Abstract
An additional tumor suppressor gene on chromosome 9p telomeric to the CDKN2A/B locus has long been postulated to exist. Using Affymetrix 250K single nucleotide polymorphism arrays to screen for copy number changes in glioblastoma multiforme (GBM), we detected a high frequency of deletions of the PTPRD gene, which encodes a receptor protein tyrosine phosphatase at chromosome 9p23-24.1. Missense and nonsense mutations of PTPRD were identified in a subset of the samples lacking deletions, including an inherited mutation with somatic loss of the wild-type allele. We then sequenced the gene in melanoma and identified 10 somatic mutations in 7 of 57 tumors (12%). Reconstitution of PTPRD expression in GBM and melanoma cells harboring deletions or mutations led to growth suppression and apoptosis that was alleviated by both the somatic and constitutional mutations. These data implicate PTPRD in the pathogenesis of tumors of neuroectodermal origin and, when taken together with other recent reports of PTPRD mutations in adenocarcinoma of the colon and lung, suggest that PTPRD may be one of a select group of tumor suppressor genes that are inactivated in a wide range of common human tumor types.
Introduction
Cancer is a genetic disease that results from the disruption of signaling pathways that regulate cellular proliferation, differentiation, and programmed cell death. Although it was originally hoped that there would be a small, finite number of genes controlling these signaling pathways whose dysregulation was common to many tumor types, most current research supports the notion that the majority of cancer-causing genes contribute to neoplasia at low frequency and in a limited tumor spectrum (1). Nonetheless, the discovery of new oncogenes and tumor suppressor genes commonly altered during tumorigenesis remains a major goal of modern cancer research because such genes and the pathways they control are the most exciting potential targets for anticancer drug development.
Constitutive activation of tyrosine phosphorylation signaling pathways is one biochemical hallmark of cancer. This is most well known to occur via activation of tyrosine kinase receptors, such as amplification of HER2/Neu in breast cancer and mutation of epidermal growth factor receptor in lung cancer. However, given the obvious importance of constitutive activation of tyrosine kinase signaling to human neoplasia, one might expect to find inactivation of protein tyrosine phosphatases (PTP) in human tumors as well. Although inactivating mutations of individual PTPs have recently been reported in human colon cancer (2), at present there is no single tyrosine phosphatase thought to play a generally important role as a tumor suppressor gene in multiple tumor types.
PTPRD is one of 21 known human receptor-type PTPs, a group of genes that are increasingly thought to be important in cancer development and progression (see refs. 3, 4 for reviews). Deletions of PTPRD in human cancer cell lines were first identified by Cox and colleagues in 2005 (5). Subsequent studies have reported homozygous deletions of PTPRD in multiple human tumor types (6–11), and missense mutations of unknown functional significance have recently been reported in adenocarcinoma of the colon and lung (11, 12).
Here, we identify frequent deletion and mutation of PTPRD in glioblastoma multiforme (GBM) and malignant melanoma and show that these mutations are inactivating. These data provide the first functional evidence that PTPRD is a tumor suppressor gene and, when taken together with other recent studies identifying mutations in adenocarcinoma of the colon and lung, suggest that inactivation of PTPRD contributes to the pathogenesis of a wide range of common human cancers.
Materials and Methods
Tumor tissues
A panel of 21 GBM cell lines was obtained from the American Type Culture Collection (U87MG, U138MG, M059J, Hs683, H4, A172, LN18, LN229, CCF-STTG1, T98G, and DBTRG-05MG), DSMZ (8MGBA, 42MGBA, DKMG, GAMG, GMS10, LN405, and SNB19), and the Japan Health Sciences Foundation Health Science Research Resources Bank (AM38, NMC-G1, and KG-1-C). Normal human astrocytes (NHA) were obtained from Clonetics and AllCells. All cell lines were grown in DMEM + 10% fetal bovine serum (FBS) at 37°C in 5% CO2.
S.c. xenografts in immunodeficient mice were obtained from the Duke University Brain Tumor Center or created in the Lombardi Comprehensive Cancer Center Animal Shared Resource from tissue taken from patients undergoing craniotomy at Georgetown University Hospital (IRB #2006-344).
