Abstract
 Parametrization of the additive all-atom CHARMM force field for acyclic polyalcohols, acyclic carbohydrates and inositol is conducted. Initial parameters were transferred from the alkanes and hexopyranose carbohydrates, with subsequent development and optimization of parameters unique to the molecules considered in this study. Using the model compounds acetone and acetaldehyde, nonbonded parameters for carbonyls were optimized targeting quantum mechanical interaction data for solute-water pairs and pure solvent thermodynamic data. Bond and angle parameters were adjusted by comparing optimized geometries to small molecule crystal survey data and by performing vibrational analyses on acetone, acetaldehyde and glycerol. C-C-C-C, C-C-C-O, C-C-OH and O-C-C-O torsional parameters for polyol chains were fit to quantum mechanical dihedral potential energy scans comprising over 1500 RIMP2/cc-pVTZ//MP2/6-31G(d) conformations using an automated Monte Carlo simulated annealing procedure. Comparison of computed condensed-phase data, including crystal lattice parameters and densities, NMR proton-proton couplings, densities and diffusion coefficients of aqueous solutions, to experimental data validated the optimized parameters. Parameter development for these compounds proved particularly challenging because of the flexibility of the acyclic sugars and polyalcohols as well as the intramolecular hydrogen bonding between vicinal hydroxyls for all of the compounds. The newly optimized additive CHARMM force field parameters are anticipated to be of utility for atomic level of detail simulations of acyclic polyalcohols, acyclic carbohydrates and inositol in solution.
Parametrization of the additive all-atom CHARMM force field for acyclic polyalcohols, acyclic carbohydrates and inositol is conducted. Initial parameters were transferred from the alkanes and hexopyranose carbohydrates, with subsequent development and optimization of parameters unique to the molecules considered in this study. Using the model compounds acetone and acetaldehyde, nonbonded parameters for carbonyls were optimized targeting quantum mechanical interaction data for solute-water pairs and pure solvent thermodynamic data. Bond and angle parameters were adjusted by comparing optimized geometries to small molecule crystal survey data and by performing vibrational analyses on acetone, acetaldehyde and glycerol. C-C-C-C, C-C-C-O, C-C-OH and O-C-C-O torsional parameters for polyol chains were fit to quantum mechanical dihedral potential energy scans comprising over 1500 RIMP2/cc-pVTZ//MP2/6-31G(d) conformations using an automated Monte Carlo simulated annealing procedure. Comparison of computed condensed-phase data, including crystal lattice parameters and densities, NMR proton-proton couplings, densities and diffusion coefficients of aqueous solutions, to experimental data validated the optimized parameters. Parameter development for these compounds proved particularly challenging because of the flexibility of the acyclic sugars and polyalcohols as well as the intramolecular hydrogen bonding between vicinal hydroxyls for all of the compounds. The newly optimized additive CHARMM force field parameters are anticipated to be of utility for atomic level of detail simulations of acyclic polyalcohols, acyclic carbohydrates and inositol in solution.
Keywords: polyol, polyhydric alcohol, sugar alcohol, alditol, empirical force field
INTRODUCTION
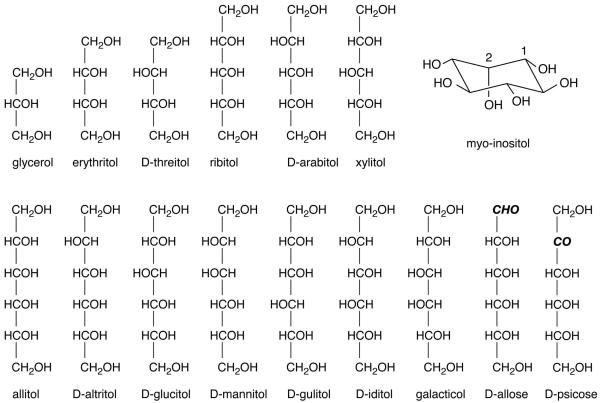
Glucose and related six-carbon monosaccharides exist in aqueous solution in equilibrium between the thermodynamically-favored cyclic pyranose form and the linear aldose form. Reduction of the aldehyde functionality in the aldose form to an alcohol yields a linear polyalcohol or polyol, also commonly referred to as an “alditol” or “sugar alcohol.” Such six-carbon sugar alcohols, as well as related cyclic and shorter-chain linear polyols (Figure 1), have both biological and industrial significance. One example is the conversion of the linear aldose sugar D-glucose to the alditol D-glucitol (also known as sorbitol) by reduction of the aldehyde group at C1 and subsequent oxidation at C2 to produce the linear ketose sugar D-fructose. Elevated conversion from glucose to sorbitol to fructose in humans is a factor in diabetes, and pharmacologic inhibition of the enzyme aldose reductase that catalyzes the reaction is a form of therapy.1-3 In addition to participating in metabolism, these compounds also participate in cell-signaling. Inositol 1,4,5-trisphosphate, a derivative of the cyclic polyol inositol (Figure 1), is produced from the hydrolysis of the membrane phospholipid phosphatidyl inositol 4,5-bisphosphate, and acts as a secondary messenger that signals a rapid release of calcium from intracellular stores.4,5 In addition to their biological significance, this class of compounds and their derivatives have industrial applications as sugar substitutes, surfactants, lubricants and even explosives, motivating their structural, dynamic, and thermodynamic characterization.
Figure 1.
Acyclic polyalcohols, acyclic carbohydrates and inositol. In the linear compounds, C1 is at the topmost position of the carbon chain; the aldehyde and ketone functionalities at C1 and C2 in D-allose and D-psicose, respectively, are in bold italics. In inositol, the C1 and C2 position are indicated with 1 and 2, respectively. Those compounds not designated with a D are meso compounds.
Molecular dynamics (MD) simulations are a powerful and flexible way of studying structure, dynamics, and thermodynamics at an atomic level of detail. However, MD simulation results of a compound are only as reliable as the force field used to describe its structural and energetic properties. Accordingly, there has been much effort toward improving carbohydrate force fields, with focus primarily on cyclic pyranoses such as glucose.6-13 Owing to the vast array of carbohydrates and their derivatives found in both biological and industrial settings, a comprehensive carbohydrate force field remains to be fully developed and validated. To maximize its utility, such a carbohydrate force field should also be compatible with available protein, lipid, and nucleic acid force fields since carbohydrates rarely participate in biological processes without interaction with these other three major biomolecular classes.
The present work describes the development of CHARMM force field parameters for linear polyalcohols, inositol, and linear sugars. These parameters have been developed for compatibility with the existing protein,14,15 lipid16,17 and nucleic acid18,19 CHARMM all-atom additive force fields20,21 and extend the library of available carbohydrates, which had been previously limited to cyclic hexopyranoses.22 Bond and angle internal parameters were transferred from previous work22 with some modifications based on a survey of the Cambridge Crystallographic Database (CSD)23 and vibrational analyses. Torsional parameters were fit to the relaxed quantum mechanical (QM) potential energy surfaces at the RIMP2/cc-pVTZ//MP2/6-31G(d) level using an automated Monte Carlo simulated annealing procedure.24 The nonbonded parameters for the linear polyols and inositol were transferred from the hexopyranoses,22 whereas the nonbonded parameters for the carbonyl in aldoses and ketoses were fit to reproduce heats of vaporization and molecular volumes for neat liquids of representative model compounds as part of the present work. Parameter validation involved pure solvent calculations to compare to experimental heats of vaporization and molecular volumes, crystal simulations to compare to crystal lattice parameters, and aqueous phase simulations to compare to experimental solution densities, diffusion constants and NMR J-coupling constants. Also addressed are the difficulties encountered during the parametrization process due to the flexibility of these compounds and the extensive intramolecular hydrogen bonding between the vicinal hydroxyl groups.
METHODS
All molecular mechanics calculations were performed with the CHARMM program.20,25 The all-atom additive CHARMM force field uses the energy potential U(r) given in Equation 1.
