Abstract
Marine nematodes that carry sulfur-oxidizing bacteria on their cuticle (Stilbonematinae, Desmodoridae) migrate between oxidized and reduced sand layers thereby supplying their symbionts with oxygen and sulfide. These symbionts, in turn, constitute the worms' major food source. Due to the accessibility, abundance and relative simplicity of this association, stilbonematids may be useful to understand symbiosis establishment. Nevertheless, only the symbiont of Laxus oneistus has been found to constitute one single phylotype within the Gammaproteobacteria. Here, we characterized the symbionts of three yet undescribed nematodes that were morphologically identified as members of the genus Robbea. They were collected at the island of Corsica, the Cayman Islands and the Belize Barrier Reef. The surface of these worms is covered by a single layer of morphologically undistinguishable bacteria. 18S rDNA-based phylogenetic analysis showed that all three species belong to the Stilbonematinae, although they do not form a distinct cluster within that subfamily. 16S rDNA-based analysis of the symbionts placed them interspersed in the cluster comprising the sulfur-oxidizing symbionts of L. oneistus and of marine gutless oligochaetes. Finally, the presence and phylogeny of the aprA gene indicated that the symbionts of all three nematodes can use reduced sulfur compounds as an energy source.
Introduction
Marine nematodes that live a few centimetres below the surface of sandy bottoms may carry sulfur-oxidizing bacteria (SOB) within their body as endosymbionts [Astomonema (Ott et al., 1982; Vidakovic and Boucher 1987; Giere et al., 1995; Musat et al., 2007) and Parastomonema (Kito, 1989)] or on their surface as ectosymbionts. The latter belong to the subfamily Stilbonematinae and consist of the genera Adelphus Ott 1997, Catanema Cobb 1920, Eubostrichus Greef 1869, Laxus Cobb 1894, Leptonemella Cobb 1920, RobbeaGerlach 1956, SquanemaGerlach 1963 and Stilbonema Cobb 1920 (reviewed in Ott et al., 2004a,b). The worms migrate between oxygenated, upper sand layers and anoxic, sulfidic, deeper ones (Ott et al., 1991) allowing the bacteria to obtain the oxygen they need as e- acceptor and the sulfur compounds (e.g. hydrogen sulfide, thiosulfate) as e- donor (Polz et al., 1992; Hentschel et al., 1999). Stable carbon isotope incorporation experiments showed that the ectosymbionts are the major components of their host diet (Ott et al., 1991).
Symbionts are probably acquired from the environment because unhatched early embryos of Laxus oneistus are symbiont-free (Silvia Bulgheresi and Joerg A. Ott, in preparation). Environmental transmission would also enable nematodes to re-establish their symbiotic coat every time they replace their cuticle with a newly synthesized one. This process, known as molting or ecdysis, occurs several times during worm development. Moreover, Robbea sp.1 and sp.3 symbiont 16S rDNAs were detected in sand and seawater by polymerase chain reaction (PCR) and fluorescence in situ hybridization (FISH) with 16S rRNA-specific primers. As for the mechanisms of symbiont recruitment from the environment, we showed that the Ca2+-dependent lectin Mermaid mediates symbiont–symbiont and worm–symbiont attachment in L. oneistus (Bulgheresi et al., 2006).
Up to the present study only the symbionts of L. oneistus have been shown to belong to one single phylotype of Gammaproteobacteria closely related to the endosymbionts of marine gutless oligochaetes (Polz et al., 1994) and of Astomonema sp. (Musat et al., 2007). Although a molecular characterization of the large, multinucleated, filamentous symbiont of Eubostrichus dianae has been attempted, its 16S rDNA could not be amplified by PCR (Polz et al., 1999).
In this study, we molecularly characterized three associations involving stilbonematids which we assigned to the genus Robbea (Gerlach, 1956; 1963) based on their morphological characteristics. We collected Robbea sp.1 in the Mediterranean Sea from a subtidal sand patch close to a Posidonia oceanica seagrass meadow near Calvi (Corsica, France), and Robbea sp.2 and Robbea sp.3 in the Caribbean Sea from shallow back-reef sandbars at Little Cayman Island (Cayman Islands) and Carrie Bow Cay (Belize) respectively. We first analysed the phylogenetic position of the worms by making clone libraries of their 18S rRNA genes. We then characterized the symbionts associated with each species by cloning their respective 16S rRNA genes. To confirm that the latter were indeed derived from the ectosymbionts, we applied FISH on whole worms. Finally, the cloning and phylogenetic analysis of a gene that is involved in sulfur metabolism support the sulfur-oxidizing nature of the Robbea symbionts.
Results and discussion
Morphological and 18S rDNA-based molecular characterization of Robbea nematodes
The genus Robbea was established by Gerlach (1956). It is characterized by a clearly set off and muscle-rich distal part (corpus) of the tripartite pharynx. Moreover, all males, except in Robbea caelestis, are provided by a row of ventromedian suckers in the postpharyngeal region which are supposed to be copulation-helping organs (J.A. Ott, unpubl. data; Fig. 1A, C, E and G and asterisks in Fig. 1F). The number of suckers is constant and species-specific. Because each of the three nematodes characterized in this study had a tripartite, muscle-rich pharynx and carried a row of ventromedian suckers, we assigned them to the genus Robbea. Nevertheless, they did not form a monophyletic lineage within the Stilbonematinae (Chromadorea) in our 18S rDNA-based phylogenetic reconstruction (Fig. 2). It is therefore conceivable that their distinctive morphological traits evolved several times independently. Alternatively, supplementary sequence information from the 28S or Internal Transcribed Spacers (ITS) rDNA or from mitochondrial genes might be needed to support the genus Robbea at the molecular level.
