Abstract
Background
Black patients with hemophilia A (factor VIII deficiency) are twice as likely as white patients to produce inhibitors against factor VIII proteins given as replacement therapy. There are six wild-type factor VIII proteins, designated H1 through H6, but only two (H1 and H2) match the recombinant factor VIII products used clinically. H1 and H2 are found in all racial groups and are the only factor VIII proteins found in the white population to date. H3, H4, and H5 have been found only in blacks. We hypothesized that mismatched factor VIII transfusions contribute to the high incidence of inhibitors among black patients.
Methods
We sequenced the factor VIII gene (F8) in black patients with hemophilia A to identify causative mutations and the background haplotypes on which they reside. Results from previous Bethesda assays and information on the baseline severity of hemophilia, age at enrollment, and biologic relationships among study patients were obtained from review of the patients' medical charts. We used multivariable logistic regression to control for these potential confounders while testing for associations between F8 haplotype and the development of inhibitors.
Results
Of the 78 black patients with hemophilia enrolled, 24% had an H3 or H4 background haplotype. The prevalence of inhibitors was higher among patients with either of these haplotypes than among patients with haplotype H1 or H2 (odds ratio, 3.6; 95% confidence interval, 1.1 to 12.3; P = 0.04), despite a similar spectrum of hemophilic mutations and degree of severity of illness in these two subgroups.
Conclusions
These preliminary results suggest that mismatched factor VIII replacement therapy may be a risk factor for the development of anti–factor VIII alloantibodies.
Infusion of plasma-derived or recombinant factor VIII is the standard method of arresting hemorrhage in patients with hemophilia A (factor VIII deficiency). Alloantibodies that neutralize the activity of the replacement molecules develop in approximately 20 to 25% of patients,1,2 however, and the treatment of patients who have these inhibitors can be costly.
The risk of formation of an inhibitor is influenced by the type of mutation in the factor VIII gene (F8).3-7 Large deletions, inversions, and nonsense mutations are associated with the highest risk, probably because the recipient's immune system recognizes the normal factor VIII replacement protein as a foreign molecule. The type of mutation also is associated with the severity of hemophilia A. Thus, the association between the type of mutation and the development of inhibitors may be confounded by variables related to the severity of illness, such as age at the first infusion of therapy8 or the cumulative number of days of replacement therapy.9
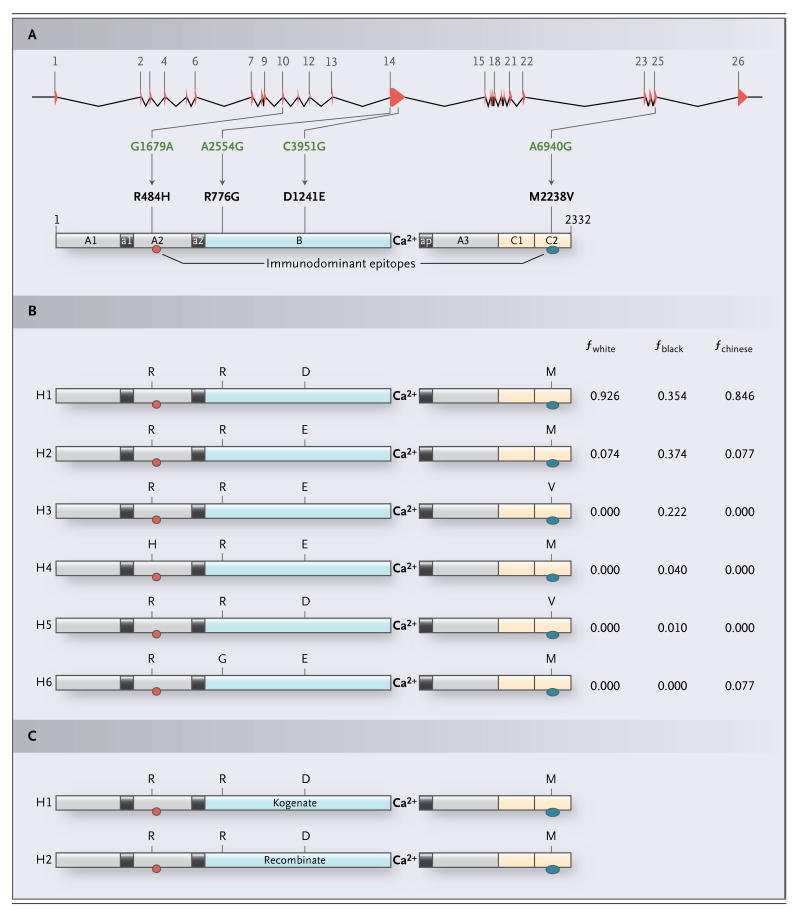
The prevalence of factor VIII inhibitors in black patients is about twice that in white patients.9-16 The mechanisms that account for this difference are unknown. In a study of F8 in 137 healthy, unrelated people from seven groups of diverse geographic origins, we identified four nonsynonymous single-nucleotide polymorphisms (SNPs) — G1679A (encoding the amino acid substitution of histidine for arginine at position 484 [R484H]), A2554G (encoding the substitution of glycine for arginine [R776G]), C3951G (encoding the substitution of glutamic acid for aspartic acid [D1241E]), and A6940G (encoding the substitution of valine for methionine [M2238V])17 — whose haplotypes (allelic combinations) encode six distinct factor VIII proteins, which we designated H1 through H6.18 Two of these proteins (H1 and H2) were found in all seven groups, but three (H3, H4, and H5) were found only in black people (16 subjects) and one (H6) was found only in Chinese people (10 subjects). (See Supplementary Appendix A, available with the full text of this article at NEJM. org, and Fig. 1.) The prevalence rates of H1 and H2 were 0.93 and 0.07, respectively, among whites in this study (86 subjects) and 0.35 and 0.37 among blacks. The prevalence rates of H3, H4, and H5 were 0.22, 0.04, and 0.01, respectively, among blacks. Kogenate (Bayer) and Recombinate (Baxter), the two full-length recombinant factor VIII products currently approved for use in persons with hemophilia A, correspond to the amino acid sequences of H1 and H2, respectively.21-24 In principle, therefore, one in four blacks with hemophilia A who require replacement therapy with recombinant factor VIII will receive products that differ from their own factor VIII protein at one or two residues, in addition to having amino acid differences attributable to the specific F8 mutation. Plasma-derived factor VIII is also a source of exposure to H1 and H2, because most blood donors are white.25-28
Figure 1. Four Nonsynonymous Single-Nucleotide Polymorphisms (SNPs) Whose Haplotypes Encode Six Distinct Factor VIII Proteins, Designated H1 through H6.