| (1) |
In Equation 1, Kb, Kθ, KUB, Kχ and Kimp are bond, valence angle, Urey-Bradley, dihedral angle, and improper dihedral angle force constants, respectively, while b, θ, S, χ and φ are the bond distance, valence angle, Urey-Bradley 1,3-distance, dihedral angle and improper dihedral angle, respectively, where the subscript zero represents the equilibrium value. In the dihedral potential energy term, n is the multiplicity and σ is the phase angle as in a Fourier series. The nonbonded interaction energy between atoms i and j is separated into two terms, the Lennard-Jones (LJ) 6-12 term and the Coulomb term. For the nonbonded terms, εij is the LJ well depth, Rmin,ij is the distance at the LJ energy minimum, qi and qj are the partial atomic charges, and rij is the distance between atoms i and j. For the LJ parameters, the Lorentz-Berthelot combining rules are applied.26
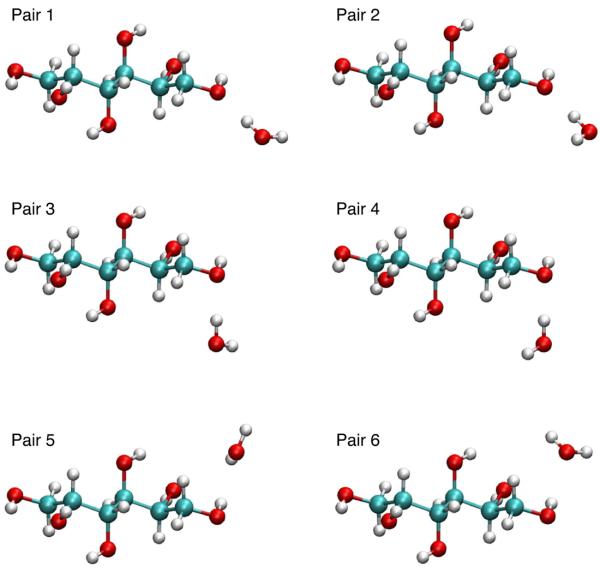
Hydrogen bonding water-solute pair interaction energies and distances were calculated using the standard additive CHARMM force field protocol, so as to maintain compatibility with existing CHARMM biomolecular force fields.19,21 Solute geometries were obtained from optimization at the MP2/6-31G(d) level of the conformation in the respective crystals obtained from the Cambridge Structural Database.23 Using the optimized geometry of the monomer, water-monomer pairs were constructed, with the water internal geometry identical to that of the TIP3P water model.27 Examples of these pair interactions are shown in Figure 2 where the water molecule is interacting with the terminal hydroxyl of allitol. In pairs 1, 2, 3 and 4 the hydroxyl oxygen is the hydrogen bond donor and in 5 and 6 the hydroxyl hydrogen is the hydrogen bond acceptor. For pairs 1, 2, 3 and 4 the hydrogen of the water molecule is directed at the COH bisector. In pairs 1 and 2 the water molecule lies in the COH plane whereas in pairs 3 and 4 the water molecule lies at a 120 degrees angle to the COH plane. Pairs 5 and 6 are different because the water molecule in these interaction pairs acts as the hydrogen bond acceptor; therefore, the COH hydroxyl is directed along the water HOH bisector. For pair 5, the HOH plane of the water is at a 90-degree angle to the COH plane, and for pair 6 the HOH and COH atoms are coplanar.
Figure 2.
Allitol-water interaction orientations used for the water interaction calculations. VMD70 is used to prepare the molecular graphics.
Reference data for comparison of molecular mechanics (MM) interaction energies and distances were generated by geometry optimization of the interaction distances at the QM HF/6-31G(d) level for each of the water-solute pairs above, with all other degrees of freedom constrained. The QM data cannot be targeted directly, but are instead empirically scaled to account for the fact that the MM force field needs to be able to account for many-body effects in the condensed phase. The CHARMM additive force field empirical scaling rules are well-established15,28 and are such that the MM target distance is the RQM − 0.2 Å and the MM target pair interaction energy (denoted “EQM”) is given by the expression 1.16*(EQM,pair − EQM,solute − EQM,water). The energy-scaling factor of 1.16 and the offset of the QM distance by 0.2 Å account for limitations in the potential energy function and in the QM level of theory, and these empirical corrections lead to good agreement with condensed phase properties, as shown in previous work.20,21
All of the C-C-C-C, C-C-C-O, O-C-C-O and C-C-O-H dihedral parameters are fit to relaxed QM potential energy scans. The Gaussian03 package29 is used to optimize geometries at the MP2/6-31G(d) level of theory followed by single point calculations performed at the RIMP2/cc-pVTZ level with the QCHEM program.30 This level of theory has previously been shown to be sufficiently accurate for a number of systems including carbohydrates.22,31 The target dihedral is scanned at 15° intervals from −180 to 165°, with the exception of inositol, which is scanned from 15 to 135° due to the constrained nature of the ring. The dihedral parameters are then fit to the QM dihedral scans using an automated Monte Carlo simulated annealing (MCSA) method.24 In the MCSA method the selected dihedral parameters are fit simultaneously to minimize the root mean squared error (RMSE),
| (2) |
where and are the QM and MM energies of conformation i, wi is a weighting factor for conformation i and c is a constant that aligns the QM and MM data to minimize the RMSE. All of the six-carbon (n=6) alditols are used in the fitting procedure (Figure 1) and all of the five-carbon (n=5) and four-carbon (n=4) alditols are used as the test set for the parameter validation. With inositol, the C-C-C-O, O-C-C-O and C-C-O-H dihedrals are transferred from the hexopyranose parameters and only the C-C-C-C is fit (independently from the n=6 alditols). For the dihedral parameters in the aldehyde and ketone groups in the linear carbohydrates D-allose and D-psicose (Figure 1), only the torsions containing non-hydrogen atoms and including the carbonyl atoms are parametrized and the other torsional parameters are transferred from the n=6 alditols.
Parameter optimization and validation of the parameters is performed via a number of condensed phase MD simulations. A cubic box containing TIP3P water molecules27,32 with periodic boundary conditions is used for all aqueous simulations. Particle Mesh Ewald (PME)33 with a 12 Å real space cutoff is used to treat the long-range Coulomb interactions and a force-switched smoothing function34 with a range of 10-12 Å is used for the Lennard-Jones interactions, with a long-range correction applied beyond the truncation distance.26 The SHAKE algorithm35 is used to constrain all hydrogen atom bonds to their equilibrium lengths and to maintain rigid water geometries. For the constant pressure – constant temperature (NPT) simulations the Nosé-Hoover thermostat36,37 is used to maintain the temperature and the Langevin piston barostat38 is used to maintain the pressure. A leapfrog integrator39 is used with a 1 fs timestep for all of the simulations.
Pure solvent simulations are performed with a periodic box of 125 solvent molecules. The box of solvent molecules is minimized and then equilibrated for 50 ps followed by five production runs performed for 1 ns. The heat of vaporization ΔHvap is calculated from the pure solvent simulation using the relation
| (3) |
Here, 〈U〉monomer is the average potential energy of the monomers calculated from five individual gas-phase simulations of all 125 molecules, with each simulation run for 500 ps. The 〈U〉box term is the average potential energy of the periodic box. N is the number of molecules in the box, R is the universal gas constant for an ideal gas and T is the temperature.
The free energy of aqueous solvation ΔGsol is calculated from the difference in free energy of a molecule in aqueous solution compared to that in the gas phase. ΔGsol is calculated from the sum of nonpolar ΔGnp and electrostatic ΔGelec free energies40:
| (4) |
ΔGnp is the sum of the repulsive and dispersive contribution, which are calculated using the Weeks, Chandler, Anderson decomposition of the LJ potential.41 The repulsion term in the LJ potential is treated using a soft-core potential.42 In the aqueous phase, free energy calculations are performed using 1 molecule centered in a water box of 250 TIP3P water molecules. The aqueous system at each window is equilibrated for 50 ps and then simulated for 200 ps in the NPT ensemble. In the gas phase, Langevin dynamics are used with an infinite non-bond cutoff.26,43 Since the gas phase energies converge much more quickly, the gas phase system is equilibrated for 10 ps and the production run is simulated for 100 ps. The simulations are performed at a temperature of 298K and a pressure of 1 atm, which is consistent with experiment. The free energy calculations are analyzed using thermodynamic integration44 and the weighted-histogram analysis method45 (WHAM). Additional details for calculating the free energy have been described previously.40,46 Unlike all other condensed phase simulations in the present work, due to software limitations, the long-range pressure correction is not part of the MD protocol for the free energy simulations. Thus, the long-range contribution (LRC) from the LJ potential to the free energy of solvation is calculated as the difference in LJ energy of the aqueous system with a nonbond cutoff of 12 Å and a nonbond cutoff of 30 Å. The LRC is calculated from a 5 ps NPT simulation trajectory of the molecules in solution using coordinates saved every 100 fs and averaged over all values.
From the pure solvent simulation trajectories, the self-diffusion coefficient Dsim incorporates a system-size dependent finite-size correction developed by Yeh and Hummer47:
| (5) |
In Equation 5, DPBC is the diffusion coefficient calculated from a simulation with periodic boundary conditions to which the correction term is added. ζ is a constant of 2.837297, kB is the Boltzmann constant, T is the temperature, η is the viscosity and L is the length of the cubic simulation box. DPBC is calculated from the slope of the mean square displacement of the C1 atom of all solute molecules in the simulation box versus time.26 For diffusion coefficients of polyols in aqueous solutions of TIP3P water, Equation 5 is further modified to take into account the low viscosity of TIP3P water relative to experiment:
| (6a) |
| (6b) |
Here, the scaling factor of 0.375 is applied to correct for the underestimation of the viscosity of water by the TIP3P model. The scaling factor is calculated from ηTIP3P /ηw where ηTIP3P = 0.35 cP and ηw = 0.93 cP, the experimental viscosity of water. Equation 6b is the viscosity of a solution with the presence of a solute estimated by the Einstein formula,48 where ηTIP3P is the viscosity of TIP3P (0.35 cP) and ϕ is the volume fraction of the solute. The method for calculating the simulation diffusion coefficient for a polyol-water mixture is similar to that previously used for a system of polyethylene oxide and polyethylene glycol.49
Complete crystal unit cells, obtained from the Cambridge Structural Database,23 are used as starting structures for crystal simulations, with periodic boundary conditions applied in accordance with the length and angle parameters of the respective crystals. Each crystal system is minimized initially to remove bad contacts and is then equilibrated for 100 ps. After equilibration, the simulation is run for 2 ns. For all of the polyol crystals, the reference temperature is set to room temperature 298K, the temperature at which the crystals were obtained, and constant pressure is maintained at 1 atm by allowing independent variation in the crystal cell length parameters.