Fig. 1.
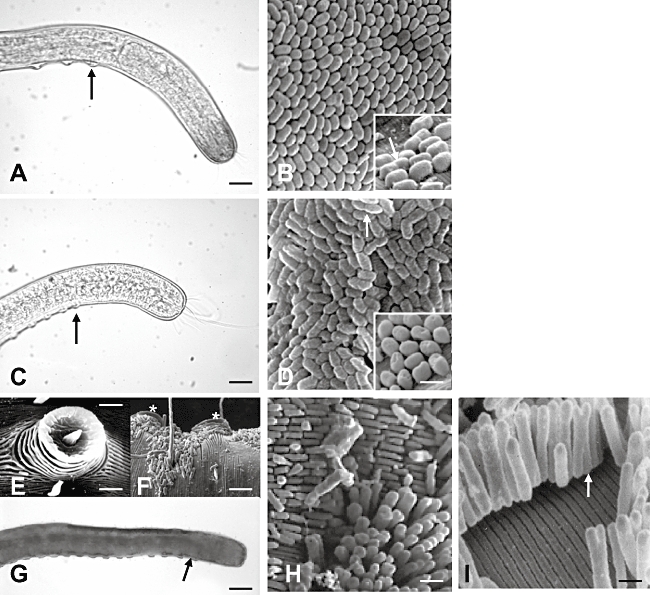
Photomicrographs of the anterior regions of fixed Robbea sp.1 (A), Robbea sp.2 (C) and Robbea sp.3 (G) and scanning electron microscopy (SEM) photographs of their respective symbionts (B, D, H and I). Black arrows point to the beginning of the suckers' row on each worm in (A), (C) and (G), while white arrows point to dividing symbionts in (B), (D) and (I). (E) and (F) are SEM photographs of one individual bacteria-free sucker, and two symbiont-coated suckers (asterisks) of Robbea sp.3, respectively. Scale bar is: 25 µm in (A) and (C); 1.5 µm in (B) and (D); 3 µm in (E); 8 µm in (F); 40 µm in (G); 2 µm in (H); 0.6 µm in (I).
Fig. 2.
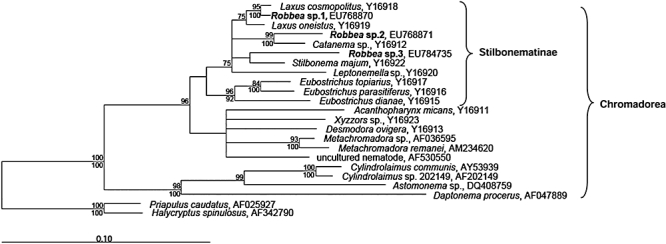
18S rDNA-based consensus phylogenetic tree based on maximum parsimony and Treepuzzle analysis showing the relationship of the Robbea worms (in bold) with other Stilbonematinae and other Chromadorea. Treepuzzle support values are depicted above the respective branches and maximum parsimony bootstrap values below the branches. Only Treepuzzle support values above 75% and parsimony bootstrap values higher than 70% are displayed. The scale bar represents 10% estimated sequence divergence.
Robbea sp.3 and Stilbonema majum were the only stilbonematids which showed the highest 18S rDNA sequence similarity with one another, while co-occurring in the same collection site, the Belize Barrier Reef. The 18S rDNAs of Robbea sp. 2, however, showed the highest sequence similarity with that of stilbonematids collected in an extremely distant geographical location.
Each nematode is covered by a single morphotype of symbionts: Robbea sp.1 and sp.2 display coccoid bacteria c. 1.5 µm wide (Fig. 1B and D respectively) whose shape and arrangement are reminiscent of kernels on a corn cob. Robbea sp.3 is covered by spindle-shaped rods c. 2 µm long (Fig. 1F). These assume different orientations with respect to the worm's surface, with some standing perpendicularly, as observed for L. oneistus, and some laying horizontally. In Robbea sp.1 and sp.2, the symbionts appear to divide transversally (arrows in Fig. 1B and D respectively). In Robbea sp.3 they divide longitudinally (arrows in Fig. 1H and I), a special mode of binary fission also exhibited by L. oneistus symbionts (Polz et al., 1992; 1994). Concerning the length of the microbial coat, only the anterior-most region of Robbea sp.3 and the very tip of the tail are symbiont-free. In Robbea sp.1 and Robbea sp.2, instead, the coat starts a short distance behind the anterior end, coinciding with a reduction in the worm diameter to accommodate the symbionts. This last feature is also displayed by L. oneistus. As in all other known stilbonematids, the Robbea symbionts are densely packed and appear bright white in incident light, probably due to inclusions of elemental sulfur (Himmel et al., 2009).