Human F8 contains four common nonsynonymous SNPs whose allelic combinations encode six distinct wild-type factor VIII proteins, only two of which have the amino acid sequences found in the recombinant factor VIII molecules used clinically. Panel A shows a schematic illustration of both F8, with its 26 exons and 25 introns indicated by red triangles and intervening lines, respectively, and factor VIII, with highlighting of its three A domains (A1, A2, and A3, shown in gray), single B domain (B, in blue), two C domains (C1 and C2, yellow), three acidic connecting peptides (a1, a2, and ap, black), and two immunodominant-inhibitor epitopes located in the A2 domain (red oval) and the C2 domain (blue oval).19 By sequencing all 26 exons of the F8 genes in 137 unrelated healthy persons from seven groups of diverse geographic origins, we identified four nonsynonymous SNPs: one in exon 10 (G1679A), two in exon 14 (A2554G and C3951G), and one in exon 25 (A6940G).17 These polymorphisms encode the following amino acid substitutions, respectively: histidine for arginine at position 484 (R484H), glycine for arginine at position 776 (R776G), glutamic acid for aspartic acid at position 1241 (D1241E), and valine for methionine at position 2238 (M2238V). The numbering systems used to designate the four nonsynonymous SNPs and the amino acid substitutions they encode are based on their nucleotide and residue locations, respectively, in the full-length F8 complementary DNA (with the use of the transcription start site found by Mansvelt et al.20) and the mature circulating form of factor VIII. Whereas R776G and D1241E are located in the B domain, R484H and M2238V are components of the A2 and C2 immunodominant epitopes, respectively, which have been mapped to residues located at epitopes R484 to I508 (isoleucine at position 508) and E2181 to V2243. Panel B shows the six structurally distinct wild-type factor VIII proteins encoded by the naturally occurring allelic combinations (haplotypes) of the F8 nonsynonymous SNPs G1679A, A2554G, C3951G, and A6940G.18 The amino acid residue at positions 484 (R or H), 776 (R or G), 1241 (D or E), and 2238 (M or V) are shown. The haplotype frequencies (f) listed for the six factor VIII proteins (H1 through H6) are based on their occurrence in 86 white (fwhite), 67 black (fblack), and 10 Chinese (fchinese) subjects.17,18 In Panel C, the two full-length recombinant factor VIII proteins used in replacement therapy, Kogenate and Recombinate, contain the same amino acid sequences found in H1 (R–R–D–M) and H2 (R–R–E–M), respectively.21-24
Alloimmunization against factor VIII can occur in white patients with F8 missense mutations29 that change only a single amino acid residue in the factor VIII protein.18 However, differences between replacement factor VIII and the patient's factor VIII at residues encoded by nonsynonymous SNPs, whose alleles do not cause hemophilia, have not been investigated as risk factors for the development of inhibitors. We hypothesized that the higher prevalence of inhibitors in black patients may be due in part to the greater degree of population-level variation that exists in their factor VIII amino acid sequence and the resultant increased probability of a mismatch with replacement factor VIII proteins.
Methods
Patients
Between November 13, 2003, and April 6, 2006, we invited black patients with hemophilia A undergoing treatment at any of four Federal Region IV South Hemophilia Treatment Centers to participate in this study during scheduled annual visits.30 The participating centers were Emory University, Atlanta; the University of Alabama at Birmingham, Birmingham; the Medical College of Georgia, Augusta; and the University of Mississippi Medical Center, Jackson. Each of the 78 enrolled patients provided a blood sample. Patients or their parents or legal guardians gave written informed consent for participation in the study. The institutional review boards of each participating center approved the protocol.
Questionnaire
A short, standardized survey was administered to all patients by each center. Information concerning self-reported race, age, baseline severity of hemophilia, results of previous testing for inhibitors, and other affected family members was obtained from medical records and interviews with patients by the nurses involved with enrollment. To take into account nonindependence of subjects due to family relationships, all patients with affected relatives were asked whether any relative was being treated at any of the participating centers and thus might be enrolled in this study.
Inhibitor Surveillance and Determination of Baseline Severity of Hemophilia
Data on inhibitors were obtained from reviews of the medical charts by the nurses. To identify inhibitors, the participating centers used the Bethesda assay31 with a Nijmegen modification32 known to improve its specificity near the cutoff for a positive test result, which was 0.6 Bethesda unit per milliliter. In general, patients were screened for inhibitors during their annual evaluations. Baseline severity of hemophilia was defined according to the initial level (in units per milliliter) of factor VIII activity as a percentage of normal. Mild hemophilia corresponded to a baseline level of factor VIII greater than 5% but less than 40% of normal, moderate hemophilia to a baseline level equal to or greater than 1% but no greater than 5% of normal, and severe hemophilia to a baseline level less than 1% of normal.33 To measure factor VIII, each center used factor VIII–deficient plasma and assessment of the activated partial-thromboplastin time.