For the aqueous phase MD simulations, a box containing 1100 waters and the number of solute molecules based on the experimental concentration is set up and then minimized using harmonic restraints with a force constant of 1*(particle mass)*kcal*mol−1*Å−2*amu−1 on only the solute molecules. The system is equilibrated for 500 ps and then the equilibrated conformation is used as the starting conformations for five different unrestrained 1ns runs, using different initial velocities for each of the runs to achieve improved statistics. The reference pressure of the glucitol and mannitol systems is 1 atm, and the reference pressure of the galacitol, xylitol, erythritol, ribitol, glycerol and myoinositol system is 3.5 atm, in accordance with the experimental conditions. The density of each system is calculated using the following equations:
| (7a) |
| (7b) |
where 〈V〉 is the average volume calculated from all five runs. Nwater , Nsolute and NAvogadro are the number of water molecules, the number of polyol solute molecules and Avogadro's number respectively. 〈MW〉 is the average molecular weight of the system. Equation 7a is also used to calculate the density for neat liquids; however, in this case N is simply the number of molecules in the periodic box.
The J coupling constants for glucitol and mannitol are also calculated from the aqueous simulations described above. However, the coupling constants for arabitol, ribitol and xylitol are calculated from aqueous phase simulations at 1 atm and a molality of 0.5 mol*kg−1 using the same protocol for the aqueous simulations. The dihedral value for the proton-proton coupling is calculated every 1 ps for each of the production runs. Moreover, the dihedral value is calculated for each of the solute molecules in the respective systems; therefore, depending on the concentration of the simulation the amount of torsional data differs. The J coupling is then calculated from the dihedral values for each snapshot using the generalized Karplus equation 50
| (8) |
where ϕ is the H-X-X-H dihedral angle. Manipulation of the Karplus equation given in Equation 8 allows the fraction of trans conformers to be calculated:50
| (9) |
In Equation 9, Jobs is the observed coupling constant.
RESULTS AND DISCUSSION
Parameter Optimization
All parameter optimization was done in a self-consistent manner so that when one parameter was changed in a molecule all other parameters were tested and reoptimized as necessary. The presented empirical force field data reflect the final set of self-consistently optimized nonbonded and bonded parameters.
Nonbonded parameters
Polyols
The non-bonded parameters for the aliphatic and hydroxyl moieties in the polyol compounds in Figure 1 were directly transferred from alkanes51 and hexopyranoses,22 and testing showed that no further optimization was required. This testing included the ability of the force field model to properly describe hydrogen bonding as compared to QM data, both in terms of hydrogen bonding strength and distance.
Taken as a group, hydrogen bonds in the water-solute pairs are well represented using the transferred nonbonded parameters. The average error over all 24 interaction pairs examined (Table 1) is −0.14 kcal/mol for the interaction energy and 0.03 Å for the interaction distance, which shows a slight systematic underestimation of the energy and overestimation of the distance. The mean absolute error for the interaction energy and distance is calculated to be 0.43 kcal/mol and 0.04 Å respectively. For allitol, both a terminal (O1) and non-terminal hydroxyl (O2) were investigated with good results for both types. Of particular note is that the interaction energies range from −2.81 kcal/mol all the way to −8.65 kcal/mol in the scaled QM representation, and the MM representation faithfully captures this diversity in hydrogen bond strength. MM water interactions for n=4 (threitol), n=5 (ribitol), n=6 (allitol and altritol) linear polyols as well as for inositol are all independently in good agreement with the QM results with the exception of a few weakly interacting pairs, i.e. pair interactions 1 and 2 for ribitol. However, in the cases where the weakly interacting pair has a large error, the strongly interacting pairs are in very good agreement. As the more favorable interactions dominate structural and dynamic properties, it is deemed more important to treat these interactions accurately. Of note is that all hydroxyls in all compounds have the same partial charges and LJ parameters assigned to them, which attests to their generality and transferability to similar compounds, and provides evidence that an accurate force field can be developed with a relatively parsimonious set of nonbonded parameters.
Table 1.
Comparison of optimized water interaction energies and O---H distance by HF/6-31G(d) QM calculations and CHARMM force field for the pair conformations shown in Figure 1.
|
EQM (kcal/mol) |
EMM (kcal/mol) |
EQM – EMM (kcal/mol) |
RQM (Å)a | RMM (Å) |
RQM – RMM (Å) |
||
|---|---|---|---|---|---|---|---|
| Allitol | O1, Pair1 | −5.84 | −5.87 | −0.03 | 1.84 | 1.87 | 0.03 |
| O1, Pair2 | −5.52 | −5.84 | −0.32 | 1.85 | 1.87 | 0.02 | |
| O1, Pair3 | −5.81 | −5.25 | 0.56 | 1.82 | 1.88 | 0.06 | |
| O1, Pair4 | −6.25 | −5.80 | 0.45 | 1.83 | 1.88 | 0.05 | |
| O2, Pair1 | −2.81 | −3.38 | −0.57 | 1.85 | 1.87 | 0.02 | |
| O2, Pair2 | −3.21 | −3.86 | −0.65 | 1.87 | 1.88 | 0.01 | |
| O2, Pair5 | −8.65 | −8.83 | −0.18 | 1.73 | 1.80 | 0.07 | |
| O2, Pair6 | −8.51 | −8.80 | −0.29 | 1.74 | 1.80 | 0.06 | |
| Altritol | Pair1 | −5.48 | −5.73 | −0.25 | 1.85 | 1.87 | 0.02 |
| Pair2 | −4.83 | −5.21 | −0.38 | 1.87 | 1.88 | 0.01 | |
| Pair3 | −5.43 | −5.12 | 0.31 | 1.85 | 1.89 | 0.04 | |
| Pair4 | −4.76 | −4.35 | 0.40 | 1.86 | 1.89 | 0.03 | |
| Ribitol | Pair1 | −2.34 | −3.83 | −1.49 | 1.93 | 1.90 | −0.03 |
| Pair2 | −2.09 | −2.88 | −0.79 | 2.13 | 1.98 | −0.15 | |
| Pair5 | −7.31 | −7.14 | 0.17 | 1.76 | 1.82 | 0.06 | |
| Pair6 | −7.04 | −7.00 | 0.04 | 1.78 | 1.82 | 0.04 | |
| Threitol | Pair1 | −5.46 | −5.63 | −0.17 | 1.85 | 1.88 | 0.03 |
| Pair2 | −5.04 | −5.54 | −0.50 | 1.87 | 1.87 | 0.00 | |
| Pair3 | −5.40 | −4.99 | 0.41 | 1.84 | 1.89 | 0.05 | |
| Pair4 | −4.83 | −4.65 | 0.18 | 1.87 | 1.90 | 0.03 | |
|
myo- Inositol |
Pair1 | −5.71 | −6.13 | −0.42 | 1.87 | 1.88 | 0.01 |
| Pair2 | −5.78 | −6.57 | −0.79 | 1.86 | 1.87 | 0.01 | |
| Pair3 | −4.47 | −3.48 | 0.99 | 2.71 | 2.91 | 0.20 | |
| Pair4 | −6.20 | −6.19 | 0.01 | 2.51 | 2.48 | −0.03 | |
| Average | −0.14 | 0.03 | |||||
0.2 has been subtracted from the RQM values
As an additional test of the hydroxyl nonbonded parameters, the pure solvent properties of glycerol were determined. The results show an error of 4.8% in the molecular volume and −14.2% in the heat of vaporization (Table 2). These errors are significantly beyond the typical error for CHARMM pure solvents, especially considering that the pure solvent properties of ethylene glycol are in good agreement with experiment.22 One possibility for the large error in the heat of vaporization is overly favorable intramolecular hydrogen bonding occurring between hydroxyl groups in the gas phase simulations resulting from the need to over polarize hydroxyls to obtain correct scaled water-solute interaction energies appropriate for a condensed phase additive force field. The resultant lowering of the value of 〈U〉monomer in Equation 3 would lead to a larger heat of vaporization. To test this possibility, the gas phase energy 〈U〉monomer was calculated using monomer trajectories extracted from the pure solvent simulation. In these trajectories it is expected that intramolecular hydrogen bonding would be diminished due to intermolecular hydrogen bonding of the monomers with surrounding molecules in the solvent environment. Using this approach to calculate 〈U〉monomer the resulting heat of vaporization is 22.34 kcal/mol, which is only a 2.0 % error with respect to experiment. Thus, it appears that the significant deviation in the heat of vaporization of neat glycerol is due to overestimation of intramolecular hydrogen bonding in the gas phase, although it should be noted that this effect will not influence the calculated molecular volume. Intramolecular hydrogen bonding in these compounds also complicates the parametrization of their conformational energetics, as discussed below.
Table 2.