Robbea symbionts belong to the marine nematode and oligochaete symbionts cluster
Robbea symbiont 16S rDNA clones were randomly picked and comparison of their complete sequences showed that they could be assigned to three distinct clone groups belonging to the Gammaproteobacteria, with a sequence similarity within each clone group ≥ 99.8%. In our 16S rDNA-based phylogenetic reconstruction (Fig. 3) the three obtained gammaproteobacterial 16S rDNAs clustered with those of the symbionts of L. oneistus, of the nematode Astomonema sp., and of all known marine gutless oligochaetes (Inanidrilus and Olavius spp.). This nematode–oligochaete symbiont cluster is most closely related to the SOB from the family Chromatiaceae (> 90%). It is intriguing that, although free-living, some of these sulfur purple bacteria engage in symbiotic associations with unrelated bacteria in phototrophic consortia (Tonolla et al., 2000; Overmann, 2002).
Fig. 3.
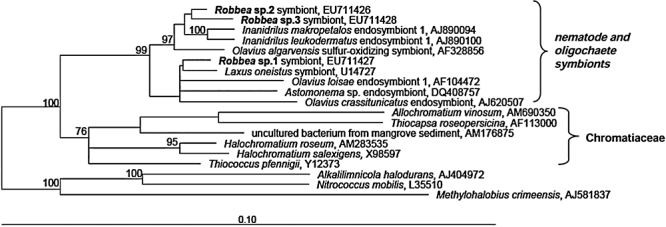
16S rDNA-based phylogenetic tree based on Treepuzzle analysis showing the relationship of the Robbea symbionts (in bold) with other stilbonematid and oligochaete symbionts, as well as other bacteria belonging to the Chromatiaceae and other vestimentiferan and mussel symbionts. Treepuzzle support values are depicted above the respective branches and maximum parsimony bootstrap values below the branches. Only Treepuzzle support values above 75% are displayed. The scale bar represents 10% estimated sequence divergence.
Our phylogenetic reconstruction shows that the three Robbea symbionts (16S rDNA sequence identity ≥ 97.1%) do not form a distinct group within the nematode–oligochaete sulfur-oxidizing symbionts cluster (16S rDNA sequence identity ≥ 95.4%). Moreover, nematode symbionts cannot be consistently grouped according to the geographical origin of their hosts and probably did not speciate in concert with their hosts. Phylogenetic incongruence between host and symbiont is typical of horizontally transmitted symbioses (Moran and Baumann, 2000), and was also observed for marine gutless oligochaetes and their sulfur-oxidizing symbionts (Dubilier et al., 2001; Blazejak et al., 2006; Musat et al., 2007).
To confirm that the gammaproteobacterial 16S rDNA sequences derived from the Robbea symbionts, we carried out FISH with the symbiont-specific probes Rca470, Rss457 and Rhs465, for Robbea sp.1, sp.2 and sp.3 respectively (Table 1). All the bacteria attached to the worms were triple stained by the eubacterial probe EUB338, by the Gammaproteobacteria-specific probe GAM42a and by the respective specific probe (Fig. 4). In contrast, no FISH signal was detectable with the negative control probe NON338 or with a Betaproteobacteria-specific probe (data not shown). This indicates that the bacteria covering each of the three Robbea species belong to one single phylotype and that no additional bacteria are present. This is consistent with the electron microscopy analysis, which shows only one bacterial morphotype on each Robbea worm, and with our highly homogeneous 16S rDNA libraries.
Table 1.
Probes used for FISH.
| Probe | Standard probe namea | Specificity | Sequence/5′ modification | Target RNA | Positionb,c | Formamide percentage/ incubation time (h)/probe concentration (ng µl−1) | Reference |
|---|---|---|---|---|---|---|---|
| EUB338 | S-*-BactV-0338-a-A-18 | Most bacteria | 5′-GCT GCC TCC CGT AGG AGT-3′/fluorescein | 16S | 338–355 | 35–40%/1.5–o.n./3 | Amann et al. (1990) |
| GAM42a | L-C-gProt-1027-a-A-17 | Gammaproteobacteria | 5′-GCC TTC CCA CAT CGT TT-3′/Cy5 | 23S | 1027–1043 | 35–40%/1.5–o.n./3 | Manz et al. (1992) |
| Rcas470 | S-Rob1s-0471-a-A-21 | Robbea sp.1 symbiont | 5′-TGC GTA ACG TCA AGA CCC TGG-3′/Cy3 | 16S | 471–491 | 25%/1.5/3.8 | This study |
| Rss456 | S-*-Rob2s-0457-a-A-21 | Robbea sp.2 symbiont, Inanidrilus leukodermatus endosymbiont 1 (GenBank Accession No. AJ890100) | 5′-ACC CTG AGC TAT TAA CCC AAG-3′/Cy3 | 16S | 457–477 | 35%/o.n./4 | This study |
| Rhs465 | S-Rob3s-a-A-21 | Robbea sp.3 symbiont | 5′-AAC GTC AGG ATC CCG AGC TAT-3′/Cy3 | 16S | 466–486 | 40%/3/2.3 | This study |
| NON338 | Not named | Negative control | 5′-ACT CCT ACG GGA GGC AGC-3′/Cy3 | 16S | 338–355 | 35–40%/1.5–o.n./3 | Wallner et al. (1993) |
| BET42a | L-C-bProt-1027-a-A-17 | Betaproteobacteria | 5′-GCC TTC CCA CTT CGT TT-3′/fluorescein | 16S | 1027–1043 | 35–40%/1.5–o.n./3 | Manz et al. (1992) |
According to Alm and colleagues (1996).