F8 Sequencing
All known functional regions of F8, including 1194 bp of the contiguous promoter sequence, all 26 exons, 50 to 100 bp of each junctional-intronic segment, and 309 bp of flanking 3′-genomic DNA, were amplified by the polymerase chain reaction (PCR) and sequenced as described by Viel et al.17 Sequencing was performed to genotype the known nonsynonymous SNPs, discover new nonsynonymous SNPs, and identify the noninversion hemophilia-causing mutations. The sequencing chromatograms were processed with Phred software (www.phrap.org)34,35 and SAS software programs written in-house17 and were then reviewed manually. Given that males have only one X chromosome, patients with hemophilia are hemizygous for F8, and thus haplotypes were constructed as a simple combination of the patient's nonsynonymous SNP alleles.
F8 Inversion Assays
To identify inversions in introns 1 and 22, we used genomic DNA samples and slightly modified versions of three PCR-based assays.36-38 Patients whose F8 mutations were not identified definitively by sequencing were evaluated for the intron 22 inversion by long-range PCR.36 Unless an intron 22 inversion was definitively identified, the intron 1 inversion assay was performed.37 Finally, unless an intron 1 inversion was definitely identified, a more robust inverse-PCR-based intron 22 inversion assay was performed.38
Statistical Analysis
Outcome, Exposure, and Covariates
We considered a patient to have an inhibitor if any screening assay ever had a value of 0.6 Bethesda unit per milliliter or higher.31,32 We designated the background wild-type form of the factor VIII protein encoded by a patient's F8 gene as the exposure on the basis of specified amino acid residues at positions 484 (R or H), 776 (R or G), 1241 (D or E), and 2238 (M or V). On the basis of the alleles of G1679A, A2554G, C3951G, and A6940G, the background F8 haplotypes identified in this study were predicted to encode four of the five wild-type factor VIII proteins observed previously in the black population, namely, H1, H2, H3, and H4 (Fig. 1). Because of the small number of subjects, we combined them into two groups: H1 with H2 (H1+H2) and H3 with H4 (H3+H4). Patients in the H1+H2 group represent nonexposed (control) subjects, since their hemophilic mutations are present in F8 haplotypes that encode the factor VIII proteins represented by or enriched in recombinant and plasma-derived replacement products. The H3+H4 group is composed of exposed (case) subjects, since their F8 mutations reside within haplotypes encoding the black-restricted factor VIII proteins H3 and H4, which are structurally distinct from, and therefore mismatched with, the recombinant (and plasma-derived) factor VIII products used clinically. In an unadjusted analysis, we tested whether the prevalence of inhibitors was significantly different among patients grouped according to their factor VIII haplotypes. We also performed logistic-regression analysis with control for age at enrollment and baseline severity of illness and repeated these analyses in the subgroup of subjects with hemophilia-causing missense mutations only. We used SAS software for Windows, version 9.1.3, for all statistical analyses.
Accounting for Nonindependence Due to Family Relationships
Because the study questionnaire identified several related patients, we were concerned that association of the development of inhibitors with F8 haplotype might be due to the fact that family members, who share the same haplotype, are also more likely to share alleles of other polymorphic loci, including those that may influence the development of inhibitors, such as the genes for tumor necrosis factor α and interleukin-10.39,40 We thus treated patients without affected relatives enrolled in the study as singletons and grouped those with reported affected relatives into pedigrees. We performed a series of both crude and adjusted subanalyses after progressing through all combinations of unrelated subjects, selecting only one member from each family that had more than one affected member, and recorded the resulting odds ratios.
Results
Seventy-eight black patients with hemophilia A were enrolled. We identified the hemophilic F8 mutation in 70 of the 78 patients (Fig. 2). Two full brothers with large gene deletions that included A6940G, the nonsynonymous SNP encoding M2238V (Table 1), were excluded from the association analyses, since they could not be classified within either haplotype group. The mean (±SD) age of the 76 remaining subjects was 17.5± 12.9 years. According to initially recorded measurements of factor VIII, 11 subjects (14%) had mild hemophilia, 17 (22%) had moderate hemophilia, and 48 (63%) had severe hemophilia; this distribution of severity is similar to that seen in cross-sectional studies of patients with hemophilia from other racial groups.42-47
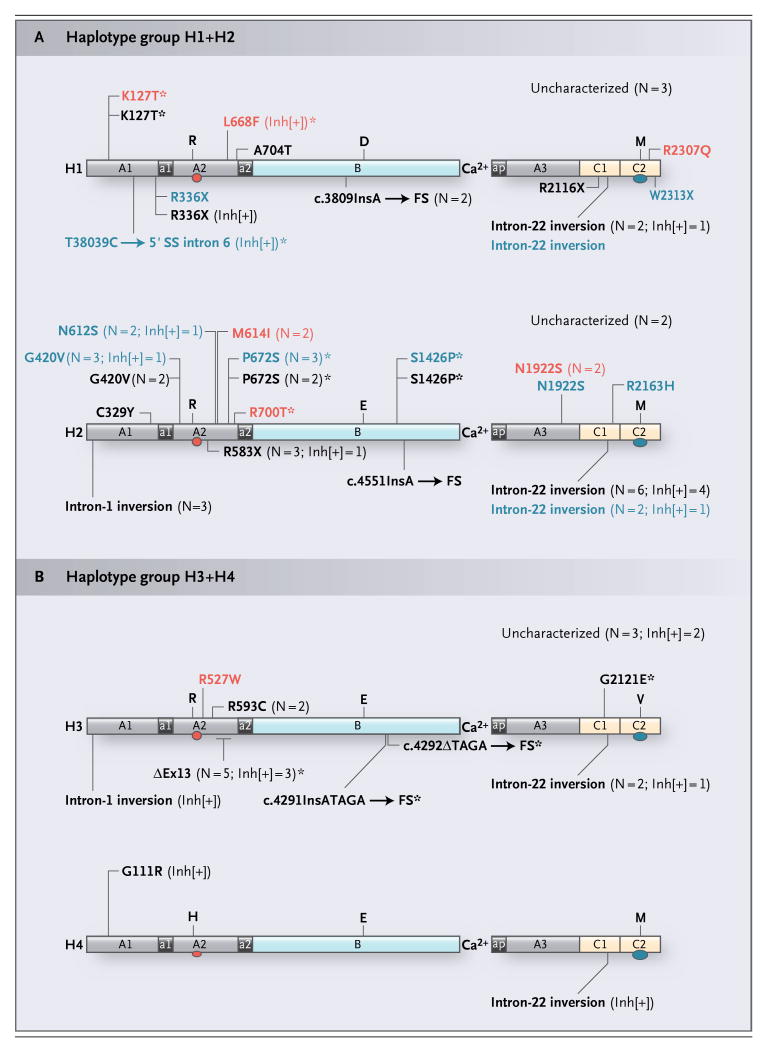
Figure 2. Hemophilic Mutations and the Four Wild-Type Factor VIII Proteins Predicted to Be Encoded by the Background F8 Haplotypes on Which They Were Identified.