Water interactions for acetone and acetaldehyde and condensed phase properties including heats of vaporization, molecular volumes, free energies of aqueous solvation and self-diffusion coefficients for neat acetaldehyde, acetone, and glycerol. Acetone/acetaldehyde-water interaction pairs are shown in Figure S1 of the supplemental materials.
| Water Interactions | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Dipole Moments | Interactions | ||||||||||
| X | Y | Z | Total | EQM | EMM |
EQM- EMM |
RQMa | RMM |
RQM- RMM |
||
| Acetone | |||||||||||
| QM | 0.00 | 2.77 | 0.00 | 2.77 | Pair1 | −5.26 | −6.49 | −1.23 | 1.89 | 1.69 | −0.2 |
| MM | 0.00 | 3.57 | 0.00 | 3.57 | Pair2 | −7.09 | −7.19 | −0.10 | 1.82 | 1.71 | −0.11 |
| Pair3 | −5.44 | −7.02 | −1.58 | 1.90 | 1.69 | −0.21 | |||||
| Acetaldehyde | |||||||||||
| QM | 2.38 | 1.10 | 0.00 | 2.62 | Pair1 | −4.76 | −4.96 | −0.20 | 1.92 | 1.83 | −0.09 |
| MM | 3.28 | 0.64 | 0.00 | 3.35 | Pair2 | −6.62 | −6.07 | 0.55 | 1.84 | 1.81 | −0.03 |
| Pair3 | −4.86 | −5.50 | −0.64 | 1.94 | 1.81 | −0.13 | |||||
| Pair4 | −2.20 | −2.50 | −0.30 | 2.36 | 2.18 | −0.18 | |||||
| Pair5 | −2.12 | −2.48 | −0.36 | 2.37 | 2.18 | −0.19 | |||||
| Condensed Phase Properties | ||||||||
| ΔHvap | % error | Vm | %error | ΔGsol (LRC) | Absolute error |
Dsim (cm2/s) |
% error | |
| Acetone | ||||||||
| Expt | 7.41 | 123.00 | −3.85 | 4.77E-5 | ||||
| Calc | 7.37 | −0.54 | 124.47 | 1.20 | −5.02 (−0.31) | −1.17 | 4.84E-5 | 1.5 |
| Acetaldehyde | ||||||||
| Expt | 6.08 | 92.85 | −3.50 | |||||
| Calc | 6.21 | 2.14 | 94.84 | 2.14 | −3.23 (−0.20) | 0.27 | ||
| Glycerol | ||||||||
| Exp | 21.90 | 121.40 | ||||||
| Calc | 18.80 (22.34)b |
−14.16 2.01 |
127.17 | 4.75 | ||||
0.2 has been subtracted from the RQM values.
Value in parantheses calculated using the monomer energy based on glycerol conformations obtained from the pure solvent simulation; see text for details. Energies in kcal/mol and molecular volumes in Å3.
Carbonyls
The nonbonded parameters for aldehyde and ketone carbonyls were explicitly optimized for application to the linear aldose and ketose forms of monosaccharides. Acetaldehyde and acetone were selected as model compounds for the parameter development process. Methyl parameters were as previously published51 and only the carbonyl C and O atoms and, with acetaldehyde, the aldehydic H LJ parameters and charges were optimized, with the charges of the adjacent bonded methyl carbons adjusted to yield a total molecular charge of zero. Target data for the parameter optimization were water-solute pair interaction energies and distances, dipole moments, heats of vaporization, and molecular volumes, while the free energies of aqueous solvation and the diffusion constant were calculated using the final optimized parameters.
Optimization of the carbonyl LJ parameters and partial charges yielded a set of parameters capable of reproducing all the target data. Overall, interaction energies with water are in satisfactory agreement with the QM target data, though in some cases the balance between the in-plane and out-of-plane orientations is not ideal (Table 2 and Figure S1 of the supplemental materials). This problem is due to limitations in the form of the energy function and can, to a large extent, be corrected using an explicit representation of lone pairs.52 However, to maintain consistency with the remainder of the CHARMM additive force fields, such an addition was not made. The final charges overestimate the QM MP2/6-31G(d) dipole moments by 28% and 29% for acetaldehyde and acetone, respectively. Such overestimation is required in the additive model to account for the implicit over-polarization required to accurately predict the condensed phase properties.21 The corresponding partial atomic charges along with the appropriate LJ parameters lead to excellent agreement for the pure solvent properties for both molecules, with the agreement within 2.2% of experiment53-59 in all cases (Table 2).
Bonded parameters
Bonds and angles
All bond and angle parameters were initially transferred from similar existing CHARMM additive force field parameter values. Optimized geometries were compared to target data from a crystal database survey of the CSD. This analysis revealed systematic errors in geometries, in particular the bond lengths associated with C-C bonds, in which both carbons are hydroxylated. Accordingly, the respective geometric force field parameters were suitably optimized. A comparison of the optimized bond lengths and angles to the crystal survey data and QM data at the MP2/6-31G(d) level for compounds acetone and acetaldehyde is given in Table S2 in the supplemental materials. Vibrational analyses for acetone, acetaldehyde and glycerol (Table S3 in supplemental materials) were done both at the QM MP2/6-31G(d) level and in the MM representation, and the MOLVIB utility in CHARMM was used to assign the bond and angle contributions to internal normal modes as described by Pulay.60 MM force constants were optimized as required to reproduce the scaled QM frequencies61 to complete the bond and angle parameter optimization.
Dihedrals
Parameters associated with methyl rotations in acetone and acetaldehyde were readily optimized to yield excellent agreement between QM and MM methyl dihedral scans (O=C-C-H) (supplemental materials, Figure S4). In the case of the C-C-C-C, C-C-C-O, O-C-C-O, and C-C-O-H dihedrals, the torsional parameters were fit to QM RIMP2/cc-pVTZ//MP2/6-31G(d) potential energy scans performed on linear polyols. These parameters were simultaneously fit to conformations with relative energies below a cutoff value of 15 kcal/mol for scan points on all of the n=6 linear polyols, yielding approximately 1500 different conformational energies. Prior to fitting, with the parameters for the targeted dihedrals set to zero, the total RMSE (Eq. 2) for all of the conformations, including those above the energy cutoff of 15 kcal/mol (∼1730 different conformations) was 4.0 kcal/mol. Following fitting, which included the 1, 2 and 3 fold terms for each dihedral, sampling the force constant from 0 to 3 kcal/mol and sampling phase angles of either 0 or 180, the RMSE was 2.5 kcal/mol. Using the dihedral parameters fit to the n=6 sugar alcohols, the RMSE of the C-C-C-C dihedral for the n=5 linear polyols is calculated to be 1.60 kcal/mol and for the n=4 polyols to be 1.87 kcal/mol, as compared to values of 2.20 kcal/mol and 3.85 kcal/mol, respectively, with the targeted dihedral parameters set to zero. These data demonstrate both that the fitting procedure leads to significant improvements in the conformational energetics of the targeted n=6 compounds and also the transferability of the dihedral parameters to the shorter chain linear polyols.
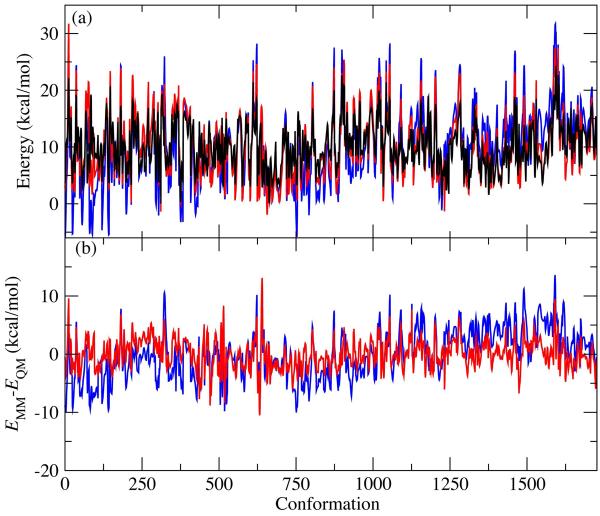
Figure 3 shows the improvement in the MM conformational energies for the n=6 target compounds. The change is visually apparent in the relative energies (Figure 3a), and especially so in the difference in QM and MM energies for both the parametrized and unparametrized energies (Figure 3b). It should be noted that all the relative energies for all the compounds are offset to a constant c, from Eq 2, minimizing the total RMSE value. Figure 4 shows the QM and MM dihedral scans, using the optimized torsional parameters for a few representative compounds, and highlights the quality of the conformational energy fitting results. In all cases, the location of the minimum using the optimized dihedral parameters reproduces the QM results to within 15 degrees. There is, however, a general trend toward overestimation of the energy barriers by the MM model. Two factors contribute to this. One is the fact that the targeted dihedral parameters are fit simultaneously to all of the n=6 compounds, as required to maximize their generality and transferability (as evidenced by their applicability to the shorter polyols). The second is due to overestimation of intramolecular hydrogen bonding, again arising from the over-polarized hydroxyl groups that are required for proper condensed phase behavior in an effective pairwise additive force field; these intramolecular hydrogen bonding interactions are broken in the region of the energy barriers, exaggerating their energy differences relative to the minima.
Figure 3.
(a) QM and MM potential energy scans for C-C-C-C, C-C-C-O, C-C-O-H, OC-C-O dihedrals for all n=6 polyols (∼1730 conformations). QM scan is black; MM scan using optimized parameters is red; MM scan using parameters set to zero is blue. QM data have been offset using the global minimum as E=0. MM scans have been root-mean square aligned with the QM scan (i.e. offset by the constant c given in Equation 2). (b) EMM-EQM using the optimized parameters (red) and parameters set to zero (blue).
Figure 4.
