Abstract
Mycobacterial UvrD1 is a DNA-dependent ATPase and a Ku-dependent 3’ to 5’ DNA helicase. The UvrD1 motor domain resembles that of the prototypal superfamily I helicases UvrD and PcrA. Here we performed a mutational analysis of UvrD1 guided by the crystal structure of a DNA-bound E. coli UvrD-ADP-MgF3 transition state mimetic. Alanine scanning and conservative substitutions identified five amino acids essential for both ATP hydrolysis and duplex unwinding, including those implicated in phosphohydrolase chemistry via transition state stabilization (Arg308, Arg648, Gln275), divalent cation coordination (Glu236), or activation of the nucleophilic water (Glu236, Gln275). Other residues important for ATPase/helicase activity include Phe280 and Phe72, which interact with the DNA 3’-single strand tail. ATP hydrolysis was uncoupled from duplex unwinding by mutations at Glu609 (in helicase motif V), which contacts the ATP ribose sugar. Introducing alanine in lieu of the adenine-binding “Q motif” glutamine (Gln24) relaxed the substrate specificity in NTP hydrolysis, e.g., eliciting a gain-of-function as a UTPase/TTPase, although the Q24A mutant still relied on ATP/dATP for duplex unwinding. Our studies highlight the role of the Q motif as a substrate filter and the contributions of adenosine-binding residues as couplers of NTP hydrolysis to motor activity. The Ku-binding function of UvrD1 lies within its C-terminal 270-aa segment. Here we found that deleting the 90-aa C-terminal domain, which is structurally uncharacterized, diminished DNA unwinding, without affecting ATP hydrolysis or binding to the DNA helicase substrate, apparently by affecting the strength of the UvrD1-Ku interaction.
Bacterial superfamily I (SF1) DNA helicases are nucleic acid-dependent NTPases that translocate along single-stranded DNA and thereby unwind the double helix (1–6). SF1 helicases play critical roles during bacterial DNA repair and recombination. Escherichia coli UvrD exemplifies a subgroup of bacterial SF1 enzymes defined by: (i) specificity for hydrolysis of ATP or dATP; (ii) 3’ to 5’ directionality of DNA translocation and unwinding; (iii) a conserved ATPase motor domain composed of four structural subdomains; and (iv) a conserved set of generic and clade-specific peptide motifs that comprise the phosphohydrolase active site and the DNA-binding interface (1–6).
We are interested in the role of DNA repair and SF1 helicases in the physiology of Mycobacterium tuberculosis, the agent of human TB, and its avirulent cousin Mycobacterium smegmatis. We had previously characterized two mycobacterial UvrD-like helicases: UvrD1 and UvrD2 (7,8). UvrD2 per se is a vigorous DNA-dependent ATPase with 3’ to 5’ helicase activity. UvrD2 has a distinctive domain architecture consisting of an N-terminal UvrD-like motor unit and a C-terminal HRDC domain. The ATPase and HRDC domains are connected by a CxxC-(14)-CxxC module that defines a newly appreciated clade of bacterial helicases found only in Actinomycetales. Initial genetic tests indicate that UvrD2 is essential for growth of M. smegmatis (8).
By contrast, the mycobacterial UvrD1 protein has a conventional SF1 domain architecture and is homologous to E. coli UvrD and Bacillus stearothermophilus PcrA throughout its entire length (Fig. 1). As anticipated from this degree of structural similarity, UvrD1 is a potent DNA-dependent ATPase (7,9). However, UvrD1 is a feeble helicase by itself, requiring a vast excess of enzyme over DNA to attain unwinding (9), but it readily catalyzes efficient ATP-dependent 3’ to 5’ unwinding under low enzyme:DNA conditions in the presence of the DNA end-binding protein Ku (7). Ku is the key agent of the mycobacterial DNA break repair via non-homologous end joining (10). Indeed, UvrD1 was identified in a mycobacterial genome-wide two-hybrid screen as a binding partner for Ku (7). UvrD1, Ku and DNA form a stable ternary complex, suggesting that Ku might serve as a processivity factor for unwinding by UvrD1. Ablation of UvrD1 has no overt effect on bacterial growth, but does sensitize M. smegmatis to killing by ultraviolet and ionizing radiation (7).
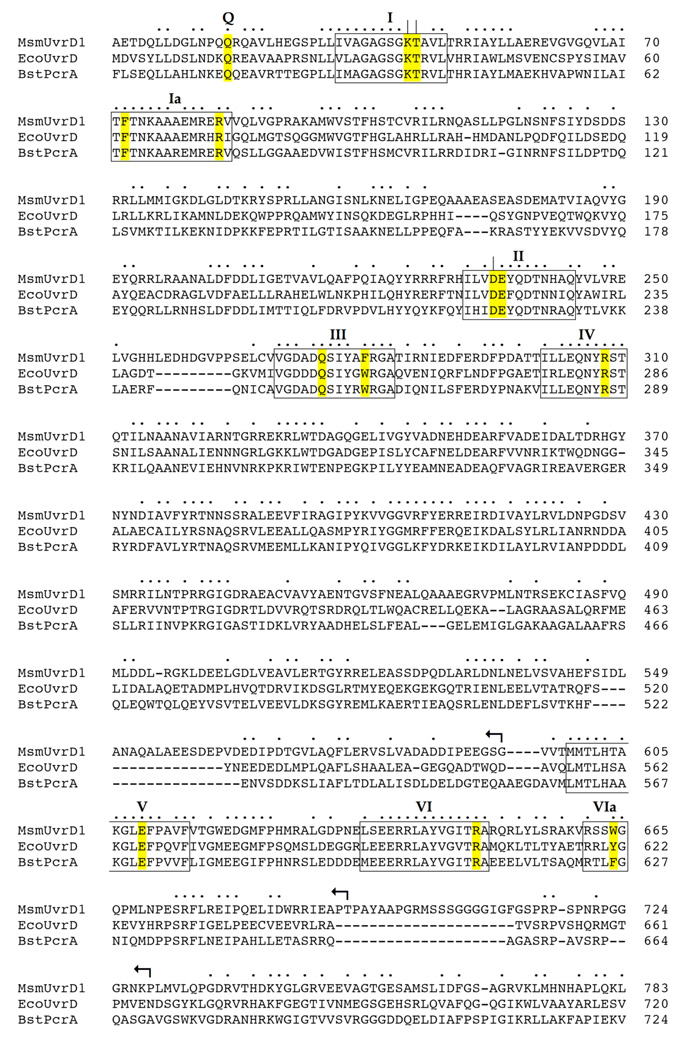
Fig. 1. Structural similarity between UvrD1, UvrD and PcrA.
The amino acid sequence of M. smegmatis (Msm) UvrD1 (accession YP_889771) is aligned to the sequences of E. coli (Eco) UvrD (accession NP_418258) and B. stearothermophilus (Bst) PcrA (accession P56255). Positions of side chain identity/similarity in all three proteins are indicated by •. Gaps in the alignment are denoted by –. The ATPase/helicase motifs are denoted in boxes and named according to Lee and Yang (4). The conserved residues comprising the ATPase active site and DNA-binding interfaces of UvrD/PcrA that were subjected to alanine scanning in UvrD1 are highlighted in yellow. Essential constituents of UvrD1 ATPase motifs I (Lys45, Thr46) and II (Asp235) that were identified previously are indicated by |. The targeted UvrD1 residues, and their equivalents in UvrD and PcrA, are listed in Table I. The C-terminal truncations of UvrD1 are denoted by ↰.
The distinctive interaction of UvrD1 with Ku, and a preliminary report that UvrD1 might play a role in persistence of M. tuberculosis infection in a murine model (9), prompt our ongoing interest in the structure and biochemical properties of this SF1 helicase. Here we exploited the available crystal structures of UvrD/PcrA in complex with ATP and DNA to guide a structure-function analysis of M. smegmatis UvrD1 by mutagenesis, especially targeting the UvrD1 equivalents of the amino acids that contact Mg2+·ATP and the single-stranded DNA tail on which the helicase loads and translocates during duplex unwinding. Our results complement and extend previous mutational studies of PcrA by Wigley and colleagues (11–13) and UvrD by the Matson lab (21–24,27) (Table I).
Table I.