16S rRNA position, Escherichia coli numbering (Brosius et al., 1978).
23S rRNA position, E. coli numbering (Brosius et al., 1981).
o.n., overnight.
Fig. 4.
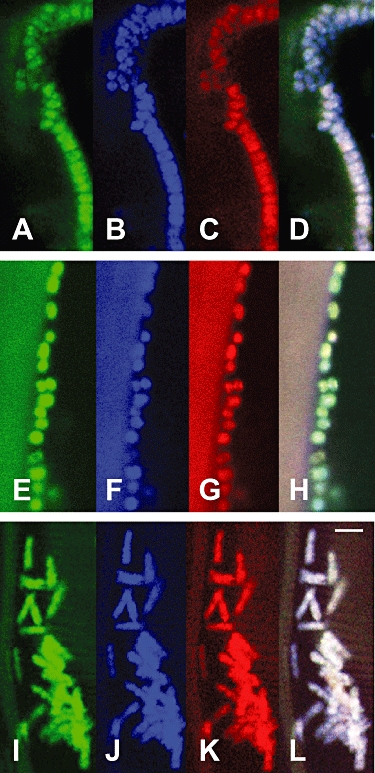
Fluorescence in situ hybridization (FISH) confocal microscope photographs of Robbea sp.1 (A–D), Robbea sp.2 (E–H) and Robbea sp.3 (I–L) symbionts attached to the worm surface. Each single symbiont is triple stained with a eubacteria-specific probe (green), a Gammaproteobacteria-specific probe (blue), and a symbiont-specific probe (red). (D), (H) and (L) are overlay pictures of (A)–(C), (E)–(G) and (I)–(K), respectively. Scale bar is 2 µm.
aprA gene analysis of stilbonematid-associated bacteria
To gain evidence that Robbea symbionts are indeed SOB, we cloned a fragment of the gene encoding for the alpha subunit of the adenosine-5′-phosphosulfate (APS) reductase (aprA), an enzyme involved in sulfur metabolism. The AprA protein reduces APS to sulfite in sulfate-reducing bacteria (SRB), but also catalyses the reverse reaction in SOB (Hipp et al., 1997; Sanchez et al., 2001; Friedrich, 2002). By using a set of aprA-specific primers, we PCR amplified and cloned a ∼1400-nt-long fragment from Robbea- and L. oneistus-associated bacteria. Several clones from each aprA library were randomly picked (see Experimental procedures) and their predicted protein sequences used for tree calculation (Fig. 5).
Fig. 5.
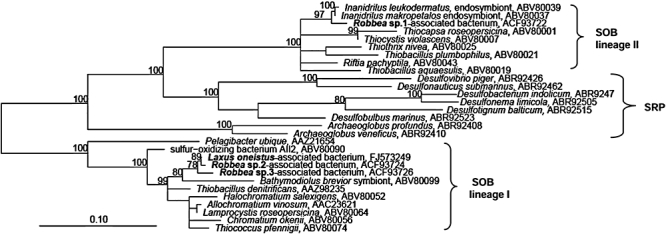
Phylogenetic reconstruction based on Treepuzzle analysis of AprA sequences from the Robbea-associated bacteria (in bold). Only support values above 75% are displayed. The scale bar represents 10% estimated sequence divergence.
The AprA sequences of bacteria associated with Robbea sp.2, sp.3 and L. oneistus clustered together with those of the Bathymodiolus brevior symbiont, and of some free-living sulfur-oxidizing gammaproteobacteria and sulfur purple bacteria [AprA-lineage I; see Meyer and Kuever (2007b) for a definition of AprA-lineages I and II]; Robbea sp.1-associated symbiont AprA, instead, clustered with those of gutless oligochaete sulfur-oxidizing symbionts (AprA-lineage II). Notably, Robbea sp.2 and sp.3 symbionts cluster together in both the 16SrDNA- and aprA-based trees.
In conclusion, all the AprA sequences obtained in this study are most closely related to SOB AprAs. This suggests that Robbea symbionts oxidize sulfur compounds as an energy source.
Conclusions
We characterized three new nematode–bacteria associations with very different geographical origins – the island of Corsica, the Cayman Islands and the Belize Barrier Reef. Although we cannot exclude that the three Robbea symbionts could stably associate with other marine organisms, our data show that each Robbea sp. is always coated by one characteristic symbiont phylotype. The basis of this conclusion is that each 16S rDNA and each aprA library was highly homogeneous and that the symbionts of each species were reproducibly stained by a symbiont 16S rDNA-specific FISH probe. Accordingly, electron microscopic analysis revealed that individuals of each Robbea sp. are always coated by the same, characteristic bacterial morphotype.
Our 18S rDNA-based tree shows that all three nematode species are stilbonematids, albeit additional worm nuclear and/or mitochondrial DNA sequence information is needed to confirm the genus Robbea at the molecular level.