QM and MM potential energy scans for dihedrals C1-C2-C3-C4, C4-C5-C6-O6, O1-C1-C2-O2 in glucitol, C1-C2-C3-C4 dihedral in erythritol, C1-C2-C3-C4 dihedral in inositol, and O1-C1-C2-C3 dihedral in allose. QM results are black. The MM results, calculated using the optimized parameters, are red.
Less extensive dihedral parameter optimization was required for the remaining compounds in Figure 1. The cyclic inositol was treated separately from the linear polyols, with parametrization of only the C-C-C-C dihedral, with the other dihedral parameters transferred from the hexopyranose force field. Using the MCSA method to fit the ring dihedral in inositol, an RMSE value of 3.27 kcal/mol was obtained for an energy scan in which the ring was deformed from the favored chair conformation, an improvement on the RMSE of 4.23 kcal/mol before optimization. Additionally, for allose and psicose, only the dihedrals involving the carbonyl atoms and heavy atoms (C-C-C=O, O-C-C=O) required parametrization, since all others could be transferred from those for the linear polyols and acetone or acetaldehyde. The RMSE for the optimized allose and psicose dihedral parameters are calculated to be 0.75 kcal/mol and 0.69 kcal/mol as compared to 1.47 kcal/mol and 2.95 kcal/mol, respectively, for the unparametrized torsions. Though only allose and psicose were considered, the current set of parameters is applicable to all related ketoses and aldoses, owing to the demonstrated generality and transferability of the polyol parameters.
Parameter validation
To validate the optimized parameters, pure solvent, crystal, and aqueous phase MD simulations were performed to calculate both condensed phase properties and conformational distributions and to compare them with experimental results. As well as testing the accuracy of the force field parameters, these calculations also gave insight into complications that arise during the parametrization process.
Acetone and acetaldehyde
Validation of the aldehyde and ketone parameters was based on the calculation of the aqueous solvation free energy ΔGsol and pure solvent diffusion coefficients for acetaldehyde and acetone. The calculated value for ΔGsol, including the long-range LJ correction, is in excellent agreement with the experimental value for acetaldehyde, with a difference of 0.3 kcal/mol (Table 2). The agreement for acetone is poorer, with the force field yielding a value that is 1.2 kcal/mol more favorable than the experimental value. While the level of agreement for acetone is not ideal, it is similar to that observed for a number of model compounds representative of amino acid sidechains.62,63 Furthermore, the diffusion coefficient for pure acetone is in excellent agreement and, combined with the water-solute interaction energy data (Table 2), indicates the model to have satisfactory energetic and dynamic properties.
Polyol Crystal Simulations
Crystal simulations were performed using crystals obtained from the CSD23 to validate nonbonded parameters and conformational properties. The compounds included a number of tetritols (n=4), pentitols (n=5) and hexitols (n=6) as well as glycerol and inositol. The selected crystals provide a thorough investigation of the crystal lattice parameters and are chosen based on diversity, purity and resolution. To be consistent with experimental conditions, all of the crystals were simulated at room temperature (298K).
The average sizes of the crystal unit cells from the simulations are systematically too large relative to the experimental values (Table 3). The average percent error of the simulated unit cell lengths A, B and C for all of the selected polyols is 2.8%, 1.5% and 4.3%, respectively, and the average unit cell volume is calculated to have an error of 7.3%. This over prediction in the average volume is consistent to what has been seen in previous studies for the hexopyranoses22 and suggests limitations in the ability of a pairwise additive condensed-phase force field designed for liquid simulations to reproduce the crystal phase. The environments surrounding a molecule in the liquid versus the crystal state are considerably different, and the inability of the force field to quantitatively model the latter environment when parametrized to the former is not entirely surprising.
Table 3.
Crystal lattice parameter and volumes calculated from crystal simulations
| Compound | CSD ID | R factor |
A | % error |
B | % error |
C | % error |
Volume | % error |
|---|---|---|---|---|---|---|---|---|---|---|
| n=6 | ||||||||||
| Allitol | ALITOL01 | 0.04 | 4.71 | 13.41 | 6.62 | 411.16 | ||||
| 4.96 | 5.35 | 13.81 | 3.03 | 6.82 | 3.14 | 445.30 | 8.30 | |||
| Altritol | JOJZOX | 0.03 | 4.90 | 5.18 | 16.26 | 409.11 | ||||
| 5.61 | 14.51 | 5.16 | −0.39 | 16.11 | −0.89 | 446.39 | 9.11 | |||
| Galacticol | GALACT | 0.05 | 8.45 | 11.50 | 9.04 | 808.64 | ||||
| 8.66 | 2.53 | 11.79 | 2.45 | 9.74 | 7.69 | 857.58 | 6.05 | |||
| Glucitol | GLUCIT01 | 0.07 | 8.68 | 9.31 | 9.73 | 785.86 | ||||
| 8.71 | 0.44 | 9.48 | 1.84 | 10.11 | 3.89 | 834.77 | 6.22 | |||
| Mannitol | DMANTL07 | 0.03 | 8.69 | 16.90 | 5.55 | 815.40 | ||||
| 8.96 | 3.07 | 17.22 | 1.91 | 5.61 | 1.03 | 865.23 | 6.11 | |||
| n=5 | ||||||||||
| Arabitol | ARABOL | 0.04 | 9.21 | 4.86 | 15.49 | 692.85 | ||||
| 9.36 | 1.59 | 4.95 | 2.00 | 15.39 | −0.64 | 713.10 | 2.92 | |||
| Ribitol | RIBTOL | 0.06 | 8.99 | 4.95 | 15.73 | 694.11 | ||||
| 9.31 | 3.60 | 5.03 | 1.64 | 15.57 | −1.04 | 720.91 | 3.86 | |||
| Xylitol | XYLTOL01 | 0.05 | 8.27 | 8.90 | 8.91 | 655.43 | ||||
| 8.37 | 1.27 | 9.39 | 5.47 | 9.18 | 3.00 | 720.74 | 9.96 | |||
| n=4 | ||||||||||
| Threitol | PAGDEG | 0.05 | 10.10 | 10.10 | 4.84 | 427.60 | ||||
| 9.72 | −3.66 | 9.72 | −3.66 | 5.85 | 20.87 | 478.68 | 11.95 | |||
| n=3 | ||||||||||
| Glycerol | GLCROL | 0.12 | 7.00 | 9.96 | 6.29 | 438.54 | ||||
| 7.06 | 0.79 | 9.88 | −0.78 | 6.82 | 8.43 | 474.91 | 8.30 | |||
| Cyclic n=6 | ||||||||||
| L-chiro- | FOPKOK | 0.03 | 6.87 | 9.13 | 6.22 | 373.68 | ||||
| Inositol | ||||||||||
| 6.93 | 0.93 | 9.44 | 3.41 | 6.33 | 1.85 | 402.09 | 7.60 | |||
| Average | 2.77 | 1.54 | 4.30 | 7.31 | ||||||
The heavy atom bonds, angles and dihedrals are monitored during the simulations and the averages reproduce the values in the crystal structure. This is particularly true for the bond lengths and angles because their equilibrium values were adjusted based on a crystal database survey. The differences between a selected set of experimental and simulated bond lengths, angles and dihedrals are given in Table S5 of the supplemental materials. However, some of the dihedrals involving hydroxyl oxygens contrast to the values found in the crystal structure. In some cases, these differences cause errors in the calculated crystal lattice parameters, as hydrogen bonding that may occur in the crystal is not maintained during the simulation. For instance, in the altritol unit cell, a terminal hydroxyl hydrogen in one of the altritol monomers is hydrogen bonded to a hydroxyl oxygen in another altritol in an adjacent unit cell; this leads to the relatively large differences in the O1-C1-C2-C3 and O1-C1-C2-O2 dihedrals. During the crystal simulation, rotation about the torsions involving the hydrogens on these hydroxyls causes a loss in hydrogen bonding capabilities. Subsequently, this causes an increase in unit cell length A leading to larger errors in the unit cell A and volume of the unit cell.
Aqueous Phase Simulations
To test the behavior of the polyols in aqueous solution, the environment in which it is anticipated the force field will primarily be applied, densities were calculated for molal solutions of glucitol, mannitol, ribitol, xylitol, galacticol, erythritol, glycerol and myo-inositol varying in concetration from dilute (0.1 mol/kg) to highly concentrated (5 mol/kg) and compared to experimental values.64,65 For consistency with experimental conditions, glucitol and mannitol were simulated at a temperature and pressure of 298K and 1 atm, while all other compounds were simulated at 298K and 3.5 atm. All of the calculated molecular densities reproduce experimental values within 3% error across the entire 50-fold difference in concentration and at ambient and elevated pressures (Table 4). Moreover, as the concentration increases there is a trend of decreasing error. The overall average error in volume, 0.64%, is much better than the overall crystal volume, emphasizing the applicability of the parameter set to heterogeneous liquid systems.
Table 4.