Summary of Mutational Effects on UvrD1 Activities
| Motif | UvrD1 | ATPase | Helicase | DNA binding |
UvrD | PcrA | ATPase | Helicase | DNA binding |
|---|---|---|---|---|---|---|---|---|---|
| Q | Gln24 | ++ | ++ | +++ | Gln14 | Gln16 | |||
| Ia | Phe72 | ++ | − | +++ | Phe62 | Phe64 | ++ | − | +++ |
| Ia | Arg83 | ++ | +++ | +++ | Arg73 | Arg75 | |||
| II | Asp235 | − | − | +++ | Asp220 | Asp223 | − | − | +++ |
| II | Glu236 | + | − | ↓↓ | Glu221 | Glu224 | − | +++ | |
| III | Gln275 | − | − | ++ | Gln251 | Gln254 | + | − | +++ |
| III | Phe280 | + | − | ↓↓ | Trp256 | Trp259 | +++ | − | ↓↓ |
| IV | Arg308 | − | − | +++ | Arg284 | Arg287 | + | + | +++ |
| V | Glu609 | +++ | − | +++ | Glu566 | Glu571 | |||
| VI | Arg648 | − | − | +++ | Arg605 | Arg610 | − | − | +++ |
| VIa | Trp664 | +++ | +++ | +++ | Tyr621 | Phe626 | +++ | ++ | +++ |
EXPERIMENTAL PROCEDURES
Recombinant proteins
Wild-type M. smegmatis UvrD1 was produced by IPTG induction at 17°C and purified from soluble lysates by Ni-agarose and DEAE-Sephacel chromatography as described previously (7). For production of mutant proteins, the UvrD1 ORF was amplified by PCR from the pET16-UvrD1 template with oligonucleotide primers that introduce BglII sites overlapping the start codon and 3’ of the stop codon. The PCR product was digested with BglII and inserted into the BamHI site of pET28-His10Smt3. The resulting pET28-His10Smt3-UvrD1 plasmid was used as a template for introducing missense mutations and for creating C-terminal truncations of the UvrD1 open reading frame. The inserts of the mutant pET28-His10Smt3-UvrD1 plasmids were sequenced to exclude the acquisition of unwanted coding changes during amplification or cloning. The expression plasmids were transformed into E. coli BL21(DE3). Cultures (250 ml) derived from single transformants were grown in Luria-Bertani medium containing 50 µg/ml kanamycin until A600 reached 0.6. The cultures were chilled on ice for 45 min and then adjusted to 0.2 mM IPTG, followed by incubation at 17°C for 16 h with constant shaking. Cells were harvested by centrifugation and the pellets were stored at −80°C. All subsequent steps were performed at 4°C. Thawed bacterial cell pellets were suspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 0.25 M NaCl, 10% sucrose). Lysis was achieved by adding lysozyme and Triton X-100 to final concentrations of 1 mg/ml and 0.1%, respectively. The lysates were sonicated to reduce viscosity and insoluble material was removed by centrifugation. The supernatants were applied to 3-ml columns of nickel-nitriloacetic acid-agarose (Qiagen) that had been equilibrated with lysis buffer. The columns were washed with 10 ml of buffer A (50 mM Tris-HCl, pH 8.0, 0.25 M NaCl, 0.05% Triton X-100, 10% glycerol) and then eluted stepwise with aliquots of buffer A containing 50, 100, 200, 500 and 1000 mM imidazole. The polypeptide compositions of the column fractions were monitored by SDS-PAGE. The His10Smt3-UvrD1 proteins were recovered predominantly in the 100 and 200 mM imidazole eluates. The peak UvrD1-containing fractions were pooled and dialyzed against buffer A with 1 mM EDTA. To deplete the preparations of residual nucleic acids, the dialysates were applied to 3-ml columns of DEAE-Sephacel (Pharmacia) that had been equilibrated with buffer A plus 1 mM EDTA. The His10Smt3-UvrD1 proteins were recovered in the flow-through fraction. This material was mixed with the Smt3-specific protease Ulp1 (at a UvrD1:Ulp1 ratio of 500:1) and the mixtures were dialyzed overnight against buffer A containing 150 mM NaCl. SDS-PAGE of the dialysates verified that the His10Smt3 tag had been cleaved. The tag-free UvrD1 proteins were separated from the His10Smt3 tag by passage over a second nickel-agarose column; UvrD1 was recovered in the flow-through fraction while the tag adhered to the nickel resin. Protein concentrations were determined by using the Bio-rad dye reagent with bovine serum albumin as the standard.
Mycobacterial Ku protein was produced in E. coli BL21(DE3) as an N-terminal His10 fusion protein and purified from a soluble lysate by Ni-agarose and DEAE-Sephacel chromatography as described previously (7).
ATPase activity
Reaction mixtures (10 µl) containing 20 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 1 mM [γ32P]ATP (Perkin-Elmer Life Sciences), 50 ng salmon sperm DNA, and varying amounts of wild-type or mutant UvrD1 proteins were incubated for 5 min at 37°C. The reactions were quenched with 2 µl of 5 M formic acid. An aliquot (2 µl) of the mixture was applied to a polyethyleneimine-cellulose TLC plate, which was developed with 0.45 M ammonium sulfate. The radiolabeled material was visualized by autoradiography and 32Pi formation was quantified by scanning the TLC plate with a Fujix BAS2500 imager. Each enzyme titration entailed five or six serial 2-fold dilutions of the wild-type or mutant UvrD1 proteins. The reaction mixtures containing wild-type UvrD1 and the catalytically active mutants received as much as 25 ng of UvrD1. For assays of the most catalytically defective mutants, as much as 1000 or 1250 ng was added to the ATPase reaction mixtures. Product formation was plotted as a function of input UvrD1 and the specific activities were determined from the slopes of the titration curves.
NTPase specificity
Reaction mixtures (20 µl) containing 20 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 50 ng salmon sperm DNA, 1 mM unlabeled NTP or dNTP substrate, and either 50 ng wild-type UvrD1 or 200 ng of the Q24A mutant were incubated for 5 min at 37°C. The reactions were quenched by adding 1 ml of malachite green reagent (BIOMOL Research Laboratories). Phosphate release was determined by measuring A620 and interpolating the value to a phosphate standard curve.
Kinetic parameters for ATP and UTP hydrolysis
Reaction mixtures (10 µl) containing 20 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 50 ng salmon sperm DNA, either 1.5 or 2.5 ng wild-type UvrD1 or 7.5 or 10 ng Q24A, and increasing concentrations of [γ32P]ATP or [α32P]UTP (in serial 2-fold increments from as low as 31 µM to as high as 4 mM) were incubated for 2, 4 or 5 min at 37°C. The products were analyzed by polyethyleneimine-cellulose TLC developed with 0.5 M LiCl, 1 M formic acid. The extents of formation of 32Pi from ATP and [α32P]UDP from UTP were quantified by scanning the TLC plates with a Fujix BAS2500 imager. Product formation was plotted as a function of ATP or UTP concentration. Kinetic parameters were calculated from each titration experiment by nonlinear regression curve fitting of the data to the Michaelis-Menten equation in Prism. The Km and kcat values cited in Results are the mean of three separate substrate titration experiments (± standard error).
DNA dependence of ATP hydrolysis
Reaction mixtures (10 µl) contained 20 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 1.5 mM [γ32P]ATP, and either 0, 0.78, 1.56, 3.12, 6.25, 12.5, 25, 50, or 100 ng of salmon sperm DNA. The amounts of UvrD1 and the incubation times were as follows: wild-type, E609A and W664A (15 ng, 5 min); R83A and F72A (50 ng, 5 min), Q24A (75 ng, 5 min); E236A (15 ng, 60 min). 32Pi formation was plotted as a function of ATP concentration. Apparent Km values for sperm DNA were obtained by nonlinear regression curve fitting of the data to the Michaelis-Menten equation in Prism.
Helicase assay
The 5’ 32P-labeled strand was prepared by reaction of a 24-mer DNA oligonucleotide with T4 polynucleotide kinase and [γ32P]ATP. The labeled DNA was purified by electrophoresis through a native 18% polyacrylamide gel. The labeled strand was annealed to 2-fold excess of complementary 44-mer DNA strand to form the 3’-tailed duplex helicase substrate shown in Fig. 3C. Helicase reaction mixtures (10 µl) containing 20 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 0.5 pmol (50 nM) 32P-labeled 3’-tailed DNA, and 100 ng UvrD1 (1.1 pmol) and 75 ng Ku (1 pmol Ku homodimer) as specified were preincubated for 5 min on ice. The reactions were initiated by adding 1 mM ATP (or other NTP where specified) and 5 pmol of an unlabeled oligonucleotide corresponding to the labeled strand of the helicase substrate. Addition of excess of unlabeled strand was necessary to prevent the spontaneous reannealing of the unwound 32P-labeled DNA strand. The reaction mixtures were incubated for 5 min at 37°C and then quenched by adding 2 µl of a solution containing 2% SDS, 200 mM EDTA, 40% glycerol, 0.3% bromophenol blue. A control reaction mixture containing no protein was heated for 5 min at 95°C. The reaction products were analyzed by electrophoresis through a 15-cm 15% polyacrylamide gel in 89 mM Tris-borate, 2.5 mM EDTA. The products were visualized by autoradiography.