Snap-frozen primary GBM tumors and paired blood samples were obtained from the Brain Tumour Tissue Bank (London Health Sciences Centre, London, Ontario, Canada) funded by the Brain Tumour Foundation of Canada. All tumors were graded by a neuropathologist as good or moderate on a scale of good to poor depending on the amount of tumor cells present (as opposed to hemorrhagic, necrotic, or fibrous tissue). All tumor samples were further categorized as “tumor center.”
A panel of 10 primary GBM cell cultures was derived from primary tumor samples at time of surgical resection at the University of Iowa Medical Center by dissociation with collagenase and then cultured in DMEM/F12 containing 15% FBS, 10 μg/mL insulin, and 5 ng/mL basic fibroblast growth factor at 37°C in 5% CO2.
A panel of 57 malignant melanoma tumor and paired blood samples was collected during surgical resection at the National Cancer Institute. The primary cell cultures 16T and 86T used for functional analysis were derived from melanoma tumor samples by dissociation with collagenase and then cultured in RPMI 1640 + 10% FBS at 37°C in 5% CO2.
Microarrays and bioinformatics
Genomic DNA derived from GBM tumor samples was interrogated with Affymetrix 250K NspI Human Gene Chip Arrays using protocols described by the manufacturer. Data processing was performed using dCHIP (13). The scanned array images and processed data sets have been deposited in the Gene Expression Omnibus.7
Western blot
Primary antibodies used were PTPRD clone C-18 (Santa Cruz Biotechnology) and α-tubulin Ab-2 clone DM1A (NeoMarkers).
DNA sequencing
Individual exons of PTPRD were PCR amplified from genomic DNA using conditions and primer pairs described by Sjoblom and colleagues (12). PCR products were purified using the Exo/SAP method followed by a Sephadex spin column. Sequencing reactions were performed using BigDye v3.1 (Applied Biosystems) using an M13F primer and analyzed on an Applied Biosystems 3730XL capillary sequencer. Sequences were analyzed using Mutation Surveyor (Softgenetics). Traces with putative mutations were reamplified and sequenced from both tumor and matched normal DNA.
PTPRD lentivirus
A wild-type PTPRD cDNA (MGC 119751) was obtained from Open Biosystems and cloned into the pCDF1-MCS2-EF1-Puro lentiviral expression vector backbone (System Biosciences). To make virus, this construct was cotransfected into 293T cells with pVSV-G (Addgene) and pFIV-34N (System Biosciences) helper plasmids using Fugene 6 (Roche) as described by the manufacturer. Virus-containing conditioned medium was harvested 48 h after transfection, filtered, and used to infect recipient cells in the presence of 8 μg/mL polybrene.
Site-directed mutagenesis
Mutations identified in GBM and melanoma tumors were engineered into the pCDF1-PTPRD construct by site-directed mutagenesis using the QuikChange II XL kit (Stratagene) as directed by the manufacturer. The coding sequence of all expression vectors was verified by DNA sequencing.
Flow cytometry
Cells were pulsed with 10 μmol/L bromodeoxyuridine (BrdUrd) for 1 h, trypsinized, and centrifuged. Cells were fixed and stained using the BrdUrd Flow kit (Pharmingen) and analyzed by flow cytometry in a BD FACSort instrument using FCS Express v.3 software (DeNovo Software).
Apoptosis quantification assay
Cells were collected by trypsinization, centrifuged, and simultaneously fixed and stained in a solution containing 3.7% formaldehyde, 0.5% Igepal, and 10 μg/mL Hoechst 33258 in PBS. Fluorescence microscopy was used to visualize and score apoptotic nuclei. At least 200 cells were counted for each determination in triplicate.
Microscopy
All imaging was performed on an Olympus BX61 light microscope with a 40× Plan-Apochromat objective.
Statistical analysis
Two-tailed unpaired t test analysis of BrdUrd incorporation data was performed using GraphPad Prism software.