Calculated and experimental64,65 densities at different concentrations of polyols in a box of water with 1100 water molecules at T = 298.15K and P = 1 atma or 3.5 atmb
| Compound | Molality (mol/kg) | Nsolute | Expt (g/cc) | Calc (g/cc) | % error |
|---|---|---|---|---|---|
| Mannitola | 0.1999 | 4 | 1.0092 | 1.0228 | 1.35 |
| 0.5998 | 12 | 1.0318 | 1.0414 | 0.93 | |
| 0.8006 | 16 | 1.0426 | 1.0503 | 0.74 | |
| 0.9995 | 20 | 1.0517 | 1.0584 | 0.64 | |
| Glucitola | 0.5085 | 10 | 1.0117 | 1.0368 | 2.48 |
| 1.9508 | 39 | 1.0987 | 1.0921 | −0.60 | |
| 4.0003 | 79 | 1.1606 | 1.1445 | −1.39 | |
| 5.9945 | 119 | 1.2052 | 1.1793 | −2.15 | |
| Galacticolb | 0.0698 | 2 | 1.0015 | 1.0179 | 1.64 |
| 0.1492 | 3 | 1.0065 | 1.0202 | 1.36 | |
| Xylitolb | 0.1000 | 2 | 1.0022 | 1.0165 | 1.43 |
| 1.0000 | 20 | 1.0427 | 1.0472 | 0.43 | |
| 2.7000 | 53 | 1.1019 | 1.0925 | −0.85 | |
| Erythritolb | 0.4998 | 10 | 1.0142 | 1.0262 | 1.18 |
| 1.0000 | 20 | 1.0298 | 1.0382 | 0.82 | |
| 3.0000 | 60 | 1.0806 | 1.0804 | −0.02 | |
| Ribitolb | 0.1042 | 2 | 1.0022 | 1.0164 | 1.42 |
| 0.5092 | 10 | 1.0208 | 1.0315 | 1.05 | |
| 3.1769 | 63 | 1.1121 | 1.1047 | −0.67 | |
| Glycerolb | 0.5002 | 10 | 1.0075 | 1.0200 | 1.24 |
| 1.0008 | 20 | 1.0171 | 1.0280 | 1.07 | |
| 3.0035 | 60 | 1.0497 | 1.0563 | 0.63 | |
| 4.9993 | 100 | 1.0750 | 1.0781 | 0.29 | |
| myo-Inositolb | 0.1000 | 2 | 1.0046 | 1.0191 | 1.44 |
| 0.2480 | 5 | 1.0163 | 1.0288 | 1.23 | |
| 0.4994 | 10 | 1.0351 | 1.0443 | 0.89 | |
| Average | 0.64 | ||||
Polyol diffusion coefficients in aqueous solution
Diffusion coefficients for polyol-water solutions show consistent correlation with experimental values (Table 5). Complicating the analysis is the fact that TIP3P water has a self-diffusion coefficient larger than the experimental value, which motivated the addition of correction terms and scaling factors to the diffusion equation. The diffusion coefficients are computed using Equation 6a, where the size-dependent correction term is calculated using the shear viscosity of TIP3P water model, which is 0.35cP66 (3.5E-4 kg/m/s), in conjunction with Equation 6b for all polyol-water mixtures. The volume fraction (〈V〉mixture −Vwater box)/Vwater box is calculated from the average volume of the box for all five runs for the polyol aqueous phase simulations. DPBC is calculated at the given concentration for each of the five production runs and is averaged. The computed results are compared to experimental results67 and the percent error is given in Table 5. The diffusion coefficients are systematically too low with a total average error of−48%. This error may be due to the fact that the DPBC varies greatly between the five different production runs with a standard deviation of the average DPBC for each polyol calculated to be ∼30%. The standard deviations are given in parentheses in Table 5. In addition, assumptions in the correction factors in Equation 6 may be limiting.
Table 5.
Diffusion coefficients for binary water-alditols solutions.
| Molality (mol/kg) |
Volume fraction |
Size correction (cm2/s) |
Expt. Dsim (cm2/s)67,69 |
Calc. Dsim (cm2/s) |
% Diff | |
|---|---|---|---|---|---|---|
| Glycerol | 0.50 | 0.02 | 5.24E-6 | 1.10E-5 | 4.51E-6 (3.15E-6) |
−59.04 |
| Galacticol | 0.15 | 0.00 | 5.48E-6 | 6.36E-6 | 3.68E-6 (1.85E-6) |
−42.08 |
| Glucitol | 0.50 | 0.05 | 4.94E-6 | 6.58E-6 | 3.12E-6 (9.12E-7) |
−52.62 |
| Mannitol | 0.60 | 0.06 | 4.82E-6 | 6.67E-6 | 3.47E-6 (1.40E-6) |
−47.90 |
|
myo- Inositol |
0.50 | 0.04 | 5.03E-6 | 6.08E-6 | 3.61E-6 (1.41E-6) |
−40.58 |
| Average | −48.45 | |||||
The standard deviation of the calculated Dsim for the five production runs is given in parentheses.
Alditol conformational sampling in aqueous solution
To investigate the ability of the model in treating the conformational properties of the alditols in solution, J coupling constants and conformer populations for all of the Hx – Hx−1 hydrogens were computed from the aqueous phase simulations of glucitol, mannitol, arabitol, ribitol, and xylitol. The dihedrals were computed for every solute molecule from snapshots taken every 1ps from the MD simulations. Since the simulations of glucitol and mannitol are run with varying concentrations, the sampling of the dihedrals is much larger. For glucitol, the number of points sampled is ∼1.2 million and for mannitol it is 260,000. The number of torsions calculated for arabitol, ribitol and xylitol is 50,000. In the case of arabitol, ribitol and xylitol, performing 5 more runs with different starting velocities and obtaining similar dihedral probability distributions tested the convergence of sampling. J coupling values are calculated for each dihedral in each sample using the Karplus equation given in Equation 8. Subsequently, the coupling constant is calculated as the average over all the J coupling values for each dihedral.
The H-H trans conformer probability was determined from the dihedral probability distributions, which were determined with a bin width of 3.6 degrees between −180 and 180 degrees. A trans conformer was considered to have any torsional value less than −135 and greater than 135 degrees. The J coupling constants and the probability of H-H trans conformers Ftrans are given in Table 6. The H-H trans conformer distributions for glucitol are shown in Figure S6 of the supporting material. As shown in Table 6, for all of the molecules studied, some of the J coupling values and Ftrans values reproduce the experimental NMR data, although the differences are larger in some cases. For example, the average difference in Ftrans involving one terminal hydrogen and one interior hydrogen (i.e. 1,2; 1',2, 5,6 and 5',6) is calculated to be 7 and the average difference involving two interior hydrogen atoms is −29. Therefore, the Ftrans are better described for those dihedrals involving the terminal hydrogen. However, it is important to note that the parameters for the associated dihedrals (terminal/non-terminal) have the same values in order to retain the transferability and simplicity of the parameters for these compounds. Therefore, the parameters overestimate the trans conformer population involving the terminal hydrogens and underestimate the trans conformer population involving only internal hydrogens. While such a compromise may represent an inherent limitation in the force field, limitations in the conversion from dihedrals to J coupling values may also contribute. Such limitations may be due to the use of a simplified generalized Karplus equation in this study (Eq. 7). Given the different environments of the terminal and nonterminal hydrogens, it may be more appropriate to use equations that are more detailed (e.g. that account for electronegativity of substituents) or are optimized for the individual types of dihedrals. However, given the large number of compounds and classes of dihedrals, such a task is significant and beyond the scope of the present study.
Table 6.
Calculated J coupling constants for alditols using the Karplus and the fraction of trans conformers Ftrans.
| Coupling | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| J(1,2) | J(1′,2) | J(2,3) | J(3,4) | J(4,5) | J(4′,5) | J(5,6) | J(5′,6) | Avg Diff |
||
| Arabitol | Expt | 5.05 | 7.57 | 2.07 | 8.45 | 3.03 | 6.44 | |||
| Calc | 1.56 | 7.44 | 2.50 | 4.30 | 2.61 | 4.24 | ||||
| Diff | −3.51 | −0.13 | 0.43 | −4.15 | −0.42 | −2.20 | −1.66 | |||
| Ribitol | Expt | 3.07 | 7.16 | 6.27 | ||||||
| Calc | 1.09 | 8.05 | 3.54 | |||||||
| Diff | −1.98 | 0.89 | −2.73 | −1.27 | ||||||
| Xylitol | Expt | 4.34 | 6.90 | 4.43 | ||||||
| Calc | 1.46 | 7.53 | 2.48 | |||||||
| Diff | −2.88 | 0.63 | −1.95 | −1.40 | ||||||
| Glucitol | Expt | 3.55 | 6.55 | 6.00 | 1.70 | 8.25 | 2.95 | 6.30 | ||
| Calc | 2.84 | 5.90 | 2.75 | 2.79 | 2.73 | 3.08 | 5.49 | |||
| Diff | −0.71 | −0.65 | 3.25 | 1.09 | −5.52 | 0.13 | −0.81 | −1.39 | ||
| Mannitol | Expt | 2.94 | 6.43 | 8.99 | 1.02 | |||||
| Calc | 1.31 | 7.68 | 2.74 | 1.74 | ||||||
| Diff | −1.63 | 1.25 | −6.25 | 0.72 | −1.48 | |||||
| Avg Diff | −2.14 | 0.40 | −2.75 | −0.78 | −3.36 | −2.20 | 0.13 | −0.81 | ||
| Fraction of trans conformers | ||||||||||
| FH1H2 | FH1′H2 | FH2H3 | FH3H4 | FH4H5 | FH4H5′ | FH5H6 | FH5H6′ | Avg | ||
| Arabitol | Expt | 32 | 71 | 0 | 85 | 0 | 54 | |||
| Calc | 4 | 86 | 1 | 39 | 0 | 3 | ||||
| Diff | −28 | 15 | 1 | −46 | 0 | −51 | −18 | |||
| Ribitol | Expt | 0 | 65 | 51 | 51 | 0 | 65 | |||
| Calc | 2 | 97 | 28 | 14 | 1 | 96 | ||||
| Diff | 2 | 32 | −23 | −37 | 1 | 31 | 1 | |||
| Xylitol | Expt | 20 | 60 | 23 | 23 | 20 | 60 | |||
| Calc | 4 | 87 | 5 | 10 | 5 | 93 | ||||
| Diff | −16 | 27 | −18 | −13 | −15 | 33 | 0 | |||
| Glucitol | Expt | 20 | 56 | 37 | 0 | 73 | 5 | 51 | ||
| Calc | 20 | 62 | 8 | 17 | 16 | 21 | 55 | |||
| Diff | 0 | 5 | −29 | 17 | −57 | 16 | 4 | −6 | ||
| Mannitol | Expt | 0 | 54 | 94 | 0 | 94 | 0 | 53 | ||
| Calc | 2 | 90 | 18 | 2 | 19 | 4 | 91 | |||
| Diff | 2 | 36 | −75 | 2 | −75 | 4 | 38 | −10 | ||
| Avg Diff | −8 | 23 | −29 | −15 | −29 | 4 | 10 | 21 | ||
Ftrans values are given in percentages.