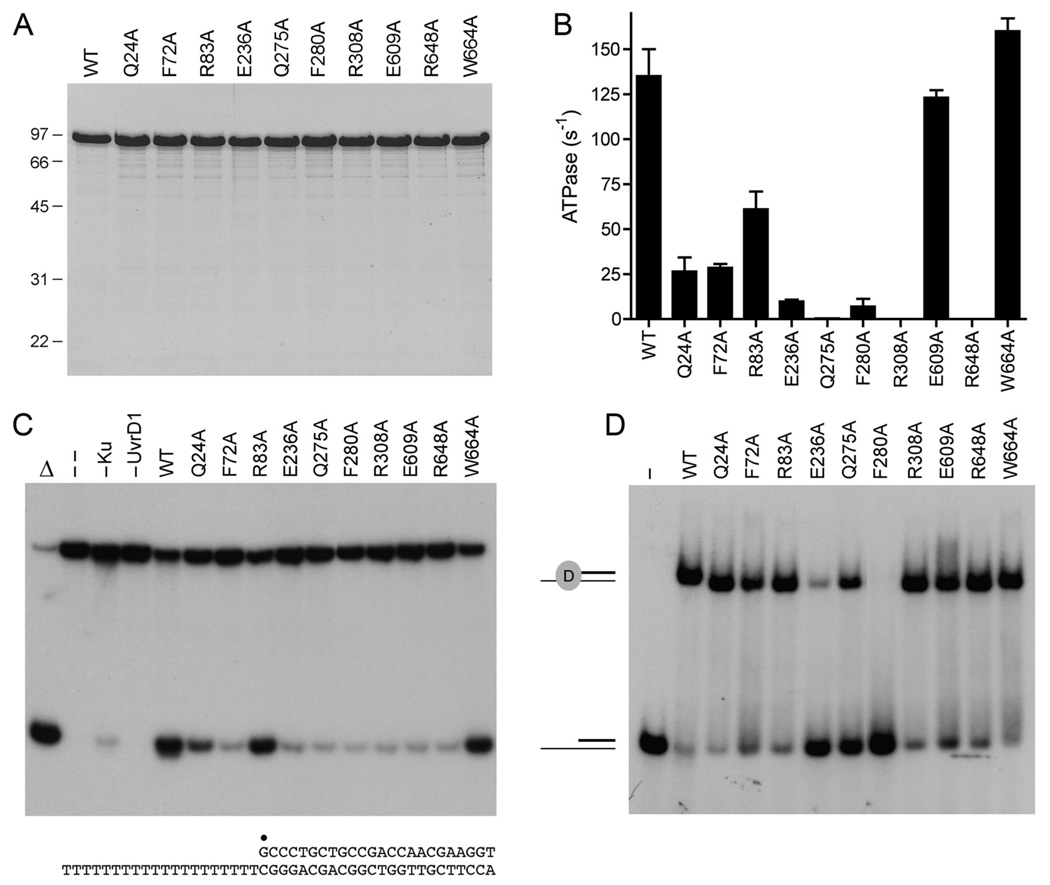
Fig. 3. Effects of alanine mutations on UvrD1 activities.
(A) Aliquots (5 µg) of recombinant wild-type (WT) UvrD1 and the indicated Ala mutants were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The sizes (kDa) and positions of marker proteins are indicated on the left. (B) ATPase specific activity was determined as specified under Methods. Each datum is the average of two separate UvrD1 titration experiments; error bars denote the mean absolute error. (C) Helicase reactions were performed as described under Methods. Complete reaction mixtures contained 1 mM ATP, 50 nM 32P-labeled tailed duplex DNA substrate, 75 ng Ku and 100 ng of wild-type or mutant UvrD1 protein as specified. The products were analyzed by native PAGE and visualized by autoradiography. Reactions without added protein (lane --), with wild-type UvrD1 only (–Ku) or with Ku only (–UvrD1) were included as controls. A reaction lacking protein that was heat denatured prior to PAGE is shown in lane Δ. The 3’-tailed duplex helicase substrate is shown at bottom with the 5’ 32P-label denoted by •. (D) Binding of UvrD1 to the helicase substrate. Binding reactions were performed as described under Methods. Complete reaction mixtures contained 1 pmol 32P-labeled tailed duplex DNA substrate and 100 ng wild-type or mutant UvrD1 protein as specified. UvrD1 was omitted from the control reaction in lane –. The products were analyzed by native PAGE and visualized by autoradiography. The positions of the free DNA and the UvrD1-DNA complex are indicated on the left.
DNA binding assay
Reaction mixtures (10 µl) containing 20 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 5% glycerol, 32P-labeled 3’-tailed DNA, and wild-type or mutant UvrD1 as specified were incubated for 10 min at 37°C. The mixtures were adjusted to 10% glycerol and then analyzed by electrophoresis through a 4% native polyacrylamide gel containing 45 mM Tris-borate, 1.2 mM EDTA. The gels were run at 200 V in the cold room. The gel was transferred to DEAE paper and dried under vacuum. The free DNA and protein-DNA complexes were visualized by autoradiography.
RESULTS
Alanine scanning of UvrD1
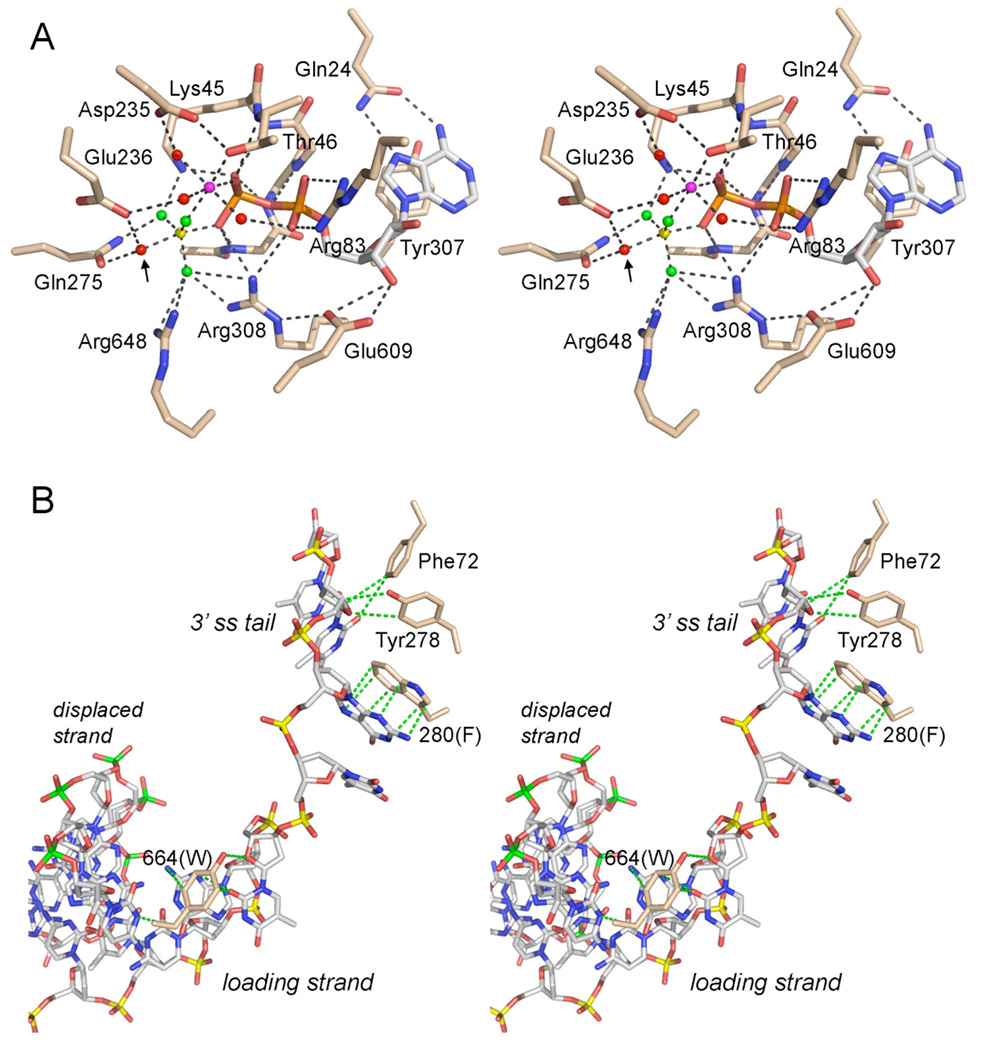
The primary structure alignment in Fig. 1 highlights 330 positions of side chain identity/similarity in M. smegmatis UvrD1, E. coli UvrD, and B. stearothermophilus PcrA. UvrD1 has apparent counterparts of all of the ATPase, DNA binding, and domain interaction motifs enumerated for E. coli UvrD (4). In our initial characterization of UvrD1 (7), we found that ATP hydrolysis was abolished by a single-alanine substitution for the metal-binding motif II aspartate (Asp235) or a double-alanine replacement of the motif I residues Lys45 (which contacts the β and γ phosphates of ATP) and Thr46 (which coordinates the metal) (Fig. 2A). Suppression of ATPase activity by the D235A change coordinately abolished duplex DNA unwinding, but did not affect the binding of UvrD1 to the 3’ tailed DNA helicase substrate (7) (Table I). Similar ablative effects of motif I and II mutations on catalysis and motor function, but not nucleic acid binding, have been reported for the PcrA DNA helicase (11) and the SF2 RNA helicase NPH-II (14), among others, which testifies to the universal importance of these two motifs found in all of the helicase superfamilies (1). The goal of the present study was to extend the mutagenesis of UvrD1 to other residues that contact Mg2+·ATP. In this effort, we were guided by the 2.2 Å structure of E. coli UvrD (4) bound to a 3’ tailed duplex DNA, with the active site occupied by Mg2+ and ADP-MgF3, comprising a mimetic of the pentacoordinate transition state of the phosphohydrolase reaction (Fig. 2A). We targeted seven candidate active site residues for alanine scanning, including the UvrD1 counterparts of: (i) those that contact the ATP adenine base (Gln24 in the “Q motif”), the ribose sugar (Glu609 in motif V), or the a phosphate (Arg83 in motif Ia); and (ii) those that coordinate the nonbridging γ phosphate oxygens, the attacking water nucleophile, or the bridging β-γ oxygen leaving group (Glu236 in motif II, Gln275 in motif III, Arg308 in motif IV, Arg648 in motif VI) (Fig. 2A). We also targeted the UvrD1 equivalents – Phe72 in motif Ia, Phe280 in motif III, and Trp664 in motif VIa – of three aromatic amino acids that directly contact the nucleosides of the 3’ single-stranded tail of the “loading strand” of the helicase substrate (Fig. 2B). This is the nucleic acid segment to which the UvrD/PcrA-type helicases initially bind and along which they translocate in response to ATP hydrolysis (1). Wild-type UvrD1 and the ten UvrD1-Ala mutants were produced in E. coli and purified from soluble bacterial extracts by serial affinity and ion-exchange column chromatography steps to achieve comparable enrichments for each of the enzyme preparations (Fig. 3A).