Results
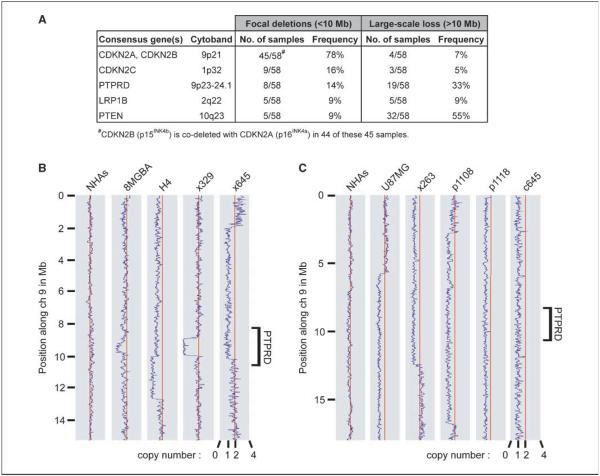
In an effort to discover genes that contribute to the pathogenesis of GBM, we used Affymetrix 250K Gene Chip Arrays to identify recurrent copy number alterations in a panel of 58 GBM tumor samples (Fig. 1A). Focal deletions of the PTPRD gene on chromosome 9p23-24.1 were among the most prevalent deletions detected, present in 14% of the GBM samples studied (Fig. 1B; Supplementary Table S1). This frequency of focal deletion is higher than that of PTEN (9%) and similar to that of CDKN2C (also named p18INK4c, 16%), a recently identified GBM tumor suppressor gene (14, 15). Larger-scale loss of the PTPRD gene was present in an additional 33% of the samples (Fig. 1C; Supplementary Table S2). Intriguingly, several studies have suggested the presence of another important tumor suppressor gene on chromosome 9p telomeric to the CDKN2A/B locus in tumor types including astrocytoma, melanoma, and lung adenocarcinoma (16–20). We therefore considered PTPRD to be an attractive candidate as a GBM tumor suppressor gene, and possibly relevant to a range of other tumor types as well.
Figure 1.
PTPRD is deleted at high frequency in GBM. A, most frequently deleted genes in 58 GBM tumor samples as determined by Affymetrix 250K SNP microarray analysis. B, copy number analysis of SNP microarray data shows focal (<10 Mb) deletions of chromosome 9p23-24.1 in 8 of 58 GBM samples (4 shown) but not in NHAs. x, primary xenograft. C, copy number analysis of SNP microarray data shows large-scale (>10 Mb) chromosomal loss of the PTPRD locus in 19 of 58 GBM samples (5 shown). p, primary tumor; c, primary culture.
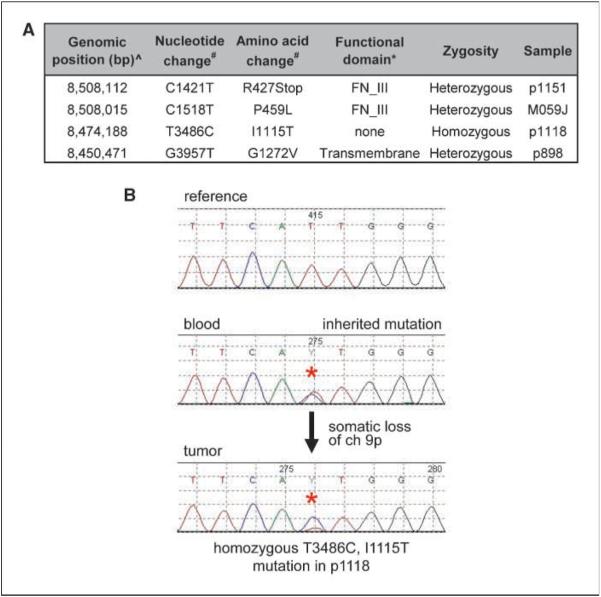
To determine if PTPRD is genetically altered by mutation during GBM tumorigenesis, we sequenced the 35 coding exons of the PTPRD gene in tumor samples lacking focal deletions of PTPRD and in corresponding normal tissue (when available). This sequence analysis identified somatic mutations of the PTPRD gene in three samples, including two missense mutations and one nonsense mutation (Fig. 2A; Supplementary Fig. S1). Additionally, we identified a heterozygous germ-line mutation that was accompanied by somatic loss of the wild-type allele in the tumor of a GBM patient with a history of multiple primary malignancies (Fig. 2A and B). This mutation is not a reported single nucleotide polymorphism (SNP) and was not present in any of >100 alleles of PTPRD sequenced during the course of this study. Together, these data show that PTPRD is altered by somatic mutation during GBM pathogenesis and raise the intriguing possibility that germ-line mutation of PTPRD might lead to a predisposition to the development of GBM and other tumor types.
Figure 2.