Intriguingly, the 16S rDNAs of the stilbonematid symbionts are tightly grouped with those of mouthless oligochaetes. One explanation is that nematodes and oligochaetes co-occur in shallow-water sandy bottoms and they are all exposed to a similar pool of environmental bacteria. This habitat potentially promoted the establishment of these associations several times in the course of the evolution and at many different geographical locations. In this scenario, nematodes and oligochaetes recruited similar bacteria from this shared habitat as prospective symbionts. Sequencing of one or more stilbonematid symbiont metagenome(s) might unveil molecular adaptations shared by the oligochaete and nematode sulfur-oxidizing symbionts.
The fact that Robbea-associated bacteria harbour SOB-like aprA genes indicates that they gain energy from oxidation of reduced sulfur compounds. Moreover, their white appearance supports their capacity to store elemental sulfur. Migration of Robbea nematodes between deep and superficial sand layers, as observed for L. oneistus (Ott et al., 1991), would alternatively supply their symbionts with reduced sulfur compounds and oxygen. In the absence of oxygen, symbionts might use nitrate to respire sulfide (Hentschel et al., 1999), while they could resort to their sulfur stores when sulfide is unavailable in the environment.
In turn, the Robbea worms might feed on their symbionts. Stable isotope incorporation experiments and electron microscope analysis of the gut microbiome indicate this to be the case for other stilbonematids (Ott et al., 1991). Cloning of other symbiont genes involved in sulfur metabolism and carbon fixation, and transmission electron microscopy of the symbionts coupled with multi-isotope imaging mass spectrometry will shed light on their physiology.
The geographical distribution of the three Robbea nematodes characterized in this study appears to be restricted to the respective collection sites. One future task will be to investigate if the stilbonematid symbionts can be found only in the host habitat, as in the case of tube worms (Harmer et al. 2008) and lucinid mussels symbionts (Gros et al., 2003) or, instead, are widely distributed throughout the oceans and can survive without their hosts.
Another key question is how specific ectosymbionts are recruited from the environment by different stilbonematid species. In this respect, we plan to identify which repertoires of Mermaid isoforms are expressed by the Robbea worms and to compare them with each other and with those of L. oneistus. An exciting outcome could be that expression of a characteristic lectin repertoire by each stilbonematid species underpins acquisition and maintenance of a specific bacterial coat.
Experimental procedures
Specimen collection
Robbea sp.1 was collected in July 2007 from a subtidal sand patch close to a P. oceanica seagrass meadow in c. 2 m depth in the harbour of the Station de Recherches Sous-Marines et Océanographiques (STARESO), Calvi, France (42°34′49″N, 8°43′27″W). Robbea sp.2 was collected in October 2006 in c. 1 m depth from a shallow water back-reef sand bar off Point of Sand Beach on Little Cayman, Cayman Islands (19°42′08″N, 79°57′46″W). Robbea sp.3 was collected in November 2007 in c. 1 m depth from a shallow water back-reef sand bar off Carrie Bow Cay, Belize (16°48′11″N, 88°04′55″W). The worms were extracted from the sand by shaking it in seawater and pouring the supernatant through a 63-µm-pore-size mesh screen. Single individuals were then picked by hand under a dissecting microscope. Robbea sp.1 and Robbea sp.3 worms were fixed either in ethanol, for DNA extraction, or in 1% osmium tetroxide in seawater, for FISH (Rinke et al., 2006), and then stored in ethanol at −80°C. Robbea sp.2 worms were flash frozen in liquid N2 and stored at −80°C either unfixed (for DNA extraction) or upon methanol fixation (for FISH).
Scanning electron microscopy
Worms were pre-fixed in a 2.5% glutaraldehyde, 0.1 M sodium cacodylate, 2% sucrose solution, rinsed with 0.1 M sodium cacodylate buffer, and post-fixed in a 1% osmium tetroxide, 0.1 M sodium cacodylate, 2% sucrose solution. After alcohol dehydration, worms were gold sputter coated and viewed through a Philips XL 20 scanning electron microscope.
DNA extraction and PCR amplification of 18S rDNA
We extracted and purified the DNA from single Robbea worms as described previously (Schizas et al., 1997) and 2 µl was used as a template for each PCR. A fragment of the Robbea sp.1 18S rRNA gene was amplified by PCR with the general eukaryotic primers 1f (5′-CTGGTTGATYCTGCCAGT-3′; Winnepenninckx et al., 1995) and 2023r (5′-GGTTCACCTACGGAAACC-3′; Pradillon et al., 2007). Cycling conditions were 94°C for 4 min; 94°C for 45 s, 49°C for 30 s, 72°C for 1 min 45 s 35×; 72°C for 10 min. The PCR product was 1779 nt. Robbea sp.2 18S rRNA was amplified with the general eukaryotic primers 1f (see above) and 18SE (5′-ATGATCCTTCCGCAGGTTCAC-3′; Perotto et al., 2000) and Robbea sp.3 18S rRNA was amplified with primers 1f and 2023r. Cycling conditions were: 95°C for 5 min; 95°C for 45 s, 48°C for 45 s, 72°C for 2 min 35×; 72°C for 10 min. The PCR product was 1755 nt for Robbea sp.2 and 1783 nt for Robbea sp.3.