Fig. 2. ATPase active site and DNA contacts.
(A) Stereo view of the phosphohydrolase active site of DNA-bound E. coli UvrD (PDB 2IS6) as a transition state mimetic in complex with Mg2+ (magenta sphere), ADP (stick model with gray carbons), and a trigonal planar MgF3 (depicted with the magnesium as a yellow sphere and the fluorines as green spheres). Waters are rendered as red spheres. The putative water nucleophile located apical to the leaving β-phosphate oxygen is indicated by an arrow. The atomic contacts of the conserved active site residues (stick models with beige carbons) are denoted by dashes lines. The residue numbers refer to the equivalent side chains in UvrD1 that were subjected to alanine scanning. (B) Stereo view of DNA contacts in the crystal structure of E. coli UvrD bound to a 3’ tailed duplex DNA (from PDB 2IS6). The 3’ tailed strand (the loading strand for initial helicase binding, on which the enzyme translocates 3’ to 5’) is depicted as a stick model with phosphorus atoms colored yellow. The complementary strand that forms the duplex segment (the displaced strand unwound by helicase translocation) is depicted with phosphorus atoms in green. Conserved aromatic residues that make base stacking and van der Waals contacts with the DNA (green dashed lines) are shown, with residue numbering referring to the equivalents in UvrD1.
Effect of active site mutations on ATPase and helicase activity
The specific activities of the wild-type and mutant UvrD1 protein in DNA-dependent ATP hydrolysis were determined by protein titration under conditions optimized previously for wild-type UvrD1 (Fig. 3B). The most severe mutational effects were elicited by alanine substitutions at Arg308, Arg648 and Gln275 – the very residues implicated in phosphohydrolase chemistry at the ATP γ phosphate (Fig. 1A). The specific activities of the R308A and R648A proteins were ≤0.05% of the wild-type value (this being the limit of sensitivity of our assays), whereas Q275A was 0.3% as active as wild-type UvrD1 (Fig. 3B). (These proteins were scored as “ATPase –” in Table I, which summarizes the mutational results.) Alanine in lieu of Glu236 (which contacts the nucleophilic water and a water in the octahedral magnesium coordination complex) reduced specific activity to 7% of wild-type (scored as “one-plus” activity in Table I, where wild-type activity is “three-plus”). By contrast, alanine substitutions at several residues that interact with the adenosine nucleoside had more benign effects on ATPase activity, ranging in magnitude from minimal (E609A, +++) to modest (R83A, Q24A, scored as ++ in Table I) (Fig. 3B).
The helicase activity of the UvrD1 proteins was assayed in the presence of Ku and ATP using a 3’-tailed duplex substrate consisting of a 24-bp duplex with a 3’ T20 tail on the loading strand and a 5’ 32P-label on the displaced strand (Fig. 3C). The assay format entails preincubation of 110 nM UvrD1, 100 nM Ku, and 50 nM labeled DNA, followed by initiation of unwinding by addition of 1 mM ATP, with simultaneous addition of a “trap” of excess unlabeled displaced strand that: (i) minimizes reannealing of any labeled 24-mer that was unwound by UvrD1 and (ii) competes with the loading strand for binding to any free UvrD1 or UvrD1 that dissociated from the labeled DNA without unwinding it. Consequently, the assay predominantly gauges a single round of strand displacement by UvrD1 bound to the labeled duplex prior to the onset of ATP hydrolysis. As noted previously (7), the combination of Ku and wild-type UvrD1 unwound the DNA to yield a radiolabeled free single strand that migrated faster than the input tailed duplex during native PAGE; the helicase reaction product comigrated with free 24-mer generated by thermal denaturation of the substrate (Fig. 3C). As expected, helicase activity was suppressed by the four alanine mutations that ablated or severely reduced ATP hydrolysis: R308A, R648A, Q275A, and E236A, (Fig. 3C; scored as “helicase –” in Table I). The R83A mutant, which retained substantial ATPase activity, also retained near wild-type helicase activity, whereas Q24A, which was less active in ATP hydrolysis, displayed concordantly reduced strand displacement activity (Fig. 3C). The remarkable finding was that the E609A mutant was as defective in duplex unwinding as any of the “deadest” ATPase-defective mutants (Fig. 3C), notwithstanding that E609A itself retained near wild-type DNA-dependent ATPase activity. This result could signify that Glu609 is required to couple ATP hydrolysis to the work-performing motor activity of UvrD1.
Effect of active site mutations on DNA binding
The ability of the UvrD1-Ala mutants to bind to the helicase substrate was gauged by native gel electrophoresis (Fig. 3D). As noted previously, wild-type UvrD1 efficiently formed a discrete protein-DNA complex of retarded electrophoretic mobility. UvrD1-DNA complexes were formed in good yield by the R83A and Q24A mutants that retained helicase activity (scored as +++ DNA binding in Table I). The instructive findings concerned DNA binding by the other active site Ala-mutants, as follows. First, the formation of a discrete UvrD1-DNA complex was unperturbed by the R308A and R648A changes that abolished ATPase and helicase activity, implying that these two arginines are direct catalysts of ATP hydrolysis by DNA-bound UvrD1. Second, the Q275A and E236A changes diminished UvrD1 binding to the helicase substrate (Fig. 3D). The reduction in DNA binding by Q275A was partial (scored as ++ DNA binding in Table I), and by no means commensurate with its ablation of ATP hydrolysis, which we presume is the dominant factor in the loss of helicase activity. It is conceivable that Q275A exerts an effect on DNA binding via local changes in motif III, especially affecting neighboring DNA-binding residues such as Tyr278 and Phe280 (Fig. 2B). The more pronounced negative effect of the E236A mutation on UvrD1 binding to the helicase substrate (scored as “↓↓” binding in Table I) precludes simple attribution of the E236A unwinding defect to a decrement in ATP hydrolysis. (Note that the loss of the vicinal Asp235 motif II side chain had no apparent effect on DNA binding in this assay (7); Table I.) The UvrD/PcrA structures suggest a possible connection between the conserved motif II glutamate and DNA binding, whereby the glutamate forms a salt bridge from Oε1 (the carboxylate oxygen not engaged to Mg2+·ATP) to a conserved motif V lysine (Lys563 in UvrD; Lys606 in UvrD1, Lys568 in PcrA). This contact appears to help tether a short protein α-helix near the DNA 3’ tail, from which a conserved motif V histidine (His560 in UvrD, His603 in UvrD1, His565 in PcrA) makes direct contact with the DNA and also donates a hydrogen bond to a main chain carbonyl in the DNA-binding segment of motif III (3,4).
The third and most informative finding from gel-shift assays of DNA binding by the active site mutants was that the E609A change had no apparent impact on stable UvrD1 binding to the helicase substrate (Fig. 3D). This signifies that Glu609 does indeed couple ATP hydrolysis to mechanical work.
As another means of assessing effects on DNA binding, we measured the extent of ATP hydrolysis as a function of increasing concentration of the input DNA cofactor. (This assay was only applicable to UvrD1 active site mutants that retained appreciable ATPase activity under standard assay conditions using 5 µg/ml salmon sperm DNA.) Wild-type UvrD1 ATPase displayed a hyperbolic dependence on DNA with an apparent Km of 0.16 µg/ml and Vmax of 170 s−1. The values for Ala-mutants were as follows: E609A (0.24 µg/ml, 170 s−1); R83A (0.34 µg/ml, 62 s−1); Q24A (0.55 µg/ml, 45 s−1); E236A (1.7 µg/ml, 8 s−1). These results fortify the inferences from the ATPase enzyme titration experiments and native gel DNA binding assays that: (i) Glu609 is not important for ATP hydrolysis or DNA binding, (ii) Arg83 and Gln24 enhance ATP hydrolysis 3- to 4-fold, and impact modestly on DNA binding, and (iii) Glu236 assists both ATP hydrolysis and DNA binding, e.g., the Km of E236A for sperm DNA was 10-fold higher than that of wild-type UvrD1.