Identification of somatic and inherited mutations of PTPRD in GBM. A, one nonsense and three missense mutations were identified in GBM samples. ^, genomic position is based on the hg18 genome assembly; #, transcript ENST00000381196 was used for annotation of the nucleotide and amino acid changes; *, assignment of functional domains was based on UniProtKB/Swiss-Prot P23468-1. FN_III, fibronectin type III domain. B, sequence traces depicting an inherited heterozygous mutation of PTPRD in constitutional DNA (blood) and somatic loss of the wild-type allele in GBM primary tumor p1118.
To determine if mutations of PTPRD were present in a second tumor type proposed to harbor an additional 9p tumor suppressor gene, we sequenced PTPRD in 57 melanoma tumor samples. Two somatic nonsense mutations and eight somatic missense mutations were identified (Table 1; Supplementary Figs. S2 and S3) in a total of seven samples. All of these mutations were C/G>T/A transversions, consistent with UV-induced DNA damage. Additionally, three of the mutations were dinucleotide CC>TT mutations caused by the formation of UV-induced cyclobutane pyrimidine dimers (Supplementary Fig. S3). Three of the seven samples harboring somatic mutations of PTPRD displayed loss of heterozygosity (LOH) of the wild-type allele. Furthermore, tumor 76T harbored four independent mutations of the gene, increasing the likelihood that both alleles of the gene had been targeted by mutation in this sample. Interestingly, five of these seven samples with PTPRD mutation also harbor activating mutations of either B-Raf or N-Ras (data not shown). This 12% mutation frequency makes PTPRD among the most commonly mutated genes in sporadic melanoma reported to date, which include B-Raf (~60%), p53 (0–25%), N-Ras (10–15%), PTEN (~10%), p16INK4a (0–5%), and PIK3CA (<1%; refs. 21, 22).
Table 1.
Identification of somatic mutations of PTPRD in malignant melanoma
| Genomic position (bp)* | Nucleotide change † | Amino acid change † | Functional domain ‡ | Zygosity | Sample |
|---|---|---|---|---|---|
| 8,626,727 | G324A | G61E | lg_C2 | Heterozygous | 34T |
| 8,508,298 | G1235A | E365K | FN_III | Heterozygous | 76T |
| 8,508,054 | G1479A | G446E | FN_III | Heterozygous | 16T |
| 8,475,256 | G3266A | E1042K | FN_III | Heterozygous | 13T |
| 8,474,156 | G3518A | V1126M | None | Heterozygous | 76T |
| 8,450,544 | G3884A | D1248N | None | Homozygous | 21T |
| 8,379,285–8,379,286 | G4474A, G4475A | W1444Stop | PTPc | Heterozygous | 76T |
| 8,331,947 | G4835A | V1565I | PTPc | Homozygous | 86T |
| 8,331,147–8,331,148 | C5210T, C5211T | P1690F | PTPc | Heterozygous | 6T |
| 8,321,724–8,321,725 | C5533T, C5534T | R1798Stop | PTPc | Heterozygous | 76T |
NOTE: Ten somatic mutations of PTPRD were identified in 7 of 57 malignant melanoma tumor samples.
Abbreviations: Ig_C2, immunoglobulin-like C2-type domain; FN_III, fibronectin type III domain.
Genomic position is based on the hg18 genome assembly.
Transcript ENST00000381196 was used for annotation of the nucleotide and amino acid changes.
Assignment of functional domains was based on UniProtKB/Swiss-Prot P23468-1.
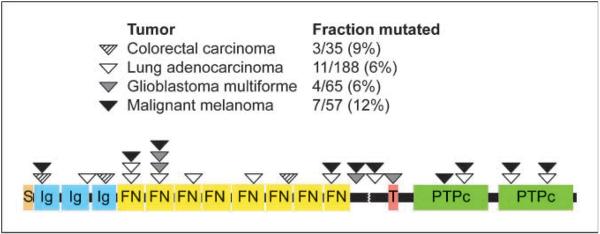
The 14 mutations of PTPRD reported here are distributed roughly evenly throughout the various extracellular and intracellular domains of the encoded PTPRD protein (Fig. 3), although there seems to be a mini-hotspot in the first and second fibronectin type III repeat.
Figure 3.
Schematic of the PTPRD protein and the location of all mutations reported to date in human cancer. The mutations in colon cancer and lung cancer were previously reported in refs. 11 and 12. S, signal peptide; Ig, immunoglobulin-like C2-type domain; FN, fibronectin type III domain. Broken line, cleavage site; T, transmembrane domain; PTPc, protein tyrosine phosphatase catalytic domain.