DNA extraction and PCR amplification of 16S rDNA
Symbionts were washed off a deep-frozen pellet of 500 Robbea sp.2 individuals with 50 µl of ddH2O. The 50 µl was then transferred to a fresh 1.5 ml tube and incubated at 94°C for 10 min. Five microlitres of this solution was directly used as a template for PCR. For Robbea sp.1 and Robbea sp.3, DNA was extracted from single worms as described (Schizas et al., 1997), and 2 µl each was used as a template for PCR. For all Robbea worms, PCR was performed using the eubacterial primers 616V (5′-AGAGTTTGATYMTGGCTC-3′; Juretschko et al., 1998) and 1492R (5′-GGYTACCTTGTTACGACTT-3′; Kane et al., 1993). The PCR programme for Robbea sp.2 and sp.3 was: 94°C for 5 min; 94°C for 45 s, 47°C for 45 s, 72°C for 1 min 30 s 35×; 72°C for 10 min. Cycling conditions for Robbea sp.1 were: 94°C for 4 min; 94°C for 45 s, 49°C for 30 s, 72°C for 1 min 45 s 35×; 72°C for 10 min. Each PCR product was 1499 nt.
DNA extraction and PCR amplification of APS reductase (aprA) gene
We extracted and purified the DNA from single Robbea worms as described previously (Schizas et al., 1997) and 2 µl was used as a template for each PCR. To amplify a c. 1400 nt aprA (adenosine phosphosulfate reductase alpha subunit) gene fragment we used the primers AprA-1-FW (5′-TGGCAGATCATGATYMAYGG-3′) and AprA-10-RV for Robbea sp.1-associated bacteria (5′-CKGWAGTAGWARCCRGGRTA-3′) and AprA-11-RV (5′-CKGYRRTAGTAKCCSGGCCA-3′) for Robbea sp.2- and Robbea sp.3-associated bacteria, as described (Meyer and Kuever, 2007a,b).
Cloning
All PCR products were gel purified and cloned into pCR2.1-TOPO using the TOPO TA Cloning Kit (Invitrogen Life Technologies, Germany).
We randomly picked and fully sequenced: 8, 7 and 6 clones of the 18S rDNA fragments obtained by Robbea sp.1 (EU768870), sp.2 (EU76887) and sp.3 (EU784735) respectively; 13, 19 and 11 clones of the 16S rDNA fragments obtained by Robbea sp.1 (EU711427), sp.2 (EU711426) and sp.3 (EU711428) respectively; 24, 31 and 21 clones of the aprA gene fragment from Robbea sp.1 (EU864035), sp.2 (EU864037) and sp.3 (EU864039), respectively. Sequences were aligned and compared with CodonCode Aligner 1.6.3 software.
Phylogenetic analysis
For each Robbea species, the sequences of the symbiont 16S rDNA and the worm 18S rDNA were compared with sequences in GenBank by using blastn, the AprA sequences by using blastp (Altschul et al., 1990). Phylogenetic analysis was carried out using the arb program package (Ludwig et al., 2004). We used TreePuzzle 5.0 to evaluate the phylogenetic position of each Robbea worm and its respective symbiont. For 18S rDNA-based phylogenetic reconstruction, we also used the maximum parsimony method and constructed a consensus tree. Similarity matrices were calculated using the similarity matrix option in the neighbour joining field of the arb software package.
For tree calculations, we applied a 50% conservation filter and we used only sequences longer than 1450 bp for host phylogeny and longer than 1325 bp for symbiont phylogeny. Sequences of Priapulus caudatus (AF025927) and Halycriptus spinulosus (AF342790) for the host 18S rDNA tree and sequences of Alkalimnicola halodurans (AJ404972), Nitrococcus mobilis (L35510) and Methylohalobius crimeensis (AJ581837) served as out-groups for the symbiont 16S rDNA tree.
For the AprA protein tree, we aligned selected members of SOB, sulfate-reducing prokaryotes (SRP) and the stilbonematid symbiont sequences using T-coffee (Notredame et al., 2000). We applied a 50% conservation insertion deletion (indel) filter for tree calculation and members of the AprA lineage I (Meyer and Kuever, 2007b) served as out-groups.
Fluorescence in situ hybridization (FISH)
We designed FISH probes by using the ARB PROBE_DESIGN tool (see Table 1) and confirmed their specificity by comparing them with all available sequences in GenBank, SILVA, Greengenes. Probes were fluorescently labelled on their 5′ end (Thermo, Germany). FISH was performed according to Manz and colleagues (1992). Briefly, fixed nematodes (n = 30) of each Robbea sp. were incubated at 46°C in hybridization buffer containing the respective FISH probes [0.9 M NaCl, 20 mM Tris·HCl (pH 8.0), 0.001% SDS; refer to Table 1 for incubation time, formamide percentage and probe concentration]. Unspecific bound probe was subsequently removed by incubating at 48°C for 15 min in appropriate washing buffer. Nematodes were mounted in DAPI Vectashield (Vector Labs) and examined using a Leica TCS-NT confocal laser scanning microscope.
Acknowledgments
We are very grateful to Janek von Byern and Gerhard Spitzer for helping with the scanning electron microscopy and Niculina Musat for sharing information about primers and protocols. Special thanks to Jan Kuever and Alexander Loy for insightfully revising our work. Uli Dirks, Harald Gruber and Katrina Vanura are acknowledged for discussion and helpful comments. This work was supported by the Austrian Science Fund (FWF) Grant P17710-B12 and the Caribbean Coral Reef Ecosystem programme of the National Museum of Natural History (Smithsonian Institution), Washington, DC, USA.