Effects of conservative mutations in the ATPase active site
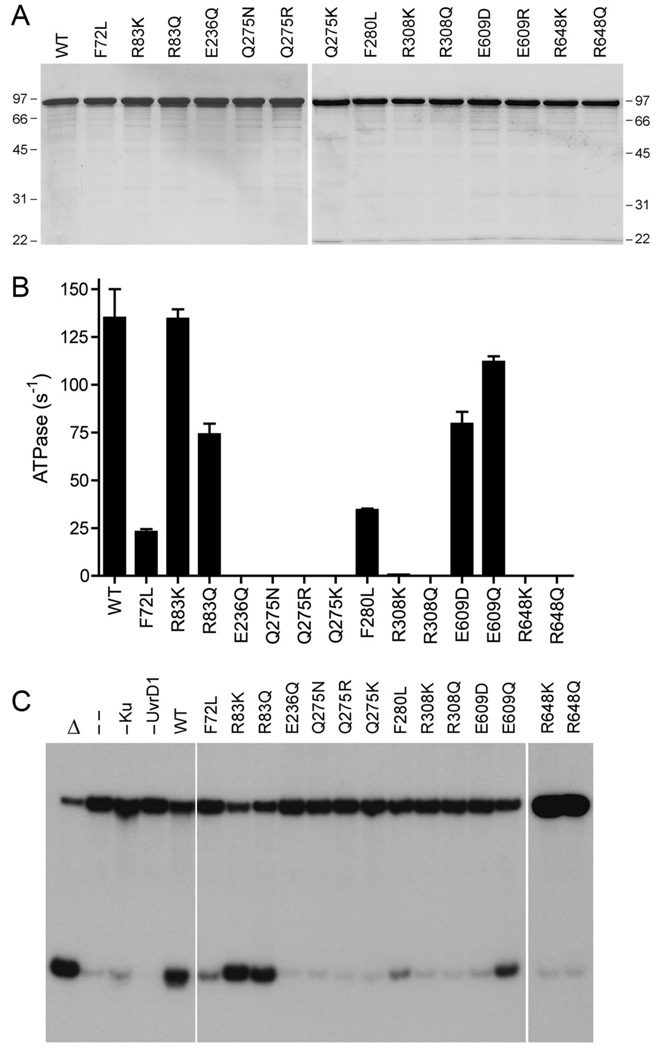
Structure-activity relationships for selected active site residues were gleaned by studying the effects of conservative amino acid substitutions. Arg83, Arg308 and Arg648 were replaced by lysine and glutamine; Glu236 was changed to glutamine; Glu609 was replaced by glutamine and aspartate; Gln275 was mutated to asparagine, lysine, and arginine. The conservative active site mutants were purified (Fig. 4A) and surveyed for DNA-dependent ATPase (Fig. 4B) and DNA unwinding (Fig. 4C) activities. We found that Arg648 was strictly essential, insofar as neither lysine nor glutamine restored ATPase or helicase activity vis a vis the catalytically defective R648A mutant (Fig. 4). Similar findings pertained to Arg308 (Fig. 4). We surmise that positive charge is not sufficient and that ATP hydrolysis requires the multidentate contacts of Arg308 and Arg648 with the ATP phosphate oxygens, especially in stabilizing the extra negative charge developed on the γ phosphate in the predicted associative transition state (Fig. 2A). Different structure-activity relations were seen at Arg83, whereby the modest defect in ATP hydrolysis by the R83A mutant was mimicked by the glutamine substitution, while activity was improved to wild-type level when lysine was introduced (Fig. 4B). We surmise that the positive charge on motif Ia residue Arg83 is the relevant factor in its helpful contact with the ATP α phosphate (Fig. 2A). As expected, the R83K and R83Q proteins retained DNA unwinding activity (Fig. 4C).
Fig. 4. Effects of conservative mutations on UvrD1 activities.
(A) Aliquots (3 µg) of recombinant wild-type (WT) UvrD1 and the indicated mutants were analyzed by SDS-PAGE. The Coomassie blue-stained gels are shown. The sizes (kDa) and positions of marker proteins are indicated. (B) ATPase specific activity was determined as specified under Methods. Each datum is the average of three separate UvrD1 titration experiments; error bars denote the standard deviation. (C) Helicase reactions were performed as described under Methods. Complete reaction mixtures contained 1 mM ATP, 50 nM 32P-labeled tailed duplex DNA substrate, 75 ng Ku and 100 ng of wild-type or mutant UvrD1 protein as specified. The products were analyzed by native PAGE and visualized by autoradiography. Reactions without added protein (lane --), with wild-type UvrD1 only (–Ku) or with Ku only (–UvrD1) were included as controls. A reaction lacking protein that was heat denatured prior to PAGE is shown in lane Δ.
Replacing Glu236 with glutamine effectively abolished ATPase activity (E236Q specific activity was ≤0.05% of the wild-type value) and was more deleterious in this regard than the E236A change. Mutation of Gln275 to asparagine elicited as severe a decrement in ATP hydrolysis as the alanine substitution, signifying that a critical distance from the main-chain to the amide functional group is required to achieve the contacts with the water nucleophile and the γ phosphate seen in the UvrD crystal structure (Fig. 2A). Lysine and arginine were equally damaging in lieu of Gln275 (Fig. 4B). These results underscored the strict reliance for ATP hydrolysis on the conserved motif III Gln and motif II Glu residues that together orient (and activate) the attacking water. As expected, the ATPase-defective Gln275 mutants were also defective for DNA unwinding (Fig. 4C).
Finally, we found that replacing Glu609 with either aspartate or glutamine preserved ATPase activity (Fig. 4B), as noted for the E609A mutant. Whereas E609D had no salutary effect on DNA unwinding, E609Q partially restored helicase activity (Fig. 4C). We infer that coupling of UvrD1 ATP hydrolysis to duplex unwinding depends on a critical distance from the main chain to the Glu609 carboxylate that allows the hydrogen bonds to the ATP ribose O3’ seen in the UvrD crystal structure (Fig. 2A). Glutamine, a bivalent hydrogen bond donor/acceptor, could in principle engage in the same ribose contacts.
The Q motif serves as an adenine nucleotide specificity filter in UvrD1
The Gln24 residue of UvrD1 comprises the “Q motif” located 21- to 24-aa upstream of the motif I lysine in a wide variety of nucleic acid-dependent NTP phosphohydrolases (15,16), including SF1 helicases UvrD, PcrA and Rep (2–4), the DExH-box protein UvrB (17), the DEAD-box RNA helicases eIF4A, Ded1, Dhh1, and Hera (15,16,18–20), and many others. In the adenine nucleotide-bound crystal structures of UvrD (4), UvrB (17) and Hera (20), the Q motif glutamine makes bidentate hydrogen bonds from Gln-Nε to the adenine N7 atom and from Gln-Oε to the adenine N6 atom (Fig. 2A). The Q motif glutamine has been touted as having a “regulatory” function in ATP hydrolysis and nucleic acid binding (15,16), but there is as yet no coherent view of what is being regulated and how, given that alanine mutations of the Q motif glutamine exert very strong suppressive effects on the phosphohydrolase activities of several different ATPase enzymes, including eIF4A (15), Ded1 (16) and reverse gyrase (21). Such findings suggest a simple requirement for the Q motif glutamine for avid binding of ATP (and hence vigorous hydrolysis), in keeping with the crystallographic contacts. It has been speculated that the Q motif glutamine might enforce specificity for adenine nucleotides (15), but there is scant evidence in support of this idea.
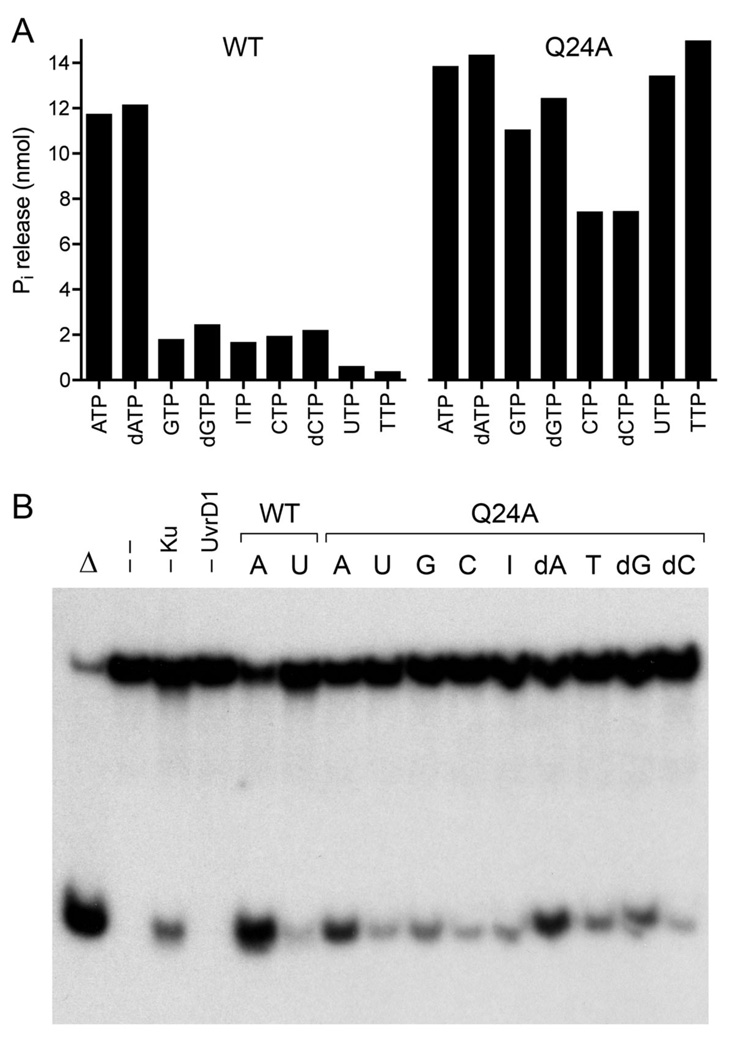
In light of our initial findings that the UvrD1 Q24A mutant retained substantial DNA-dependent ATPase activity (one fourth that of wild-type), we viewed UvrD1 as a promising model to test the regulatory potential of the Q motif. Mycobacterial UvrD1 specifically utilizes adenosine nucleotides for the phosphohydrolase and helicase reactions (7,9). Wild-type UvrD1 readily hydrolyzed either ATP or dATP in the presence of salmon sperm DNA; other rNTPs and dNTPs were hydrolyzed poorly (Fig. 5A). By contrast, the Q24A mutant was able to hydrolyze all rNTP and dNTPs (Fig. 5A). Thus, the loss of the glutamine contacts to the adenine base elicited a gain-of-function, e.g., whereby UTP and TTP (the worst substrates for wild-type UvrD1) were now on a par with ATP and dATP as substrates for Q24A.