For factor VIII, the two immunodominant-inhibitor epitopes located in the A2 domain (red oval) and the C2 domain (blue oval) are shown. Mutations found in patients with either an H1 or an H2 haplotype (H1+H2) are shown in Panel A, and mutations found in patients with either an H3 or an H4 haplotype (H3+H4) are shown in Panel B. For all haplotypes, missense mutations are shown above the appropriate factor VIII protein, and the other mutation types are shown below. Missense and nonsense mutations are indicated by their residue positions in the mature factor VIII protein. The point mutation T38039C, which occurs at position +2 of the 5′ splice site (SS) of intron 6, is designated according to the genomic nucleotide numbering system used for the F8 reference sequence.17 The positions of four frameshift (FS)-inducing small deletions and insertions are numbered according to their locations in the full-length F8 complementary DNA (c) with respect to the transcription start site.24 Specifically, one deletion (c.4292ΔTAGA) and three insertions (c.3809InsA, c.4551InsA, and c.4291InsATAGA) are indicated by the number of the wild-type nucleotide positioned immediately 5′ of the mutation site. ΔEx13 indicates an in-frame deletion of the 210-bp exon 13 sequence and an unknown amount of flanking nonexonic sequences from introns 12 and 13. For those mutations that occurred in more than one patient, whether or not the patients were related, the number of times any given abnormality was observed (N) is indicated in parentheses. All previously unknown mutations are indicated with an asterisk. The baseline severity of hemophilia for each patient is shown by the color of the text defining his mutation, with black, blue, and red indicating severe, moderate, and mild disease, respectively. For those mutations found in at least one inhibitor-positive (Inh[+]) patient, the number of patients with a given abnormality in whom inhibitors developed is also indicated in parentheses. A 3′-terminal partial gene deletion involving exons 24, 25, and 26 in two inhibitor-positive brothers is not shown. D, E, H, M, R, and V denote the amino acids aspartic acid, glutamic acid, histidine, methionine, arginine, and valine, respectively.
Table 1. Factor VIII Gene Mutation Type According to Background Factor VIII Protein Haplotype in 78 Black Patients with Hemophilia A*.
| Mutation Type† | Reported Inhibitor Prevalence‡ % |
Total§ | H1+H2§ no. (%) |
H3+H4§ |
|---|---|---|---|---|
| Higher risk | Not known | 57 (73) | 40 (70)¶ | 15 (79)¶ |
| Large deletions of ≥2 exons (>1 domain)* | 68.2–88.0 | 2 (3) | 0 | 0 |
| Nonsense mutations (light chain) | 40.0–50.0 | 2 (3) | 2 (4) | 0 |
| Intragenic inversions (intron 22) | 21.0–35.0 | 14 (18) | 11 (19) | 3 (16) |
| Large deletions of ≤1 exon (≤1 domain)‖ | 11.9–25.0 | 5 (6) | 0 | 5 (26) |
| Small insertions or deletions (non–A-run) | 20.6–21.0 | 2 (3) | 0 | 2 (11) |
| Nonsense mutations (heavy chain) | 14.3–17.0 | 5 (6) | 5 (9) | 0 |
| Intragenic inversions (intron 1) | 10.0–17.0 | 4 (5) | 3 (5) | 1 (5) |
| Missense mutations (A2, C1, and C2 domains) | 10.0–12.0 | 23 (29) | 19 (33) | 4 (21) |
| Lower risk | Not known | 13 (17) | 12 (21) | 1 (5) |
| Splice-site mutations | 2.2–17.0 | 1 (1) | 1 (2) | 0 |
| Small insertions or deletions (A-run) | 3.0–6.0 | 3 (4) | 3 (5) | 0 |
| Missense mutations (other regions) | 3.0–3.9 | 9 (12) | 8 (14) | 1 (5) |
| Not identified | Not applicable | 8 (10) | 5 (9) | 3 (16) |
| Total | Not applicable | 78 | 57 | 19 |
Two inhibitor-positive brothers with a deletion of exons 24, 25, and 26 (a large deletion) could not be assigned to either the H1+H2 or the H3+H4 group, since A6940G, which encodes M2238V, is located in exon 25.
We identified 11 different mutation types.
The number and percentage of patients with any given mutation type in the overall study cohort and either the nonexposed (H1+H2) or the exposed (H3+H4) group is given.
The proportion of patients with higher-risk mutation types does not differ significantly between the H1+H2 and the H3+H4 haplotype groups (P = 0.27 by two-sided Fisher's exact test).
An in-frame deletion of exon 13 (ΔEx13) is predicted in five patients on the basis of a repeated failure of multiple independent polymerase chain reactions to generate the appropriate amplicon only when genomic DNAs from these patients were used.