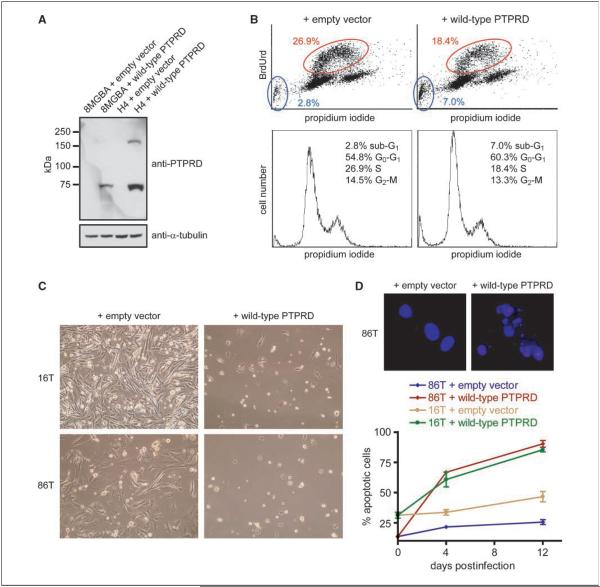
Despite its potential importance, functional data implicating PTPRD deletion or mutation in tumorigenesis are lacking. To determine if PTPRD has the growth-suppressing properties expected of a broad-spectrum tumor suppressor gene, we examined the functional consequences of reconstituting PTPRD expression in GBM and melanoma cells. A 5.1-kb human PTPRD cDNA was obtained, cloned into a lentiviral expression vector, and packaged into infectious lentivirus as described in Materials and Methods. Infection of H4 cells that harbor biallelic deletion of PTPRD (Fig. 1B; Supplementary Table S1) with lenti-PTPRD led to expression of both the PTPRD proprotein and its mature cleavage products (which then reassemble at the cell membrane to form a heterodimer; Fig. 4A; ref. 23). Infection of H4 cells with lenti-PTPRD but not vector alone led to a transient growth arrest evidenced by a reduction in BrdUrd incorporation and an increase in both G1 and sub-G1 cell populations (Fig. 4B). Infection with lenti-PTPRD had a similar effect on 8MGBA cells, which also harbor a focal deletion of PTPRD (Fig. 1B; data not shown).
Figure 4.
Expression of PTPRD in GBM and melanoma cells harboring deletions and mutations causes growth suppression and apoptosis. A, Western blot for PTPRD shows reconstitution of PTPRD expression in 8MGBA and H4 GBM cells infected with lenti-PTPRD. Both cell lines harbor focal deletions of PTPRD (Fig. 1B), and no endogenous PTPRD protein was detected in either cell line. The C-18 antibody recognizes a COOH-terminal epitope present on both the full-length PTPRD proprotein (~175 kDa) and one of the two mature cleavage products (~75 kDa). B, flow cytometry of H4 GBM cells at 48 h after infection reveals that lenti-PTPRD causes a 32% reduction in BrdUrd incorporation and a 2.5-fold increase in sub-G1 cells. Cell cycle distributions are shown. C, phase-contrast microscopy of cells 10 d after infection shows that reconstitution of PTPRD expression leads to frank cell death in 16T and 86T melanoma cells harboring homozygous missense mutations of PTPRD (Table 1). D, lenti-PTPRD causes a time-dependent increase in apoptosis in 16T and 86T cells. Top, Hoechst-stained nuclei of cells undergoing apoptosis after infection with lenti-PTPRD; bottom, quantification of apoptotic cells.
We next infected two primary melanoma cell cultures harboring homozygous missense mutations of PTPRD [16T and 86T with G446E in the second fibronectin type III domain and V1565I in the first PTP catalytic (PTPc) domain, respectively]. Infection of both primary cell cultures with wild-type PTPRD but not vector alone led to significant growth inhibition and decrease in cell viability (Fig. 4C), as well as a substantial, time-dependent increase in apoptotic cells (Fig. 4D). These are the first reported data indicating that PTPRD has growth-suppressive properties when expressed in human cancer cells, supporting the hypothesis that PTPRD is a bona fide human tumor suppressor gene.