Fig. 5. Role of the Q motif (Gln24) in NTP substrate specifity.
(A) Phosphohydrolase reaction mixtures contained 1 mM of the indicted ribonucleoside or deoxyribonucleoside triphosphate and either 50 ng wild-type UvrD1 or 200 ng of the Q24A mutant. The extents of phosphate production are shown. (B) Complete helicase reaction mixtures contained 50 nM 32P-labeled tailed duplex DNA substrate, 75 ng Ku, 100 ng of wild-type UvrD1 or Q24A, and 1 mM of the indicated NTP or dNTP. The products were analyzed by native PAGE and visualized by autoradiography. Reactions without added protein (lane --), with wild-type UvrD1 only (–Ku) or with Ku only (–UvrD1) were included as controls. A reaction mixture lacking protein that was heat denatured prior to PAGE is shown in lane Δ.
These inferences from single-point assays were fortified by nucleotide titration experiments. The steady-state kinetic parameters of wild-type UvrD1 for ATP hydrolysis were: Km 0.26 ± 0.04 mM ATP, kcat 265 ± 22 s−1. The values for the Q24A mutant were: Km 0.69 ± 0.05 mM ATP, kcat 95 ± 3 s−1. Thus, the Q motif glutamine enhances affinity of UvrD1 for ATP by a mere 3-fold, with a similar contribution to the rate of catalysis. The steady state parameters for UTP hydrolysis by Q24A were: Km 1.21 ± 0.13 mM UTP, kcat 86 ± 5 s−1. There was scant difference in the binding or hydrolysis of ATP versus UTP by the Q24A enzyme. These results provide evidence of a regulatory role for the Q motif as a substrate specificity filter.
Assays of helicase activity suggested an additional level of control by the Q motif. Wild-type UvrD1 unwound the tailed duplex in the presence of ATP, but not UTP (Fig. 5B). Q24A again had lower than wild-type strand displacement activity in the presence of ATP or dATP (Fig. 5B). The instructive findings were that Q24A retained its specificity for adenosine nucleotides in the helicase reaction, notwithstanding its relaxed nucleotide specificity in the phosphohydrolase reaction. In particular, Q24A, like wild-type UvrD1, was ineffective as a helicase in the presence of UTP (Fig. 5B). We infer that the Q motif glutamine plays a role in coupling NTP hydrolysis to mechanical work.
Effects of mutations of putative DNA-binding residues Phe72, Phe280 and Trp664
Alanine mutations were introduced at the UvrD1 counterparts of aromatic amino acids in UvrD/PcrA that contact the nucleosides of the 3’ tail of the loading strand (Fig. 2B). The recombinant F72A, F280A, and W664A proteins (Fig. 3A) exerted disparate effects on ATP hydrolysis, ranging from comparatively severe (F280A) to modest (F72A) to none (W664A) (Fig. 3B, Table I). We measured the extent of ATP hydrolysis by W664A as a function of increasing concentrations of DNA, as described above for the active site mutants, and observed an apparent Km of 0.47 µg/ml salmon sperm DNA and a Vmax of 180 s−1 (values quite similar to wild-type UvrD1 in the same assay format). W664A was able to unwind the 3’ tailed duplex (Fig. 3C) and formed a stable complex with the helicase substrate (Fig. 3D). We surmise that the putative contacts of Trp664 with the loading strand near the single-strand/duplex junction are not crucial for any of the UvrD1 activities tested. Alternatively, it is possible that UvrD1 Trp664 does not contact the DNA in the manner surmised from the E. coli UvrD crystal structure.
The F280A mutant hydrolyzed ATP with an apparent Km of 2.3 µg/ml salmon sperm DNA (15-fold higher than wild-type UvrD1) and a Vmax of 4 s−1. F280A was defective in duplex unwinding (Fig. 3C) and failed to form a stable complex with the helicase substrate in the native gel-shift assay (Fig. 3D). The aromatic motif III residue equivalent to Phe280 in UvrD1/PcrA (Fig. 1; Table I) forms a π stack on one of the bases of the loading strand (Fig. 2B). A key tenet of the translocation mechanism proposed for SF1 helicases (1,3) entails ATP-driven flipping of the single-stranded bases of the loading strand between sandwiching aromatic residues, especially those of motif III. Replacing UvrD1 Phe280 with leucine resulted in a partial gain of function in ATP hydrolysis compared to the Ala-mutant (to about one-fourth of wild-type specific activity; Fig. 4B) that was accompanied by a slight recovery of DNA unwinding activity (Fig. 4C).
F72A hydrolyzed ATP with an apparent Km of 0.23 µg/ml salmon sperm DNA and a Vmax of 33 s−1. F72A formed a stable complex with the helicase substrate (Fig. 3D), yet was defective in duplex unwinding (Fig. 3C). The F72A helicase defect was apparently more pronounced than the decrement in ATPase activity (e.g., F72A and Q24A had similar activities in ATP hydrolysis and DNA binding, yet only Q24A had partial unwinding activity), which suggests that Phe72 aids in coupling ATP consumption to motor function. Replacing Phe72 with leucine afforded no improvement in ATPase activity compared to F72A (Fig. 4B), but did allow a trace amount of DNA unwinding (Fig. 4C).
Effects of C-terminal deletions of UvrD1
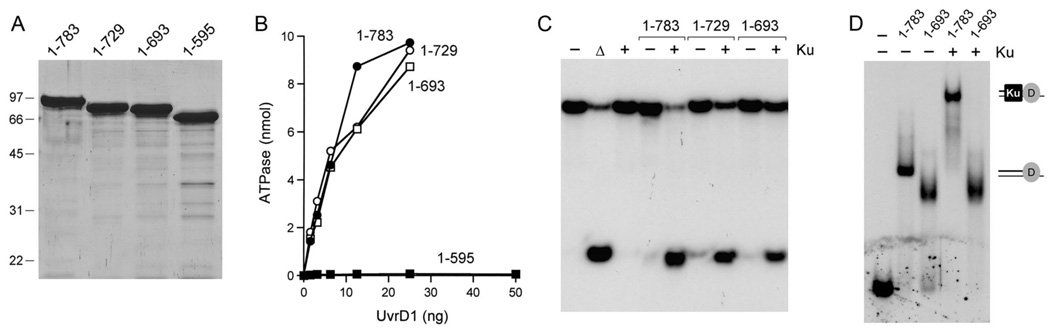
The C-terminal domains (∼70-aa) of UvrD and PcrA are either disordered or missing in the crystal structures of the respective DNA-bound enzymes (3,4). In the case of E. coli UvrD, deletion of 102-aa from the C-terminus ablated all its biochemical activities, whereas a 40-aa C-terminal truncation did not affect the ATPase and helicase functions (27). Here we studied the effects of three incremental C-terminal deletions on the activities of mycobacterial UvrD1. The full-length UvrD1-(1–783) protein was compared to C-terminal truncation mutants UvrD1-(1–729), UvrD1-(1–693) and UvrD1-(1–595). SDS-PAGE verified the anticipated decrements in the apparent sizes of the truncated polypeptides (Fig. 6A). UvrD1-(1–729) and UvrD1-(1–693) were as active as full-length UvrD1 in DNA-dependent ATP hydrolysis (Fig. 6B). By contrast, UvrD1-(1–595) was virtually catalytically inert (Fig. 6B); this was expected, given that this truncation mutant lacks the essential active site Arg648 residue of motif VI (Fig. 1). Although, the UvrD1-(1–729) and UvrD1-(1–693) proteins retained strand displacement activity in the presence of Ku, the extents of unwinding in the single-turnover helicase assay decreased progressively with each deletion increment (Fig. 6C, reflected in the higher levels of residual unwound duplex). The full-length UvrD1-(1–783) and truncated UvrD1-(1–693) proteins were equally adept at binding stably to the 3’-tailed helicase substrate in the native gel-shift assay (Fig. 6D). The binary DNA-UvrD1-(1–693) complex migrated more rapidly than that of the full-length enzyme, consistent with the reduced mass of the truncated UvrD1 protein. The instructive finding was that whereas full-length UvrD1-(1–783) formed a stable supershifted DNA-UvrD1-Ku ternary complex when Ku was included in the binding reaction, UvrD1-(1–693) did not (Fig. 6D). UvrD1-(1–729) behaved similarly to UvrD1-(1–693) in the DNA binding assay (not shown). We surmise that that the C-terminus of UvrD1 enhances its stable interaction with Ku, although it is not strictly required for functional Ku-UvrD1 interactions, insofar as UvrD1-(1–693) helicase activity is still responsive to Ku stimulation.
Fig. 6. Effects of C-terminal deletions.
(A) Aliquots (5 µg) of full-length UvrD1-(1–783) and the C-terminal truncation mutants UvrD1-(1–729), UvrD1-(1–693) and UvrD1-(1–595) were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The sizes (kDa) and positions of marker proteins are indicated on the left. (B) ATPase assays were performed as described in Methods. 32Pi release from 1 mM [γ32P]ATP is plotted as a function of input UvrD1 for each enzyme assayed. (C) Complete helicase reaction mixtures contained 1 mM ATP, 50 nM 32P-labeled tailed duplex DNA substrate, 75 ng Ku (where indicated by +) and 100 ng UvrD1 protein as specified. The products were analyzed by native PAGE and visualized by autoradiography. A reaction lacking protein that was heat-denatured prior to PAGE is shown in lane Δ. (D) DNA binding reaction mixtures contained 0.5 pmol 32P-labeled 3’-tailed DNA, 100 ng UvrD1-(1–783) or UvrD1-(1–693), and 75 ng Ku (where indicated by +). The free DNA and protein-DNA complexes (depicted on the right) were resolved by native PAGE and visualized by autoradiography.