In the black patients with hemophilia, haplotypes H1, H2, H3, and H4 were identified, but not the infrequent H5 haplotype.18 Two patients had one additional, previously unknown nonsynonymous SNP, neither of which was predicted to cause hemophilia (see Supplementary Appendix B). The frequencies of mild, moderate, and severe hemophilia did not differ significantly according to the four background haplotypes (P = 0.11). Table 2 shows the relationship between haplotype group and the prevalence of inhibitors. The odds of having a factor VIII inhibitor were significantly higher among patients with an H3 or H4 haplotype than among those with an H1 or H2 haplotype (odds ratio, 3.4; 95% confidence interval [CI], 1.1 to 10.2; P = 0.03). This association remained when we controlled for age at enrollment and baseline severity of hemophilia in a multivariable logistic regression (odds ratio, 3.6; 95% CI, 1.1 to 12.3; P = 0.04).
Table 2. Development of Inhibitors to Factor VIII According to Factor VIII Haplotype among 76 Black Patients with Hemophilia A.
| Variable | Development of Factor VIII Inhibitor | Odds Ratio (95% CI) | |
|---|---|---|---|
| Yes | No | ||
| Factor VIII haplotype | |||
| H4 | 2 | 0 | Undefined* |
| H3 | 7 | 10 | 2.5 (0.6–10.7)† |
| H2 | 8 | 31 | 0.9 (0.2–3.5)† |
| H1 | 4 | 14 | Reference group |
| Haplotype group | |||
| H3+H4 | 9 | 10 | 3.4 (1.1–10.2)‡ |
| H1+H2 | 12 | 45 | |
P = 0.79 by two-sided Fisher's exact test.
The odds of having a factor VIII inhibitor were not significantly higher among the H3 or H2 patients alone than among the H1 patients.
The odds of having a factor VIII inhibitor were significantly higher among patients in the H3+H4 group than among those in the H1+H2 group.
We excluded the two patients whose F8 genes had different background haplotypes because of the presence of one additional nonsynonymous SNP each. Of the remaining 74 patients, 51 had no reported relative among the study participants. The other 23 patients were members of 11 families (Table 1 in the Supplementary Appendix). When a single patient was selected from each of these families, the sample size for the subanalysis was 62 patients. In analyses of all 3072 possible combinations of 62 unrelated persons, the median odds ratios for the development of factor VIII inhibitors were 2.5 and 2.6 in the unadjusted and adjusted analyses, respectively. The maximum and minimum odds ratios observed in any single sub-sample of unrelated persons were 4.3 and 1.5, respectively, in the unadjusted analysis and 4.4 and 1.5 in the adjusted analysis.
Table 1 shows that 11 different categories of hemophilic mutation types were identified in the 78 black patients. These 11 mutation categories consisted of 31 distinct loss-of-function F8 alleles, 9 of which were previously unknown (Fig. 2).29 This large degree of allelic heterogeneity is similar to what has been observed in previous cross-sectional studies to identify the mutational spectrums in patients from other racial groups.42-44,46,47 Furthermore, among the 70 patients with identified F8 mutations, no difference was observed between the H1+H2 and the H3+H4 haplotype comparison groups in the proportion of patients with higher-risk or lower-risk types of mutation (P = 0.27) (Table 1). To reduce the heterogeneity of the unknown effects of different mutation types, we conducted a subanalysis among patients with missense mutations (the only category large enough to yield a meaningful result), using multivariable logistic regression to control for age at enrollment and baseline severity of illness. The prevalence of inhibitor development was higher in those whose missense mutations resided on a haplotype encoding H3 or H4 (odds ratio, 4.3), although the confidence interval in this small sub-sample of 31 patients included the possibility of a null effect (95% CI, 0.2 to 101.1).
Discussion
We investigated a potential mechanism underlying the observation that the incidence of factor VIII inhibitors in black patients with hemophilia A is about twice that in whites.9-16 Our previous investigations of nonhemophilic populations (Fig. 1)17,18 led us to predict that the causative hemophilic mutations in approximately 27% of black patients would be present on background F8 haplotypes encoding either the H3, H4, or H5 wild-type forms of factor VIII.30 These haplotypes differ from the H1 and H2 proteins.17,18 H1 and H2 proteins constitute the currently available recombinant factor VIII products21-24 and are enriched in plasma-derived factor VIII concentrates, since blood donors in the United States are predominantly white.25-28 We found that patients with either an H3 or an H4 background haplotype were more likely to have an inhibitor (at some point in their lives) than were patients whose haplotypes were either H1 or H2, a result consistent with our previous findings.48 In this preliminary study, we focused only on black patients to reduce the magnitude of any potential confounding variables due to population stratification across the haplotype comparison groups.49-51
Because the sample size was small, we combined patients into two groups, H1+H2 and H3+H4. The amino acid sequences of the background H1 and H2 proteins correspond to the full-length recombinant factor VIII molecules (Fig. 1C)21-24 and the two factor VIII proteins predicted to predominate in existing plasma-derived products.25-28 We did not have sufficient information about the brands of concentrate to which the patients had been exposed to compare the prevalence of inhibitor development between patients with an H1 haplotype who had been treated with Kogenate (the H1 molecule) and those treated with Recombinate (the H2 molecule), or, conversely, between patients with an H2 haplotype who had been treated with one or the other concentrate. Thus, in our study, the inclusion of all H1 and H2 patients in the reference group could result in a bias toward the null (i.e., it could bring the odds ratio closer to 1 than its true value). Of the three nonsynonymous SNPs whose encoded amino acid residues distinguish H3 and H4 from H1 and H2, two are located in immunodominant epitopes (R484 to I508 [isoleucine at position 508] and E2181 to V2243), sites at which neutralizing factor VIII alloantibodies from most patients with inhibitors interact.2 In our multivariable regression analysis, we did not have sufficient data to control for some potentially important variables, such as previous exposure to plasma-derived or recombinant factor VIII products (or both),9 cumulative days of exposure,9,52 age at first infusion,8 or whether the inhibitors that developed were transient or permanent and of low or high titer.2 We also did not compare the distribution of allelic variants of immune-response genes associated with the development of inhibitors39,40,53 in the two haplotype groups. We did control for age at enrollment and baseline severity of hemophilia, but these are at best poor surrogates for age at first infusion and cumulative days of exposure. We used a conservative approach to account for the effect of related patients, by selecting only one member from each family and progressing through all possible combinations of unrelated persons. The average odds ratios in these analyses differed little from those found for the overall sample. Thus, the presence of some related patients in the study was probably not a source of bias.
