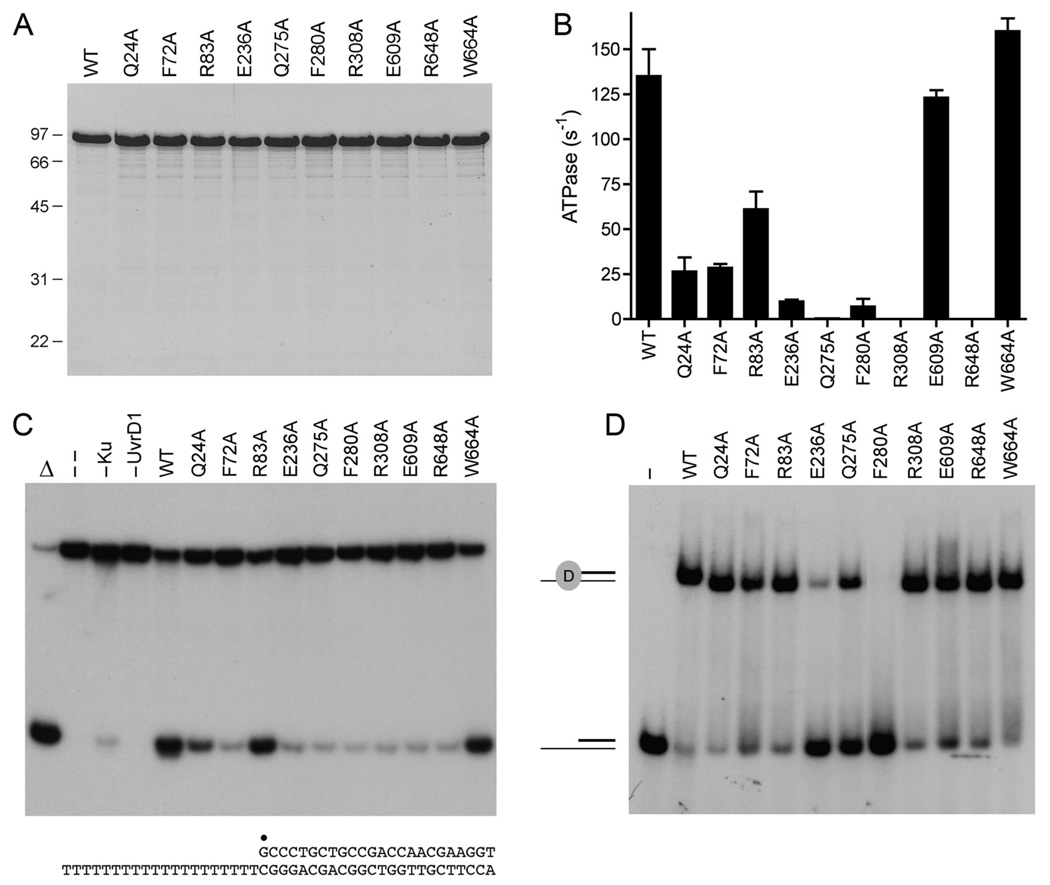
Fig. 3. Effects of alanine mutations on UvrD1 activities.
(A) Aliquots (5 µg) of recombinant wild-type (WT) UvrD1 and the indicated Ala mutants were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The sizes (kDa) and positions of marker proteins are indicated on the left. (B) ATPase specific activity was determined as specified under Methods. Each datum is the average of two separate UvrD1 titration experiments; error bars denote the mean absolute error. (C) Helicase reactions were performed as described under Methods. Complete reaction mixtures contained 1 mM ATP, 50 nM 32P-labeled tailed duplex DNA substrate, 75 ng Ku and 100 ng of wild-type or mutant UvrD1 protein as specified. The products were analyzed by native PAGE and visualized by autoradiography. Reactions without added protein (lane --), with wild-type UvrD1 only (–Ku) or with Ku only (–UvrD1) were included as controls. A reaction lacking protein that was heat denatured prior to PAGE is shown in lane Δ. The 3’-tailed duplex helicase substrate is shown at bottom with the 5’ 32P-label denoted by •. (D) Binding of UvrD1 to the helicase substrate. Binding reactions were performed as described under Methods. Complete reaction mixtures contained 1 pmol 32P-labeled tailed duplex DNA substrate and 100 ng wild-type or mutant UvrD1 protein as specified. UvrD1 was omitted from the control reaction in lane –. The products were analyzed by native PAGE and visualized by autoradiography. The positions of the free DNA and the UvrD1-DNA complex are indicated on the left.