]
- 29.Jensen DR, Solomon H. J. Am. Stat. Assoc. 1972;67:898. [Google Scholar]
- 30.Jouan-Rimbaud D, Bouveresse E, Massart DL, de Noord OE. Anal. Chim. Acta. 1999;388:283. [Google Scholar]
- 31.Jackson JE. A User's Guide to Principal Components. New York, NY, USA: John Wiley & Sons, Inc.; 1991. p. 21. [Google Scholar]
- 32.Fang F, Chu SG, Hong CS. Anal. Chem. 2006;78:5412. doi: 10.1021/ac0604742. [DOI] [PubMed] [Google Scholar]
- 33.De Beer TRM, Baeyens WRG, Ouyang J, Vervaet C, Remon JP. Vol. 131. Cambridge, UK: Analyst; 2006. p. 1137. [DOI] [PubMed] [Google Scholar]
- 34.Geladi P, Kowalski BR. Anal. Chim. Acta. 1986;185:1. [Google Scholar]
- 35.Faber NM, Rajko R. Anal. Chim. Acta. 2007;295:98. doi: 10.1016/j.aca.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 36.Thomas EV, Haaland DM. Anal. Chem. 1990;62:1091. [Google Scholar]
- 37.Wentzell PD, Montoto LV. Chemom. Intell. Lab. Syst. 2003;65:257. [Google Scholar]
- 38.Hendler RW, Shrager RI. J. Biochem. Biophys. Methods. 1994;28:1. doi: 10.1016/0165-022x(94)90061-2. [DOI] [PubMed] [Google Scholar]