CONCLUSIONS
The all-atom additive CHARMM force field for acyclic carbohydrates and polyalcohols and inositol is presented. A full list of the topology and parameter information is included in Table S7 of the supplemental material. The nonbonded parameters for the polyols are transferred from the hexopyranoses and are tested using QM water-polyol interactions. The parameters for the carbonyl groups in linear aldoses and ketoses are optimized using the model compounds acetaldehyde and acetone and target QM data, model-compound-water interactions and pure solvent properties. For all of the compounds in this study, the MM water interaction distances and energies are in good agreement with the scaled QM values. For the bond and angle parameters, initial optimization was performed using acetone, acetaldehyde, and glycerol, with the optimized parameters transferred to the larger acyclic carbohydrates. Concurrently, selected equilibrium bond distances were adjusted to fit CSD survey data. The dihedral parameters are optimized to fit an extensive set of QM dihedral energy scans on n=6 linear polyols. The resulting parameters were then shown to be directly transferable to the n=4 and n=5 polyols. Validation of the optimized parameters was performed using condensed phase simulations, including crystal simulations and aqueous phase simulations. In the crystal simulations, all heavy atom bond, angle and dihedral values reproduced the experimental crystal values. Crystal volumes calculated from the simulations are systematically too large by approximately 7%, consistent with a similar trend observed in crystals of hexopyanoses22. Although the crystal volumes are too large, molecular densities calculated from aqueous phase simulations concentrations ranging all the way from 0.1 mol/kg to 5 mol/kg reproduce experimental data very well to within approximately 1%. As the parameters are anticipated to be used primarily for investigations of these compounds in aqueous solution, reproduction of the solution phase results is deemed more important. Concerning the conformational properties in solution, trans conformer populations calculated using a generalized Karplus equation are overestimated for terminal dihedrals while being systematically underestimated for nonterminal dihedrals. Limitations in the applied Karplus equation may limit this analysis.
All of the compounds considered in the present study (Figure 1) are flexible molecules with multiple hydroxyls or carbonyl moieties that allow for extensive intramolecular hydrogen bonding. Such hydrogen bonding leads to difficulties in parametrizing a force field for these compounds. This is due to the need to overestimate the partial atomic charges in additive force field so as to account for the polarization of molecules that occurs in the condensed phase. Such increased charges, or over-polarization, is particulary problematic with these compounds, as it leads to the extensive gas-phase intramolecular hydrogen bonding in the molecules being systematically overestimated; in the condensed phase it is assumed that competition between inter- and intramolecular hydrogen bonding yields a proper representation of interactions with the environment. Overestimation of intramolecular hydrogen bonding leads to several problems presented above. With glycerol, calculation of the heat of vaporization is confounded as the gas phase intramolecular hydrogen bonding will tend to make the gas phase energy too favorable, leading to the heat of vaporization being systematically too unfavorable. Calculation of the heat of vaporization using the gas phase energy determined using the monomer conformations from the condensed phase simulation (which is equivalent to determination of ΔHvap based on only the intermolecular interactions in the condensed phase) yields much better agreement. This is consistent with previous results for ethylene glycol, where both the heat of vaporization and molecular volume were in good agreement with experiment.22 However, it does not explain the molecular volume of glycerol being too large by 4.8%, which may be due to limitations in the nonbond parameters or the subtle balance in the competition of the inter- and intramolecular hydrogen bonding not being ideal. Another limitation due to overestimation of intramolecular hydrogen bonding is its impact on the dihedral energy scans. This leads to the low energy conformations, which maximize intramolecular hydrogen bonds, being artificially too favorable, which manifests itself in the MM energy surfaces overestimating the barrier height in the plots (Figure 4). This limitation leads to the RMSE for the energy surfaces to be higher than anticipated and may impact the resulting conformational properties of these molecules in solution, leading to poorer agreement with the NMR data. As this problem is due to inherent assumptions in the additive force field used in this study, it cannot be solved, though we have attempted to alleviate this problem via targeting and validating the force field against a large body of target data. It is anticipated that polarizable force fields, in which the need to over polarize the gas phase charge distribution is eliminated,68 may overcome these problems.
Supplementary Material
Acknowledgements
Financial support from the NIH (R01GM070855 (ADM) and F32CA1197712 (OG)) and computational support from the Department of Defense High Performance Computing and the National Cancer Institute Advanced Biomedical Computing Center are acknowledged.
Footnotes
Supporting Information Available.
Acetone and acetaldehyde water interaction orientations; acetone and acetaldehyde geometric data; vibrational analysis for acetone, acetaldehyde and glycerol; acetone and acetaldehyde dihedral scans; comparison of crystal and calculated bond lengths, angle and dihedrals; conformer distributions of glucitol; topology and parameter files. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Costantino L, Rastelli G, Vescovini K, Cignarella G, Vianello P, DelCorso A, Cappiello M, Mura U, Barlocco D. J. Med. Chem. 1996;39:4396. doi: 10.1021/jm960124f. [DOI] [PubMed] [Google Scholar]
- 2.Singh SB, Malamas MS, Hohman TC, Nilakantan R, Carper DA, Kitchen D. J. Med. Chem. 2000;43:1062. doi: 10.1021/jm990168z. [DOI] [PubMed] [Google Scholar]
- 3.de la Fuente JA, Manzanaro S, Martin MJ, de Quesada TG, Reymundo I, Luengo SM, Gago F. J. Med. Chem. 2003;46:5208. doi: 10.1021/jm030957n. [DOI] [PubMed] [Google Scholar]
- 4.Berridge MJ, Irvine RF. Nature. 1984;312:315. doi: 10.1038/312315a0. [DOI] [PubMed] [Google Scholar]
- 5.Volpe P, Salviati G, Di Virgilio F, Pozzan T. Nature. 1985;316:347. doi: 10.1038/316347a0. [DOI] [PubMed] [Google Scholar]
- 6.Momany FA, Willett JL. Carbohydr. Res. 2000;326:194. doi: 10.1016/s0008-6215(00)00042-2. [DOI] [PubMed] [Google Scholar]
- 7.Kirschner KN, Woods RJ. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10541. doi: 10.1073/pnas.191362798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuttel M, Brady JW, Naidoo KJ. J. Comput. Chem. 2002;23:1236. doi: 10.1002/jcc.10119. [DOI] [PubMed] [Google Scholar]
- 9.Lii JH, Chen KH, Allinger NL. J. Comput. Chem. 2003;24:1504. doi: 10.1002/jcc.10271. [DOI] [PubMed] [Google Scholar]
- 10.Kony D, Damm W, Stoll S, van Gunsteren WF. J. Comput. Chem. 2002;23:1416. doi: 10.1002/jcc.10139. [DOI] [PubMed] [Google Scholar]
- 11.Lins RD, Hünenberger PH. J. Comput. Chem. 2005;26:1400. doi: 10.1002/jcc.20275. [DOI] [PubMed] [Google Scholar]
- 12.Sixou B, Faivre A, David L, G V. Mol. Phys. 2001;99:1845. [Google Scholar]