We next sought to examine the consequences of tumor-derived mutations on PTPRD function in these assays. To do this, five tumor-derived mutations were introduced into lenti-PTPRD as described in Materials and Methods, including two mutations in the second fibronectin type III domain mini-hotspot (one each from GBM and melanoma), one mutation in the first PTPc domain (melanoma), and two mutations flanking the proprotein cleavage site (one each from GBM and melanoma). Initially, H4 cells were infected with wild-type and mutant lenti-PTPRD, protein lysates were prepared, and PTPRD expression was documented by Western blot. As shown in Fig. 5A, infection of H4 cells with lentivirus expressing either wild-type or mutant PTPRD resulted in similar levels of protein expression. However, there was a marked decrease in growth inhibition as measured by BrdUrd incorporation, indicating that each of the five tumor-derived mutants alleviated the growth suppression activity of PTPRD, albeit to differing extents (Fig. 5B). Next, 16T melanoma cells were similarly infected with wild-type and mutant PTPRD lentiviruses. As depicted in Fig. 5C, wild-type PTPRD led to apoptosis of ~75% of the cells at 10 days after infection, whereas the mutant PTPRD lentiviruses led to a substantially reduced fraction of cells that had undergone programmed cell death. When taken together, these experiments show that tumor-derived mutations of PTPRD attenuate its function, confirming that the mutations of PTPRD are likely to be pathogenic.
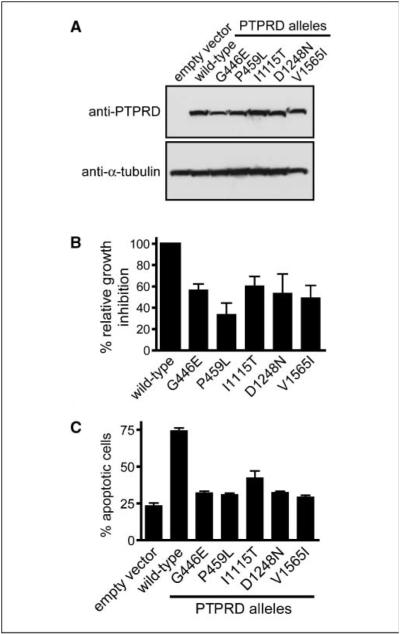
Figure 5.
Tumor-derived mutations compromise the growth-suppressive function of PTPRD in GBM and melanoma cells. Five tumor-derived mutations were introduced into lenti-PTPRD. A, Western blot analysis for PTPRD protein shows equivalent expression of wild-type and mutant proteins in infected H4 GBM cells. B, infection of H4 cells with wild-type PTPRD led to growth suppression, as measured by BrdUrd incorporation using a 1-h pulse 48 h after infection. In contrast, ectopic expression of the PTPRD cDNA harboring tumor-derived mutations led to less potent growth suppression, albeit to varying extents (25–60% of the activity relative to wild-type). This experiment was performed in triplicate, and the inactivation of growth-suppressive activity was shown to be statistically significant (P < 0.05 for each of the mutants, except D1248N with P = 0.06). C, infection of 16T melanoma cells with wild-type PTPRD led to apoptosis of ~75% of the infected cells at 10 d after infection, whereas the mutant PTPRD lentiviruses led to a substantially reduced fraction of cells that had undergone programmed cell death.
Discussion
Aberrant regulation of signaling pathways governed by tyrosine phosphorylation is common to virtually all types of human cancer. Whereas activation of tyrosine kinase growth factor receptors by amplification and/or mutation is well established as a major mechanism leading to constitutive tyrosine phosphorylation, the role of inactivation of PTPs in tumorigenesis is comparatively poorly understood. Here, we implicate one such PTP as a tumor suppressor gene in two major tumor types and show that its reconstitution in human cancer cells harboring deletions and mutations leads to cell cycle arrest and apoptosis.
Receptor-type PTPs are transmembrane proteins whose extracellular immunoglobulin-like and fibronectin domains are known to promote cell-cell adhesions (3, 4, 24, 25). It is postulated that these PTPs transduce information about cell contacts across the membrane to the intracellular phosphatase domains, which control cytoplasmic tyrosine phosphorylation levels appropriate for the current state of cell adhesion. PTPRD is highly expressed in the neuroepithelium during early development of the nervous system (26)—an environment where cell-to-cell contacts are essential for patterning and directing appropriate synaptic connections (27). Accordingly, homozygous knockout of PTPRD in the mouse results in impaired learning, enhanced hippocampal long-term potentiation, and early postnatal lethality due to insufficient food intake (28). Whether heterozygous knockout mice are tumor prone has not yet been established.