Influence of 3’ tail length on DNA unwinding by UvrD1
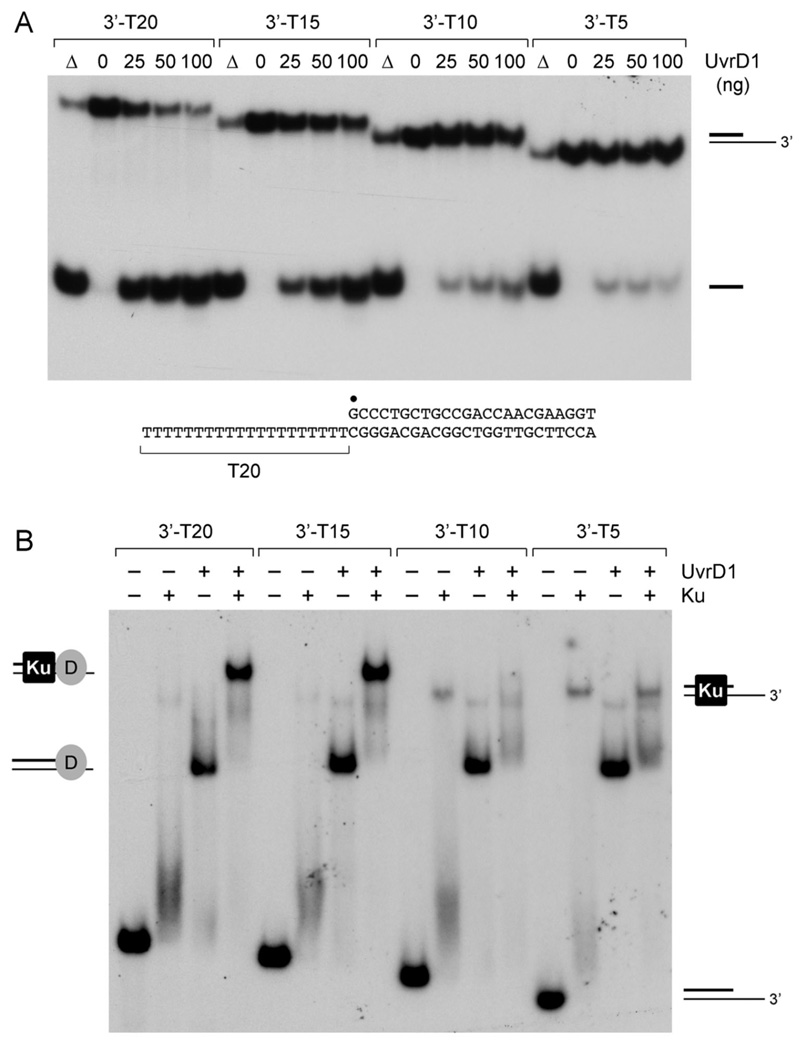
In the UvrD/PcrA structures, the helicase contacts a five-nucleotide segment of the 3’ single-stranded tail of the loading DNA strand and a 14–16 bp duplex DNA segment (2,3). To probe the 3’-tail requirement for duplex unwinding by mycobacterial UvrD1, we tested a series of helicase substrates with 3’-oligo(dT) tails of varying length (20, 15, 10, or 5 nucleotides) attached to an identical 24-bp duplex segment. We found that UvrD1-mediated displacement of the radiolabeled 24-mer strand in the presence of Ku was optimal when the 3’ tail length was 20 nucleotides (Fig. 7A). Helicase activity decreased slightly when the 3’ tail was retracted to 15 nucleotides, and then declined acutely when the tail was shortened to only 10 or 5 nucleotides. Analysis of UvrD1 binding to the helicase substrates by native gel electrophoresis revealed efficient formation of stable DNA-UvrD1 binary complexes whether the tail was 20, 15, 10 or 5 nucleotides (Fig. 7B). As noted above, inclusion of Ku in the binding reaction mixture resulted in the formation of a stable supershifted DNA-Ku-UvrD1 ternary complex (Fig. 7B). By contrast, the binding of Ku alone to the tailed duplex was metastable, yielding scant amounts of a slowly migrating Ku-DNA binary complex (indicated at right in Fig. 7B), plus a diffuse smear of radiolabeled material migrating just above the free DNA. As suggested previously (7), the Ku-DNA binary complex is prone to dissociate during electrophoresis as the ringshaped Ku dimer (28) slides off the short duplex segment of the DNA ligand. The pertinent findings here were that UvrD1 formed a discrete supershifted ternary complex with Ku when the 3’ single-stranded tail was shortened to 15 nucleotides, but not when the tail was further recessed to 10 or 5 nucleotides (Fig. 7B). In the latter two cases, we see that Ku converted the discrete DNA-UvrD1 binary complexes to a diffuse species migrating just slower than the binary complex. We surmise that the ternary complex is less stable when the 3’ tail is 10 or 5 nucleotides. Therefore, the decrement in duplex unwinding upon shortening the 3’ tail is explained by an attenuation of Ku stimulation, not a primary effect on UvrD1 binding to the helicase substrate. We speculate that UvrD1 might bind closer to the duplex segment of the DNA when the 3’ single-strand tail is shortened, thus providing less room for Ku to embrace the duplex segment.
Fig. 7. Effect of 3’ tail length on helicase activity and DNA binding.
(A) Helicase reaction mixtures contained 50 nM duplex DNA substrate with 3’-T20, 3’-T15, 3’-T10 or 3’-T5 tails, 1 mM ATP, 75 ng Ku, and 0, 25, 50 or 100 ng UvrD1. The products were analyzed by native PAGE and visualized by autoradiography. A control reaction lacking enzyme that was heat denatured prior to PAGE is shown in lane Δ. The 3’-T20 substrate is shown at bottom with the 5’ 32P-label denoted by •. (B) DNA binding reaction mixtures contained 0.5 pmol 32P-labeled 3’-tailed DNAs as specified, 100 ng UvrD1 (where indicated by +), and 75 ng Ku (where indicated by +). The products were analyzed by native gel electrophoresis. The free DNAs and various protein-DNA complexes (depicted at left and right) were visualized by autoradiography.
DISCUSSION
The present structure-guided mutational analysis of UvrD1 contributes to our understanding of SF1 motor proteins in the following respects: (i) by demonstrating the essentiality and structure-activity relationships of catalytic residues in motifs II, III, IV, and VI that support the phosphohydrolase transition state; (ii) by underscoring the importance of motif III aromatic contacts with the 3’ tail of the loading strand; (iii) by verifying the role of the conserved motif Ia phenylalanine in coupling ATP hydrolysis to strand displacement; (iv) by identifying a conserved motif V glutamate as a novel coupler of ATP hydrolysis to motor activity; and (v) by providing new insights to the role of the Q motif as a substrate specificity filter.
The effects of UvrD1 single-alanine mutations on ATPase, helicase and DNA binding functions are summarized in Table I (with +++ indicating wild-type or near wild-type level of activity in the assays applied). The corresponding amino acids of UvrD and PcrA are listed in Table I, along with a summary of alanine scanning data for eight of the equivalent residues of PcrA from the Wigley lab (11–13). The UvrD1 and PcrA results are in good accord, and generally agree with available mutational data for E. coli UvrD from the Matson lab (21–24). Together, the results testify to a common structural basis for ATP hydrolysis and motor function among homologous SF1 proteins from widely different bacterial phyla.
To our knowledge, the conserved motif Ia arginine and motif V glutamate of an SF1 helicase have not been subjected to mutational analysis. We found that loss of the putative contacts of motif Ia Arg83 with the ATP α phosphate (Fig. 2A) had relatively little impact on ATP hydrolysis or duplex unwinding, e.g., compared to mutations of the two arginines that coordinate the γ phosphate. This could reflect the unimportance of contacts at a phosphate that does not participate directly in reaction chemistry. Alternatively, UvrD1 might rely on contacts with the α phosphate, but this requirement is fulfilled (in large part) by Arg308. The Arg308 equivalent coordinates all three ATP phosphates in the UvrD structure and is probably the key agent in attaining a proper conformation of the triphosphate bridge for catalysis.
Motif V Glu609 fulfills the criteria for a chemo-mechanical coupling determinant. Alanine substitution at Glu609 eliminated duplex unwinding while having virtually no impact on ATP hydrolysis or UvrD1 binding to the helicase substrate. Uncoupling mutations have been documented previously for several exemplary SF1 and SF2 helicases, including PcrA (12,13,25) and the RNA helicase NPH2 (26). The uncoupling mutations typically involve residues that either: (i) bind the nucleic acid substrate, or (ii) trigger or transmit protein conformational changes that drive protein movement and strand separation. Glu609 is a remarkable instance of a coupling residue that makes direct contact with the adenosine ribose.