- 13.Kirschner KN, Yongye AB, Tschampel SM, González-Outeiriño J, Daniels CR, Foley BL, Woods RJ. J. Comput. Chem. 2008;29:622. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacKerell AD, Jr., Feig M, Brooks CL., III J. Comput. Chem. 2004;25:1400. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 15.MacKerell AD, Jr., Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. J. Phys. Chem. B. 1998;102:3586. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 16.Feller SE, MacKerell AD., Jr. J. Phys. Chem. B. 2000;104:7510. [Google Scholar]
- 17.Feller SE, Gawrisch K, MacKerell AD., Jr. J. Am. Chem. Soc. 2002;124:318. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 18.Foloppe N, MacKerell AD., Jr. J. Comput. Chem. 2000;21:86. [Google Scholar]
- 19.MacKerell AD, Jr., Banavali NK. J. Comput. Chem. 2000;21:105. [Google Scholar]
- 20.MacKerell AD, Jr., Brooks B, Brooks CL, III, Nilsson L, Roux B, Won Y, Karplus M. CHARMM: The energy function and its paramerization with an overview of the program. In: Schleyer P. v. R., Allinger NL, Clark T, Gasteiger J, Kollman PA, Schaefer HF, III, Schreiner PR., editors. Encyclopedia of Computational Chemistry. Vol. 1. John Wiley & Sons; Chichester: 1998. p. 271. [Google Scholar]
- 21.MacKerell AD., Jr. J. Comput. Chem. 2004;25:1584. doi: 10.1002/jcc.20082. [DOI] [PubMed] [Google Scholar]
- 22.Guvench O, Greene SN, Kamath G, Brady JW, Venable RM, Pastor RW, MacKerell AD., Jr. J. Comput. Chem. 2008;29:2543. doi: 10.1002/jcc.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allen FH. Acta Crystallogr. Sect. B-Struct. Sci. 2002;58:380. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- 24.Guvench O, MacKerell AD. J. Mol. Model. 2008;14:667. doi: 10.1007/s00894-008-0305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J. Comput. Chem. 1983;4:187. [Google Scholar]
- 26.Allen MP, Tildesley DJ. Computer Simulation of Liquids. Oxford University Press; Oxford: 1987. [Google Scholar]
- 27.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J. Chem. Phys. 1983;79:926. [Google Scholar]
- 28.MacKerell AD, Jr., Karplus M. J. Phys. Chem. 1991;95:10559. [Google Scholar]
- 29.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Vreven T, Jr., Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda K, Kitao O, Nakai H, Klene M, Li TW, Knox JE, Hratchian HP, Cross JB, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03. Revision B.04 ed. Gaussian, Inc.; Pittsburgh, PA: 2003. [Google Scholar]
- 30.Shao Y, Molnar LF, Jung Y, Kussmann J, Ochsenfeld C, Brown ST, Gilbert ATB, Slipchenko LV, Levchenko SV, O'Neill DP, DiStasio RA, Lochan RC, Wang T, Beran GJO, Besley NA, Herbert JM, Lin CY, Van Voorhis T, Chien SH, Sodt A, Steele RP, Rassolov VA, Maslen PE, Korambath PP, Adamson RD, Austin B, Baker J, Byrd EFC, Dachsel H, Doerksen RJ, Dreuw A, Dunietz BD, Dutoi AD, Furlani TR, Gwaltney SR, Heyden A, Hirata S, Hsu CP, Kedziora G, Khalliulin RZ, Klunzinger P, Lee AM, Lee MS, Liang W, Lotan I, Nair N, Peters B, Proynov EI, Pieniazek PA, Rhee YM, Ritchie J, Rosta E, Sherrill CD, Simmonett AC, Subotnik JE, Woodcock HL, Zhang W, Bell AT, Chakraborty AK, Chipman DM, Keil FJ, Warshel A, Hehre WJ, Schaefer HF, Kong J, Krylov AI, Gill PMW, Head-Gordon M. Phys. Chem. Chem. Phys. 2006;8:3172. doi: 10.1039/b517914a. [DOI] [PubMed] [Google Scholar]
- 31.Guvench O, MacKerell AD., Jr. J. Phys. Chem. A. 2006;110:9934. doi: 10.1021/jp0623241. [DOI] [PubMed] [Google Scholar]
- 32.Durell SR, Brooks BR, Ben-Naim A. J. Phys. Chem. 1994;98:2198. [Google Scholar]
- 33.Darden T, York D, Pedersen L. J. Chem. Phys. 1993;98:10089. [Google Scholar]
- 34.Steinbach PJ, Brooks BR. J. Comput. Chem. 1994;15:667. [Google Scholar]
- 35.Ryckaert JP, Ciccotti G, Berendsen HJC. J. Comput. Phys. 1977;23:327. [Google Scholar]
- 36.Nosé S. Mol. Phys. 1984;52:255. [Google Scholar]
- 37.Hoover WG. Phys. Rev. A. 1985;31:1695. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 38.Feller SE, Zhang YH, Pastor RW, Brooks BR. J. Chem. Phys. 1995;103:4613. [Google Scholar]
- 39.Hockney RW. The potential calculation and some applications. In: Alder B, Fernbach S, Rotenberg M, editors. Methods in Computational Physics. Vol. 9. Academic Press; New York: 1970. p. 136. [Google Scholar]
- 40.Deng YQ, Roux B. J. Chem. Phys. 2008;128:8. doi: 10.1063/1.2842080. [DOI] [PubMed] [Google Scholar]
- 41.Weeks JD, Chandler D, Andersen HC. J. Chem. Phys. 1971;54:5237. [Google Scholar]
- 42.Zacharias M, Straatsma TP, McCammon JA. J. Chem. Phys. 1994;100:9025. [Google Scholar]
- 43.Pastor RW. Techniques and Applications of Langevin Dynamics Simulations. In: Luckhurst GR, Veracini CA, editors. The Molecular Dynamics of Liquid Crystals. Kluwer Academic Publishers; The Netherlands: 1994. p. 85. [Google Scholar]
- 44.Simonson T. Free energy calculations. In: Becker OM, MacKerell AD, Roux B, Watanabe M, editors. Computational biochemistry and biophysics. Marcel Dekker, Inc.; New York: 2001. p. 169. [Google Scholar]
- 45.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. J. Comput. Chem. 1992;13:1011. [Google Scholar]
- 46.Anisimov VM, Lamoureux G, Vorobyov IV, Huang N, Roux B, MacKerell AD. J. Chem. Theory. Comput. 2005;1:153. doi: 10.1021/ct049930p. [DOI] [PubMed] [Google Scholar]
- 47.Yeh IC, Hummer G. J. Phys. Chem. B. 2004;108:15873. [Google Scholar]
- 48.Tanford C. Physical chemistry of macromolecules. John Wiley and Sons; New York: 1961. [Google Scholar]
- 49.Lee H, Venable RM, MacKerell AD, Pastor RW. Biophys. J. 2008;95:1590. doi: 10.1529/biophysj.108.133025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franks F, Dadok J, Ying S, Kay RL, Grigera JR. J. CHEM. SOC. FARADAY TRANS. 1991;87:579. [Google Scholar]
- 51.Vorobyov IV, Anisimov VM, MacKerell AD., Jr. J. Phys. Chem. B. 2005;109:18988. doi: 10.1021/jp053182y. [DOI] [PubMed] [Google Scholar]
- 52.Kollman P. Chem. Rev. 1993;93:2395. [Google Scholar]
- 53.Vargaftik NB. Handbook of Physical Properties of Liquids and Gases: Pure Substances and Mixtures. 2nd ed. Hemisphere; Bristol, PA: 1983. [Google Scholar]
- 54.Hawkins GD, Cramer CJ, Truhlar DG. J. Phys. Chem. B. 1998;102:3257. [Google Scholar]
- 55.Smith BD, Srivastava R. Thermodynamic Data for Pure Compounds: Part A: Hydrocarbons and Ketones. Elsevier; New York: 1986. [Google Scholar]
- 56.McCall DW, Douglass DC, Anderson EW. The Journal of Chemical Physics. 1959;31:1555. [Google Scholar]
- 57.Ross GR, Heideger WJ. J. Chem. Eng. Data. 1962;7:505. [Google Scholar]
- 58.Smith BD, Srivastava R. Thermodynamic Data for Pure Compounds: Part B: Halogenated Hydrocarbons and Alcohols. Elsevier; New York: 1986. [Google Scholar]
- 59.Stejskal EO, Tanner JE. The Journal of Chemical Physics. 1965;42:288. [Google Scholar]
- 60.Pulay P, Fogarasi G, Pang F, Boggs JE. J. Am. Chem. Soc. 1979;101:2550. [Google Scholar]
- 61.Scott AP, Radom L. J. Phys. Chem. 1996;100:16502. [Google Scholar]
- 62.Shirts MR, Pitera JW, Swope WC, Pande VS. J. Chem. Phys. 2003;119:5740. [Google Scholar]
- 63.Deng YQ, Roux B. J. Phys. Chem. B. 2004;108:16567. [Google Scholar]
- 64.Hu YF, Zhang ZX, Zhang YH, Fan SS, Liang DQ. J. Chem. Eng. Data. 2006;51:438. [Google Scholar]
- 65.Blodgett MB, Ziemer SP, Brown BR, Niederhauser TL, Woolley EM. J. Chem. Thermodyn. 2007;39:627. [Google Scholar]
- 66.Feller SE, Pastor RW, Rojnuckarin A, Bogusz S, Brooks BR. J. Phys. Chem. 1996;100:17011. [Google Scholar]
- 67.Sartorio R, Wurzburger S, Guarino G, Borriello G. J. Solution Chem. 1986;15:1041. [Google Scholar]
- 68.Anisimov VM, Vorobyov IV, Roux B, MacKerell AD. J. Chem. Theory. Comput. 2007;3:1927. doi: 10.1021/ct700100a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lide DR, editor. CRC Handbook of Chemistry and Physics. 84 ed. CRC Press; Boca Raton, FL: 2003. [Google Scholar]
- 70.Humphrey W, Dalke A, Schulten K. J. Mol. Graph. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.