Interestingly, PTPRD is also known to be expressed in the adult, with highest levels present in the brain and kidney (23). The function of PTPRD in these tissues after completion of development has not yet been described. Current evidence suggests that PTPRD is a homophilic cell adhesion molecule, but it is also possible that its extracellular domain binds to other as yet unidentified ligands present in the extracellular space. Furthermore, the substrate(s) of its phosphatase domains remains unidentified (29).
Other than the fact that inactivation of PTPRD is predicted to increase tyrosine phosphorylation of signaling molecules, it is not clear at this time how PTPRD suppresses cancer development and/or progression. It is possible that in the hypercellular microenvironment of a neoplastic lesion, PTPRD senses an increasing abundance of receptor-type PTP molecules present on neighboring neoplastic cells causing activation of its phosphatase domains resulting in decreased tyrosine phosphorylation of signaling molecules that promote cellular proliferation. In this hypothesized model, it is easy to speculate how inactivation of PTPRD tumor-suppressive function through deletion or mutation might promote tumorigenesis. It is also possible that PTPRD more simply functions to maintain a normal homeostasis of intracellular tyrosine phosphorylation levels in tissues of the adult organism. In this model, inactivation of PTPRD would lead to the increase in tyrosine phosphorylation of as yet unidentified signaling molecule(s) in a way that promotes proliferation and neoplasia.
In summary, we believe that the identification of deletions and mutations of PTPRD in the wide spectrum of human tumors reported to date, the presence of an inherited mutation in a patient with LOH of the wild-type allele in the tumor, and the functional data presented herein provide strong evidence that PTPRD is a bona fide human tumor suppressor gene. Future studies seem warranted to identify the complete range of tumors in which PTPRD is inactivated, to determine if PTPRD plays a more general role in inherited cancer predisposition syndromes, and to identify and pharmacologically inhibit the signaling pathway(s) that is activated in cells harboring deletions and mutations of this receptor-type PTP.
Supplementary Material
Acknowledgments
Grant support: T. Waldman is funded by the National Cancer Institute and the American Cancer Society. Y. Samuels is funded by the National Human Genome Research Institute and The Harry J. Lloyd Charitable Trust.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
We thank Marcela White (Brain Tumor Tissue Bank, London Health Sciences Centre, London, Ontario, Canada) for her assistance with the procurement of high-quality GBM specimens and Karen Creswell for assistance with flow cytometry.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed..
References
- 1.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 2.Wang Z, Shen D, Parsons DW, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–6. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 3.Ostman A, Hellberg C, Bohmer FD. Protein-tyrosine phosphatases and cancer. Nature Rev Cancer. 2006;6:307–20. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- 4.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–46. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 5.Cox C, Bignell G, Greenman C, et al. A survey of homozygous deletions in human cancer genomes. Proc Natl Acad Sci U S A. 2005;102:4542–7. doi: 10.1073/pnas.0408593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato M, Takahashi K, Nagayama K, et al. Identification of chromosome arm 9p as the most frequent target of homozygous deletions in lung cancer. Genes Chromosomes Cancer. 2005;44:405–14. doi: 10.1002/gcc.20253. [DOI] [PubMed] [Google Scholar]
- 7.Stallings RL, Nair P, Maris JM, et al. High-resolution analysis of chromosomal breakpoints and genomic instability identifies PTPRD as a candidate tumor suppressor gene in neuroblastoma. Cancer Res. 2006;66:3673–80. doi: 10.1158/0008-5472.CAN-05-4154. [DOI] [PubMed] [Google Scholar]
- 8.Stark M, Hayward N. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2007;67:2632–42. doi: 10.1158/0008-5472.CAN-06-4152. [DOI] [PubMed] [Google Scholar]