We acknowledge that our study has limited statistical power because of the small number of patients and that the results require confirmation. The importance of independent replication is that it would strengthen the evidence that mismatched factor VIII replacement proteins are a risk factor for the development of inhibitors. If our findings are confirmed, the possibility would arise that recombinant DNA technology could be used to develop additional replacement products that vary from endogenous factor VIII proteins only at a residue or residues required to correct clotting-factor deficits.
Supplementary Material
Acknowledgments
Supported in part by grants from the National Institutes of Health (HL-71130 and HL-72533, to Dr. Howard) and (HL-70751, to Dr. Almasy).
We thank all the patients who participated in this study, as well as the nurses who enrolled them, including Cara Brown, Valerie Crenshaw, Mary Katherine Noa, April Morris, and Elizabeth Meagher; Deepa Machiah, Thuy Tran, Marjorie Britten, Xiaopeng Mung, and Ming Shen for excellent technical assistance; Pam Bryant for her invaluable role in the administrative aspects of this study; and Bruce Evatt, John Blangero, Vincent La Terza, Johnny Mahlunga, and Georg Lemm for helpful discussions, review of an earlier version of the manuscript, or both.
Footnotes
Presented in part at the 48th Annual Meeting of the American Society of Hematology, Orlando, FL, December 9 to 12, 2006.
Dr. Abshire reports serving on the advisory boards for CSL Behring, Novo Nordisk, and Bayer; Dr. Kasper, serving on a data and safety monitoring board for Wyeth and receiving grant support from CSL Behring; and Dr. Thompson, participating as a site investigator for Baxter, serving as a consultant for Ipsen, and receiving grant support from Bayer. Dr. Howard is the co-founder of Haplomics, which owns a patent application with claims for novel proteins and diagnostic methods that may be useful in treating patients with hemophilia A. No other potential conflict of interest relevant to this article was reported.
References
- 1.Wight J, Paisley S. The epidemiology of inhibitors in haemophilia A: a systematic review. Haemophilia. 2003;9:418–35. doi: 10.1046/j.1365-2516.2003.00780.x. [DOI] [PubMed] [Google Scholar]
- 2.Lollar P. Pathogenic antibodies to coagulation factors. Part one: factor VIII and factor IX. J Thromb Haemost. 2004;2:1082–95. doi: 10.1111/j.1538-7836.2004.00802.x. [DOI] [PubMed] [Google Scholar]
- 3.Goodeve A. The incidence of inhibitor development according to specific mutations — and treatment? Blood Coagul Fibrinolysis. 2003;14 1:S17–S21. doi: 10.1097/00001721-200306001-00005. [DOI] [PubMed] [Google Scholar]
- 4.Oldenburg J, Grimm T, Becker J, Olek K, Brackmann HH, Schwaab R. Mutations in severe hemophilia A: distribution within the factor VIII gene, origin and influence on inhibitor development. Beitr Infusionsther Transfusionsmed. 1997;34:224–30. [PubMed] [Google Scholar]
- 5.Oldenburg J, Schröder J, Brackmann HH, Müller-Reible C, Schwaab R, Tuddenham E. Environmental and genetic factors influencing inhibitor development. Semin Hematol. 2004;41:82–8. doi: 10.1053/j.seminhematol.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 6.Schröder J, El-Maarri O, Schwaab R, Müller CR, Oldenburg J. Factor VIII intron-1 inversion: frequency and inhibitor prevalence. J Thromb Haemost. 2006;4:1141–3. doi: 10.1111/j.1538-7836.2006.01884.x. [DOI] [PubMed] [Google Scholar]
- 7.Schwaab R, Brackmann HH, Meyer C, et al. Haemophilia A: mutation type determines risk of inhibitor formation. Thromb Haemost. 1995;74:1402–6. [PubMed] [Google Scholar]
- 8.Lorenzo JI, López A, Altisent C, Aznar JA. Incidence of factor VIII inhibitors in severe haemophilia: the importance of patient age. Br J Haematol. 2001;113:600–3. doi: 10.1046/j.1365-2141.2001.02828.x. [DOI] [PubMed] [Google Scholar]
- 9.Goudemand J, Rothschild C, Demiguel V, et al. Influence of the type of factor VIII concentrate on the incidence of factor VIII inhibitors in previously untreated patients with severe hemophilia A. Blood. 2006;107:46–51. doi: 10.1182/blood-2005-04-1371. [DOI] [PubMed] [Google Scholar]
- 10.Addiego JE, Jr, Kasper C, Abildgaard C, et al. Increased frequency of inhibitors in African American hemophilia A patients. Blood. 1994;84(Suppl):239a. abstract. [Google Scholar]
- 11.Aledort LM, Dimichele DM. Inhibitors occur more frequently in African-American and Latino haemophiliacs. Haemophilia. 1998;4:68. doi: 10.1046/j.1365-2516.1998.0146c.x. [DOI] [PubMed] [Google Scholar]
- 12.Astermark J, Berntorp E, White GC, Kroner BL. The Malmö International Brother Study (MIBS): further support for genetic predisposition to inhibitor development in hemophilia patients. Haemophilia. 2001;7:267–72. doi: 10.1046/j.1365-2516.2001.00510.x. [DOI] [PubMed] [Google Scholar]