References
- Alm EW, Oerther DB, Larsen N, Stahl DA, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amann RI, Krumholz L, Stahl DA. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazejak A, Kuever J, Erseus C, Amann R, Dubilier N. Phylogeny of 16S rRNA, ribulose 1,5-bisphosphate carboxylase/oxygenase, and adenosine 5′-phosphosulfate reductase genes from gamma- and alphaproteobacterial symbionts in gutless marine worms (oligochaeta) from Bermuda and the Bahamas. Appl Environ Microbiol. 2006;72:5527–5536. doi: 10.1128/AEM.02441-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J, Palmer ML, Kennedy PJ, Noller HF. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J, Dull TJ, Sleeter DD, Noller HF. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Bulgheresi S, Schabussova I, Chen T, Mullin NP, Maizels RM, Ott JA. A new C-type lectin similar to the human immunoreceptor DC-SIGN mediates symbiont acquisition by a marine nematode. Appl Environ Microbiol. 2006;72:2950–2956. doi: 10.1128/AEM.72.4.2950-2956.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubilier N, Mulders C, Ferdelman T, de Beer D, Pernthaler A, Klein M, et al. Endosymbiotic sulphate-reducing and sulphide-oxidizing bacteria in an oligochaete worm. Nature. 2001;411:298–302. doi: 10.1038/35077067. [DOI] [PubMed] [Google Scholar]
- Friedrich MW. Phylogenetic analysis reveals multiple lateral transfers of adenosine-5′-phosphosulfate reductase genes among sulfate-reducing microorganisms. J Bacteriol. 2002;184:278–289. doi: 10.1128/JB.184.1.278-289.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach SA. Die Nematodenbesiedlung des tropischen Brandungsstrandes von Pernambuco. Kieler Meeresforschg. 1956;12:202–218. [Google Scholar]
- Gerlach SA. Robbea tenax sp. n., ein merkwürdiger mariner Nematode von den Malediven. Int Rev Ges Hydrobiol. 1963;48:153–158. [Google Scholar]
- Giere O, Windoffer R, Southward EC. The bacterial ectosymbiosis of the gutless nematode, Astomonema southwardorum: ultrastructural aspects. J Mar Biol Assoc UK. 1995;75:153–164. [Google Scholar]
- Gros O, Liberge M, Heddi A, Khatchadourian C, Felbeck H. Detection of the free-living forms of sulfide-oxidizing gill endosymbionts in the lucinid habitat (Thalassia testudinum environment) Appl Environ Microbiol. 2003;69:6264–6267. doi: 10.1128/AEM.69.10.6264-6267.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer TL, Rotjan RD, Nussbaumer AD, Bright M, Ng AW, DeChaine EG, Cavanaugh CM. Free-living tube worm endosymbionts found at deep-sea vents. Appl Environ Microbiol. 2008;74:3895–3898. doi: 10.1128/AEM.02470-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel U, Berger EC, Bright M, Felbeck H, Ott JA. Metabolism of nitrogen and sulfur in ectosymbiotic bacteria of marine nematodes (Nematoda, Stilbonematinae) Mar Ecol Prog Ser. 1999;183:149–158. [Google Scholar]
- Himmel D, Maurin LC, Gros O, Mansot JL. Raman microspectrometry sulfur detection and characterization in the marine ectosymbiotic nematode Eubostrichus dianae (Desmodoridae, Stilbonematidae) Biol Cell. 2009;101:43–54. doi: 10.1042/BC20080051. [DOI] [PubMed] [Google Scholar]
- Hipp WM, Pott AS, Thum-Schmitz N, Faath I, Dahl C, Trüper HG. Towards the phylogeny of APS reductases and sirohaem sulfite reductases in sulfate-reducing and sulfur-oxidizing prokaryotes. Microbiology. 1997;143:2891–2902. doi: 10.1099/00221287-143-9-2891. [DOI] [PubMed] [Google Scholar]
- Juretschko S, Timmermann G, Schmid M, Schleifer KH, Pommerening-Roser A, Koops HP, Wagner M. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol. 1998;64:3042–3051. doi: 10.1128/aem.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane MD, Poulsen LK, Stahl DA. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl Environ Microbiol. 1993;59:682–686. doi: 10.1128/aem.59.3.682-686.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kito K. A new mouthless marine nematode from Fiji. J Nat Hist. 1989;23:635–642. [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. arb: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. ) Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- Meyer B, Kuever J. Phylogeny of the alpha and beta subunits of the dissimilatory adenosine-59-phosphosulfate (APS) reductase from sulfate-reducing prokaryotes – origin and evolution of the dissimilatory sulfate-reduction pathway. Microbiology. 2007a;153:2026–2044. doi: 10.1099/mic.0.2006/003152-0. [DOI] [PubMed] [Google Scholar]