The Q motif glutamine plays a dual role in dictating phosphohydrolase substrate specificity and in coupling unwinding to the hydrolysis of adenosine nucleoside triphosphates. We provide clear evidence of a gain-of-function for the UvrD1 Q24A mutant as a DNA-dependent consumer of nonadenosine NTPs and dNTPs. Loss of the glutamine reduces ATP affinity only modestly; the Q24A enzyme binds and hydrolyzes ATP and UTP with comparable efficacy. Thus, for UvrD1, the bidentate contacts of the glutamine with the adenine base act as a gatekeeper to exclude nonadenosine nucleotides. This contrasts sharply with the effects of analogous Q-to-A mutations in other Q-motif proteins, e.g., the Km for ATP of the Q169A mutant of the RNA-dependent ATPase Ded1 is increased by two orders of magnitude (to 33 mM ATP) compared to wild-type Ded1, to an extent that Ded1-Q169A is inactive at physiological ATP concentrations (16). It is conceivable that UvrD1 tolerates the Q24A mutation because it relies on other functional groups to interact with the nucleoside base. The likely candidate is Tyr308, a conserved motif IV residue (Fig. 1) that forms a π stack under the adenine ring (Fig. 2A). [Preliminary characterization of a UvrD1 Y307A mutant revealed that it had 50-fold lower ATPase specific activity than wild-type UvrD1 (K.M.S and S.S., unpublished).]
The finding that Q24A still relies on ATP to promote its duplex unwinding activity, and is not activated in this regard by UTP, which it readily hydrolyses, underscores the theme that adenosine contacts are relevant to chemomechanical coupling, be it to the ribose (via Glu609) or adenine (via the Q motif). A thorough account of the coupling roles of the motif V glutamate and Q motif glutamine residues will ultimately hinge on obtaining crystal structures of UvrD1 bound to its nucleic acid and nucleotide substrates (with and without Ku) at discrete functional states along the reaction pathway. Investigators interested in other SF1 DNA helicases might be prompted by the present results to evaluate the roles of these residues in their favorite enzyme, keeping in mind that functional inferences from mutational effects are not always portable from one helicase to another. This appears to be the case for the Q motif, as discussed above and underscored by recent mutational studies of a squadron of putative yeast DExD/Hbox helicases involved in ribosomal subunit biogenesis – in which the Q motif glutamine was found to be unconditionally essential for helicase function in vivo in only three of the nine proteins subjected to glutamine mutation (29,30). Indeed, even among the structurally defined SF1 helicases UvrD and PcrA that have a Q motif glutamine, there are major differences in NTP specificity – whereby UvrD is strongly selective for ATP/dATP while PcrA utilizes ATP, dATP, CTP, GTP, dGTP, and etheno-ATP with equal facility (31,32) – that hint that the mere presence of the glutamine does not guarantee its relevance as a substrate filter.
Acknowledgments
† This research was supported by grant AI64693 from the U.S. National Institutes of Health.
Abbreviations
- EDTA
ethylenediaminetetraacetic acid
- DTT
dithiothreitol
- TLC
thin layer chromatography
- ATPase
adenosine triphosphatase
- UTPase
uridine triphosphatase
- PCR
polymerase chain reaction
- SF1
superfamily 1
- SF2
superfamily 2.
REFERENCES
- 1.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 2.Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 3.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structure of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;87:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 4.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dillingham MS, Wigley DB, Webb MR. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry. 2000;39:205–212. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- 6.Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM. A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol. Cell. 2007;26:335–347. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinha KM, Stephanou NC, Gao F, Glickman MS, Shuman S. Mycobacterial UvrD1 is a Ku-dependent DNA helicase that plays a role in multiple DNA repair events, including double-strand break repair. J. Biol. Chem. 2007;282:15114–15125. doi: 10.1074/jbc.M701167200. [DOI] [PubMed] [Google Scholar]
- 8.Sinha KM, Stephanou NC, Unciuleac MC, Glickman MS, Shuman S. Domain requirements for DNA unwinding by mycobacterial UvrD2, an essential DNA helicase. Biochemistry. 2008;47:9355–9364. doi: 10.1021/bi800725q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curti E, Smerdon SJ, Davis EO. Characterization of the helicase activity and substrate specificity of Mycobacterium tuberculosis UvrD. J. Bacteriol. 2007;189:1542–1555. doi: 10.1128/JB.01421-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aniukwu J, Glickman MS, Shuman S. The pathways and outcomes of mycobacterial NHEJ depend on the structure of the broken DNA ends. Genes Dev. 2008;22:512–527. doi: 10.1101/gad.1631908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soultanas P, Dillingham MS, Velankar SS, Wigley DB. DNA binding mediates conformational changes and metal ion coordination in the active site of PcrA helicase. J. Mol. Biol. 1999;290:137–148. doi: 10.1006/jmbi.1999.2873. [DOI] [PubMed] [Google Scholar]
- 12.Dillingham MS, Soultanas P, Wigley DB. Site-directed mutagenesis of motif III in PcrA helicase reveals a role in coupling ATP hydrolysis to strand separation. Nucleic Acids Res. 1999;27:3310–3317. doi: 10.1093/nar/27.16.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dillingham MS, Soultanas P, Wiley P, Webb MR, Wigley DB. Defining the roles of individual residues in the single-stranded DNA binding site of PcrA helicase. Proc. Natl. Acad. Sci USA. 2001;98:8381–8387. doi: 10.1073/pnas.131009598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gross CH, Shuman S. Mutational analysis of vaccinia virus nucleoside triphosphate phosphohydrolase-II, a DExH box RNA helicase. J. Virol. 1995;69:4727–4736. doi: 10.1128/jvi.69.8.4727-4736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanner NK, Cordin O, Banroques J, Doere M, Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 16.Cordin O, Tanner NK, Moere M, Linder P, Banroques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 2004;23:2478–2487. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Theis K, Chen PJ, Skorvaga M, Van Houten B, Kisker C. Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. EMBO J. 1999;18:6899–6907. doi: 10.1093/emboj/18.24.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benz J, Trachsel H, Baumann U. Crystal structure of the ATPase domain of translation initiation factor 4A from Saccharomyces cerevisiae – the prototype of the DEAD box protein family. Structure. 1999;7:671–679. doi: 10.1016/s0969-2126(99)80088-4. [DOI] [PubMed] [Google Scholar]
- 19.Cheng Z, Coller J, Parker R, Song H. Crystal structure and functional analysis of the DEAD-box protein Dhh1p. RNA. 2005;11:1258–1270. doi: 10.1261/rna.2920905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudolph MG, Heissmann R, Wittmann JG, Klostermeier D. Crystal structure and nucleotide binding of the Thermus thermophilus RNA helicase Hera. J. Mol. Biol. 2006;361:731–743. doi: 10.1016/j.jmb.2006.06.065. [DOI] [PubMed] [Google Scholar]
- 21.Brosh RM, Matson SW. Mutations in motif II of Escherichia coli helicase II render the enzyme nonfunctional in both mismatch repair and excision repair with different effects on the unwinding reaction. J. Bacteriol. 1995;177:5612–5621. doi: 10.1128/jb.177.19.5612-5621.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brosh RM, Matson SW. A point mutation in Escherichia coli helicase II renders the enzyme nonfunctional in two DNA repair pathways. J. Biol Chem. 1997;272:572–579. doi: 10.1074/jbc.272.1.572. [DOI] [PubMed] [Google Scholar]
- 23.Hall MC, Matson SW. Mutations of a highly conserved arginine in motif IV of Escherichia coli DNA helicase II results in an ATP-binding defect. J. Biol. Chem. 1997;272:18614–18620. doi: 10.1074/jbc.272.30.18614. [DOI] [PubMed] [Google Scholar]
- 24.Hall MC, Özsoy AZ, Matson SW. Site-directed mutations in motif VI of Escherichia coli DNA helicase II result in multiple biochemical defects: evidence for the involvement of motif VI in the coupling of ATPase and DNA binding activities via conformational changes. J. Mol. Biol. 1998;277:257–271. doi: 10.1006/jmbi.1997.1614. [DOI] [PubMed] [Google Scholar]
- 25.Soultanas P, Dillingham MS, Wiley P, Webb MR, Wigley DB. Uncoupling DNA translocation and helicase activity in PcrA: direct evidence for an active mechanism. EMBO J. 2000;19:3799–3810. doi: 10.1093/emboj/19.14.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gross CH, Shuman S. The nucleoside triphosphatase and helicase activities of vaccinia virus NPH-II are essential for virus replication. J. Virol. 1998;72:4729–4736. doi: 10.1128/jvi.72.6.4729-4736.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mechanic LE, Latta ME, Matson SW. A region near the C-terminal end of Escherichia coli helicase II is required for single-stranded DNA binding. J. Bacteriol. 1999;181:2519–2526. doi: 10.1128/jb.181.8.2519-2526.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 29.Granneman S, Bernstein KA, Bleichert F, Baserga SJ. Comprehensive mutational analysis of yeast DEXD/H box RNA helicases required for small ribosomal subunit synthesis. Mol. Cell. Biol. 2006;26:1183–1194. doi: 10.1128/MCB.26.4.1183-1194.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernstein KA, Granneman S, Lee AV, Manickam S, Baserga SJ. Comprehensive mutational analysis of yeast DEXD/H box RNA helicases involved in large ribosomal subunit biogenesis. Mol. Cell. Biol. 2006;26:1195–1208. doi: 10.1128/MCB.26.4.1195-1208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matson SW, George JW. DNA helicase II of Escherichia coli: characterization of the single-stranded DNA-dependent NTPase and helicase activities. J. Biol. Chem. 1987;262:2066–2076. [PubMed] [Google Scholar]
- 32.Bird LE, Brannigan JA, Subramanya HS, Wigley DB. Characterisation of Bacillus stearothermophilus PcrA helicase: evidence against an active rolling mechanism. Nucleic Acids Res. 1998;26:2686–2693. doi: 10.1093/nar/26.11.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]