- 13.Bray GL, Gomperts ED, Courter S, et al. A multicenter study of recombinant factor VIII (recombinate): safety, efficacy, and inhibitor risk in previously untreated patients with hemophilia A. Blood. 1994;83:2428–35. [PubMed] [Google Scholar]
- 14.Gill FM. The natural history of factor VIII inhibitors in patients with hemophilia A. Prog Clin Biol Res. 1984;150:19–29. [PubMed] [Google Scholar]
- 15.Lusher JM, Arkin S, Abildgaard CF, Schwartz RS. Recombinant factor VIII for the treatment of previously untreated patients with hemophilia A: safety, efficacy, and development of inhibitors. N Engl J Med. 1993;328:453–9. doi: 10.1056/NEJM199302183280701. [DOI] [PubMed] [Google Scholar]
- 16.Rothschild C, Laurian Y, Satre EP, et al. French previously untreated patients with severe hemophilia A after exposure to recombinant factor VIII: incidence of inhibitor and evaluation of immune tolerance. Thromb Haemost. 1998;80:779–83. [PubMed] [Google Scholar]
- 17.Viel KR, Machiah DK, Warren DM, et al. A sequence variation scan of the coagulation factor VIII (FVIII) structural gene and associations with plasma FVIII activity levels. Blood. 2007;109:3713–24. doi: 10.1182/blood-2006-06-026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howard TE, Machiah DK, Tran TT, et al. African-Americans express multiple haplotypic forms of the wildtype factor VIII (FVIII) protein: a possible role for pharmacogenetics in FVIII inhibitor development? Blood. 2004;104:113a. abstract. [Google Scholar]
- 19.Thompson AR. Structure and function of the factor VIII gene and protein. Semin Thromb Hemost. 2003;29:11–22. doi: 10.1055/s-2003-37935. [DOI] [PubMed] [Google Scholar]
- 20.Mansvelt EP, Laffan M, McVey JH, Tuddenham EG. Analysis of the F8 gene in individuals with high plasma factor VIII: C levels and associated venous thrombosis. Thromb Haemost. 1998;80:561–5. [PubMed] [Google Scholar]
- 21.Gitschier J. Remembrances of factor VIII. 1. The race to the gene. J Thromb Haemost. 2004;2:383–7. doi: 10.1111/j.1538-7933.2004.00614.x. [DOI] [PubMed] [Google Scholar]
- 22.Gitschier J, Wood WI, Goralka TM, et al. Characterization of the human factor VIII gene. Nature. 1984;312:326–30. doi: 10.1038/312326a0. [DOI] [PubMed] [Google Scholar]
- 23.Toole JJ, Knopf JL, Wozney JM, et al. Molecular cloning of a cDNA encoding human antihaemophilic factor. Nature. 1984;312:342–7. doi: 10.1038/312342a0. [DOI] [PubMed] [Google Scholar]
- 24.Vehar GA, Keyt B, Eaton D, et al. Structure of human factor VIII. Nature. 1984;312:337–42. doi: 10.1038/312337a0. [DOI] [PubMed] [Google Scholar]
- 25.Benjamin RJ, Busch MP, Fang CT, et al. Human immunodeficiency virus-1 infection correlates strongly with herpes simplex virus-2 (genital herpes) seropositivity in South African and United States blood donations. Transfusion. 2008;48:295–303. doi: 10.1111/j.1537-2995.2007.01523.x. [DOI] [PubMed] [Google Scholar]
- 26.Garratty G, Glynn SA, McEntire R. ABO and Rh(D) phenotype frequencies of different racial/ethnic groups in the United States. Transfusion. 2004;44:703–6. doi: 10.1111/j.1537-2995.2004.03338.x. [DOI] [PubMed] [Google Scholar]
- 27.Glynn SA, Schreiber GB, Murphy EL, et al. Factors influencing the decision to donate: racial and ethnic comparisons. Transfusion. 2006;46:980–90. doi: 10.1111/j.1537-2995.2006.00831.x. [DOI] [PubMed] [Google Scholar]
- 28.Wang B, Higgins MJ, Kleinman S, et al. Comparison of demographic and donation profiles and transfusion-transmissible disease markers and risk rates in previously transfused and nontransfused blood donors. Transfusion. 2004;44:1243–51. doi: 10.1111/j.1537-2995.2004.04034.x. [DOI] [PubMed] [Google Scholar]
- 29.Kemball-Cook G, Tuddenham EG, Wacey AI. The factor VIII structure and mutation resource site: HAMSTeRS version 4. Nucleic Acids Res. 1998;26:216–9. doi: 10.1093/nar/26.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howard TE, Machiah DK, Viel KR, et al. The Pharmacogenetics and Inhibitor Risk (PIR) Study: establishing the spectrum of factor (F)VIII gene (F8) mutations in African-American hemophilia A patients. Blood. 2005;106:896a. abstract. [Google Scholar]
- 31.Kasper CK, Aledort L, Aronson D, et al. Proceedings: a more uniform measurement of factor VIII inhibitors. Thromb Diath Haemorrh. 1975;34:612. [PubMed] [Google Scholar]
- 32.Verbruggen B, Novakova I, Wessels H, Boezeman J, van den Berg M, Mauser-Bunschoten E. The Nijmegen modification of the Bethesda assay for factor VIII:C inhibitors: improved specificity and reliability. Thromb Haemost. 1995;73:247–51. [PubMed] [Google Scholar]
- 33.White GC, II, Rosendaal F, Aledort LM, Lusher JM, Rothschild C, Ingerslev J. Definitions in hemophilia: recommendation of the scientific subcommittee on factor VIII and factor IX of the scientific and standardization committee of the International Society on Thrombosis and Haemostasis. Thromb Haemost. 2001;85:560. [PubMed] [Google Scholar]