- 9.Purdie KJ, Lambert SR, Teh MT, et al. Allelic imbalances and microdeletions affecting the PTPRD gene in cutaneous squamous cell carcinomas detected using single nucleotide polymorphism microarray analysis. Genes Chromosomes Cancer. 2007;46:661–9. doi: 10.1002/gcc.20447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagayama K, Kohno T, Sato M, Arai Y, Minna JD, Yokota J. Homozygous deletion scanning of the lung cancer genome at 100-kb resolution. Genes Chromosomes Cancer. 2007;46:1000–10. doi: 10.1002/gcc.20485. [DOI] [PubMed] [Google Scholar]
- 11.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–8. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 13.Li C, Wong WH. Model-based analysis of oligonucle-otide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–6. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solomon DA, Kim JS, Jenkins S, et al. Identification of p18INK4c as a tumor suppressor gene in glioblastoma multiforme. Cancer Res. 2008;68:2564–9. doi: 10.1158/0008-5472.CAN-07-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiedemeyer R, Brennan C, Heffernan TP, et al. Feedback circuit among INK4 tumor suppressors constrains human glioblastoma development. Cancer Cell. 2008;13:355–64. doi: 10.1016/j.ccr.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueki K, Rubio MP, Ramesh V, et al. MTS1/CDKN2 gene mutations are rare in primary human astrocytomas with allelic loss of chromosome 9p. Hum Mol Genet. 1994;3:1841–5. doi: 10.1093/hmg/3.10.1841. [DOI] [PubMed] [Google Scholar]
- 17.Puig S, Ruiz A, Lázaro C, et al. Chromosome 9p deletions in cutaneous malignant melanoma tumors: the minimal deleted region involves markers outside the p16 (CDKN2) gene. Am J Hum Genet. 1995;57:395–402. [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SK, Ro JY, Kemp BL, et al. Identification of three distinct tumor suppressor loci on the short arm of chromosome 9 in small cell lung cancer. Cancer Res. 1997;57:400–3. [PubMed] [Google Scholar]
- 19.Pollock PM, Welch J, Hayward NK. Evidence for three tumor suppressor loci on chromosome 9p involved in melanoma development. Cancer Res. 2001;61:1154–61. [PubMed] [Google Scholar]
- 20.Cook AL, Pollock PM, Welch J, et al. CDKN2A is not the principal target of deletions on the short arm of chromosome 9 in neuroendocrine (Merkel cell) carcinoma of the skin. Int J Cancer. 2001;93:361–7. doi: 10.1002/ijc.1352. [DOI] [PubMed] [Google Scholar]
- 21.Fecher LA, Cummings SD, Keefe MJ, Alani RM. Toward a molecular classification of melanomas. J Clin Oncol. 2007;25:1606–20. doi: 10.1200/JCO.2006.06.0442. [DOI] [PubMed] [Google Scholar]
- 22.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;355:2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 23.Pulido R, Krueger NX, Serra-Pages C, Saito H, Streuli M. Molecular characterization of the human transmembrane protein-tyrosine phosphatase δ. Evidence for tissue-specific expression of alternative human transmembrane protein-tyrosine phosphatase δ isoforms. J Biol Chem. 1995;270:6722–8. doi: 10.1074/jbc.270.12.6722. [DOI] [PubMed] [Google Scholar]
- 24.Aricescu AR, Siebold C, Choudhuri K, et al. Structure of a tyrosine phosphatase adhesive interaction reveals a spacer-clamp mechanism. Science. 2007;317:1217–20. doi: 10.1126/science.1144646. [DOI] [PubMed] [Google Scholar]
- 25.Gebbink MF, Zondag GC, Wubbolts RW, Beijersbergen RL, van Etten I, Moolenaar WH. Cell-cell adhesion mediated by a receptor-like protein tyrosine phosphatase. J Biol Chem. 1993;268:16101–4. [PubMed] [Google Scholar]
- 26.Sommer L, Rao M, Anderson DJ. RPTPδ and the novel protein tyrosine phosphatase RPTPψ are expressed in restricted regions of the developing central nervous system. Dev Dyn. 1997;208:48–61. doi: 10.1002/(SICI)1097-0177(199701)208:1<48::AID-AJA5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 27.Thiery JP. Cell adhesion in development: a complex signaling network. Curr Opin Genet Dev. 2003;13:365–71. doi: 10.1016/s0959-437x(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 28.Uetani N, Kato K, Ogura H, et al. Impaired learning with enhanced hippocampus long-term potentiation in PTPδ-deficient mice. EMBO J. 2000;19:2775–85. doi: 10.1093/emboj/19.12.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Bixby JL. Receptor tyrosine phosphatase-δ is a homophilic, neurite-promoting cell adhesion molecule for CNS neurons. Mol Cell Neurosci. 1999;14:370–84. doi: 10.1006/mcne.1999.0789. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.