- Meyer B, Kuever J. Molecular analysis of the distribution and phylogeny of dissimilatory adenosine-5′-phosphosulfate reductase-encoding genes (aprBA) among sulfur-oxidizing prokaryotes. Microbiology. 2007b;153:3478–3498. doi: 10.1099/mic.0.2007/008250-0. [DOI] [PubMed] [Google Scholar]
- Moran N, Baumann P. Bacterial endosymbionts in animals. Curr Opin Microbiol. 2000;3:270–275. doi: 10.1016/s1369-5274(00)00088-6. [DOI] [PubMed] [Google Scholar]
- Musat N, Giere O, Gieseke A, Thiermann F, Amann R, Dubilier N. Molecular and morphological characterization of the association between bacterial endosymbionts and the marine nematode Astomonema sp. from the Bahamas. Environ Microbiol. 2007;9:1345–1353. doi: 10.1111/j.1462-2920.2006.01232.x. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Ott J, Rieger G, Rieger G, Enders F. New mouthless interstitial worms from the sulfide system: symbiosis with prokaryotes. PSZN I: Mar Ecol. 1982;3:313–333. [Google Scholar]
- Ott J, Novak R, Schiemer F, Hentschel U, Nebelsick M, Polz M. Tackling the sulfide gradient: a novel strategy involving marine nematodes and chemoautotrophic ectosymbionts. PSZN I: Mar Ecol. 1991;12:261–279. [Google Scholar]
- Ott JA, Bright M, Bulgheresi S. Symbiosis between marine nematodes and sulfur-oxidizing chemoautotrophic bacteria. Symbiosis. 2004a;36:103–126. [Google Scholar]
- Ott JA, Bright M, Bulgheresi S. Marine microbial thiotrophic ectosymbioses. Oceanogr Mar Biol Ann rev. 2004b;42:95–118. [Google Scholar]
- Overmann J. Phototrophic consortia: a tight cooperation between non-related eubacteria. In: Sechback J, editor. Symbiosis: Mechanisms and Model Systems. Dordrecht, the Netherlands: Kluwer Academic Publishers; 2002. pp. 239–255. [Google Scholar]
- Perotto S, Nepote-Fus P, Saletta L, Bandi C, Young JP. A diverse population of introns in the nuclear ribosomal genes of ericoid mycorrhizal fungi includes elements with sequence similarity to endonuclease-coding genes. Mol Biol Evol. 2000;17:44–59. doi: 10.1093/oxfordjournals.molbev.a026237. [DOI] [PubMed] [Google Scholar]
- Polz MF, Felbeck H, Novak R, Nebelsick M, Ott JA. Chemoautotrophic, sulphur-oxidizing bacteria on marine nematodes: morphological and biochemical characterization. Microb Ecol. 1992;24:313–319. doi: 10.1007/BF00167789. [DOI] [PubMed] [Google Scholar]
- Polz MF, Distel DL, Zarda B, Amann R, Felbeck H, Ott JA, Cavanaugh CM. Phylogenetic analysis of a highly specific association between ectosymbiotic, sulfur-oxidizing bacteria and a marine nematode. Appl Environ Microbiol. 1994;60:4461–4467. doi: 10.1128/aem.60.12.4461-4467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polz MF, Harbison C, Cavanaugh CM. Diversity and heterogeneity of epibiotic bacterial communities on the marine nematode Eubostrichus dianae. Appl Environ Microbiol. 1999;65:4271–4275. doi: 10.1128/aem.65.9.4271-4275.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradillon F, Schmidt A, Peplies J, Dubilier N. Identification of early life history stages of marine invertebrate species with whole-larvae in situ hybridization. Mar Ecol Prog Ser. 2007;333:103–116. [Google Scholar]
- Rinke C, Schmitz-Esser S, Stoecker K, Nussbaumer AD, Molnár DA, Vanura K, et al. Candidatus Thiobios zoothamnicoli’, an ectosymbiotic bacterium covering the giant marine ciliate Zoothamnium niveum. Appl Environ Microbiol. 2006;72:2014–2021. doi: 10.1128/AEM.72.3.2014-2021.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez O, Ferrera I, Dahl C, Mas J. In vivo role of adenosine-5′-phosphosulfate reductase in the purple sulfur bacterium Allochromatium vinosum. Arch Microbiol. 2001;176:301–305. doi: 10.1007/s002030100327. [DOI] [PubMed] [Google Scholar]
- Schizas NV, Street GT, Coull BC, Chandler GT, Quattro JM. An efficient DNA extraction method for small metazoans. Mol Mar Biol Biotechnol. 1997;6:381–383. [PubMed] [Google Scholar]
- Tonolla M, Demarta A, Peduzzi S, Hahn D, Peduzzi R. In situ analysis of sulfate-reducing bacteria related to Desulfocapsa thiozymogenes in the chemocline of meromictic Lake Cadagno (Switzerland) Appl Environ Microbiol. 2000;66:820–824. doi: 10.1128/aem.66.2.820-824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidakovic J, Boucher G. Gutless marine nematodes of the genus Astomonema. Cah Biol Mar. 1987;28:111–120. [Google Scholar]
- Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- Winnepenninckx B, Backeljau T, Mackey LY, Brooks JM, De Wachter R, Kumar S, Garey JR. 18S rRNA data indicate that Aschelminthes are polyphyletic in origin and consist of at least three distinct clades. Mol Biol Evol. 1995;12:1132–1137. doi: 10.1093/oxfordjournals.molbev.a040287. [DOI] [PubMed] [Google Scholar]