- 34.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–85. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 35.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–94. [PubMed] [Google Scholar]
- 36.Bagnall RD, Giannelli F, Green PM. Int22h-related inversions causing hemophilia A: a novel insight into their origin and a new more discriminant PCR test for their detection. J Thromb Haemost. 2006;4:591–8. doi: 10.1111/j.1538-7836.2006.01840.x. [DOI] [PubMed] [Google Scholar]
- 37.Bagnall RD, Waseem N, Green PM, Giannelli F. Recurrent inversion breaking intron 1 of the factor VIII gene is a frequent cause of severe hemophilia A. Blood. 2002;99:168–74. doi: 10.1182/blood.v99.1.168. [DOI] [PubMed] [Google Scholar]
- 38.Rossetti LC, Radic CP, Larripa IB, De Brasi CD. Genotyping the hemophilia inversion hotspot by use of inverse PCR. Clin Chem. 2005;51:1154–8. doi: 10.1373/clinchem.2004.046490. [DOI] [PubMed] [Google Scholar]
- 39.Astermark J, Oldenburg J, Carlson J, et al. Polymorphisms in the TNFA gene and the risk of inhibitor development in patients with hemophilia A. Blood. 2006;108:3739–45. doi: 10.1182/blood-2006-05-024711. [DOI] [PubMed] [Google Scholar]
- 40.Astermark J, Oldenburg J, Pavlova A, Berntorp E, Lefvert AK. Polymorphisms in the IL10 but not in the IL1beta and IL4 genes are associated with inhibitor development in patients with hemophilia A. Blood. 2006;107:3167–72. doi: 10.1182/blood-2005-09-3918. [DOI] [PubMed] [Google Scholar]
- 41.Oldenburg J, Pavlova A. Genetic risk factors for inhibitors to factors VIII and IX. Haemophilia. 2006;12 6:15–22. doi: 10.1111/j.1365-2516.2006.01361.x. [DOI] [PubMed] [Google Scholar]
- 42.Jayandharan G, Shaji RV, Baidya S, Nair SC, Chandy M, Srivastava A. Identification of factor VIII gene mutations in 101 patients with haemophilia A: mutation analysis by inversion screening and multiplex PCR and CSGE and molecular modelling of 10 novel missense substitutions. Haemophilia. 2005;11:481–91. doi: 10.1111/j.1365-2516.2005.01121.x. [DOI] [PubMed] [Google Scholar]
- 43.Repessé Y, Slaoui M, Ferrandiz D, et al. Factor VIII (FVIII) gene mutations in 120 patients with hemophilia A: detection of 26 novel mutations and correlation with FVIII inhibitor development. J Thromb Haemost. 2007;5:1469–76. doi: 10.1111/j.1538-7836.2007.02591.x. [DOI] [PubMed] [Google Scholar]
- 44.Santacroce R, Acquila M, Belvini D, et al. Identification of 217 unreported mutations in the F8 gene in a group of 1,410 unselected Italian patients with hemophilia A. J Hum Genet. 2008;53:275–84. doi: 10.1007/s10038-007-0238-y. [DOI] [PubMed] [Google Scholar]
- 45.Ma GC, Chang SP, Chen M, Kuo SJ, Chang CS, Shen MC. The spectrum of the factor 8 (F8) defects in Taiwanese patients with haemophilia A. Haemophilia. 2008;14:787–95. doi: 10.1111/j.1365-2516.2008.01687.x. [DOI] [PubMed] [Google Scholar]
- 46.Castaman G, Giacomelli SH, Ghiotto R, et al. Spectrum of mutations in Albanian patients with haemophilia A: identification of ten novel mutations in the factor VIII gene. Haemophilia. 2007;13:311–6. doi: 10.1111/j.1365-2516.2007.01459.x. [DOI] [PubMed] [Google Scholar]
- 47.David D, Ventura C, Moreira I, et al. The spectrum of mutations and molecular pathogenesis of hemophilia A in 181 Portuguese patients. Haematologica. 2006;91:840–3. [PubMed] [Google Scholar]
- 48.Howard TE, Viel KR, Fernstrom KM, et al. Allelically mismatched replacement therapy due to common African-restricted haplotypes of the factor (F)VIII protein may underlie the increased incidence of FVIII inhibitors observed in hemophilia-A patients of African-descent. Blood. 2006;108:230a. abstract. [Google Scholar]
- 49.Kittles RA, Chen W, Panguluri RK, et al. CYP3A4-V and prostate cancer in African Americans: causal or confounding association because of population stratification? Hum Genet. 2002;110:553–60. doi: 10.1007/s00439-002-0731-5. [DOI] [PubMed] [Google Scholar]
- 50.Redden DT, Allison DB. The effect of assortative mating upon genetic association studies: spurious associations and population substructure in the absence of admixture. Behav Genet. 2006;36:678–86. doi: 10.1007/s10519-006-9060-0. [DOI] [PubMed] [Google Scholar]
- 51.Tsai HJ, Kho JY, Shaikh N, et al. Admixture-matched case-control study: a practical approach for genetic association studies in admixed populations. Hum Genet. 2006;118:626–39. doi: 10.1007/s00439-005-0080-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kempton CL, Soucie JM, Abshire TC. Incidence of inhibitors in a cohort of 838 males with hemophilia A previously treated with factor VIII concentrates. J Thromb Haemost. 2006;4:2576–81. doi: 10.1111/j.1538-7836.2006.02233.x. [DOI] [PubMed] [Google Scholar]
- 53.Astermark J, Wang X, Oldenburg J, Berntorp E, Lefvert AK. Polymorphisms in the CTLA-4 gene and inhibitor development in patients with severe hemophilia A. J Thromb Haemost. 2007;5:263–5. doi: 10.1111/j.1538-7836.2007.02290.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.