Abstract
Toll-like receptor 3 (TLR3) recognizes dsRNA, a molecular signature of most viruses, and triggers inflammatory responses that prevent viral spread. TLR3 ectodomains (ECD) dimerize on oligonucleotides of at least 40–50 bp in length, the minimal length required for signal transduction. To establish the molecular basis for ligand binding and signaling, we determined the crystal structure of a complex between two mouse TLR3-ECDs and dsRNA at 3.4 Å resolution. Each TLR3-ECD binds dsRNA at two sites located at opposite ends of the TLR3 “horseshoe”, and an intermolecular contact between the two TLR3-ECD C-terminal domains coordinates and stabilizes the dimer. This juxtaposition could mediate downstream signaling by dimerizing the cytoplasmic Toll IL-1 Receptor (TIR) domains. The overall shape of the TLR3-ECD does not change upon binding to dsRNA.
The Toll-like receptor (TLR) family comprises 10–12 type I integral membrane receptor paralogs that recognize pathogen associated molecular signatures and initiate inflammatory responses (1–3). TLR3-ECD binds double-stranded RNA (dsRNA) (4), a viral replication intermediate, and recruits the adaptor protein, TRIF, to its cytoplasmic TIR domain, thereby initiating a signaling cascade that results in the secretion of type I interferons and other inflammatory cytokines (5). Although the role of TLR3 in controlling infections is not fully understood, it is clear that TLR3 non-redundantly contributes to the prevention of herpes simplex encephalitis in children (6). The structure of human (h)TLR3-ECD has been determined by two laboratories (7, 8) and consists of a solenoid with 23 leucine rich repeats bent into a horseshoe shape, capped at each end by specialized structures known as Leucine-Rich Repeat N-terminal (LRR-NT) and C-terminal (LRR-CT) domains. TLR3-ECD binds dsRNA only at acidic pH (pH 6.5 and below) (9, 10), reflecting its endosomal location in most cell types (11). Although TLR3-ECD is monomeric in solution, it binds as dimers to dsRNA and requires a dsRNA length of at least 40–50 bp to bind a single dimer and induce signaling (10). However, the molecular basis for dimerization and signalling remains unknown. We have therefore isolated and crystallized a complex containing two mouse (m)TLR3-ECD molecules bound to a 46 bp dsRNA.
We first determined the apo-mTLR3-ECD structure by molecular replacement with the hTLR3-ECD structure (Table S1). mTLR3-ECD has 78% sequence identity with hTLR3-ECD (Fig S1), and as expected the structures of the two ECDs are also highly homologous. mTLR3-ECD superimposes with hTLR3-ECD with an r.m.s.d. of 1.36 Å (for 659 Cα atoms, pdb code 2A0Z) and 0.93 Å (for 613 Cα atoms, pdb code 1ZIW) (Fig S2). As in hTLR3-ECD, a horseshoe-shaped solenoid is formed by 23 LRRs, with N-terminal and C-terminal capping motifs. A large parallel β-sheet defines the concave surface, and two insertions in LRRs 12 and 20 protrude from the convex surface. Similar to hTLR3-ECD, the mTLR3-ECD molecule has a glycan-free surface. Both molecules contain 15 predicted N-linked glycosylation sites, but two of these are in different positions in the mouse and human proteins (Fig S1). Eleven glycan moieties are visible in the mTLR3-ECD electron density maps.
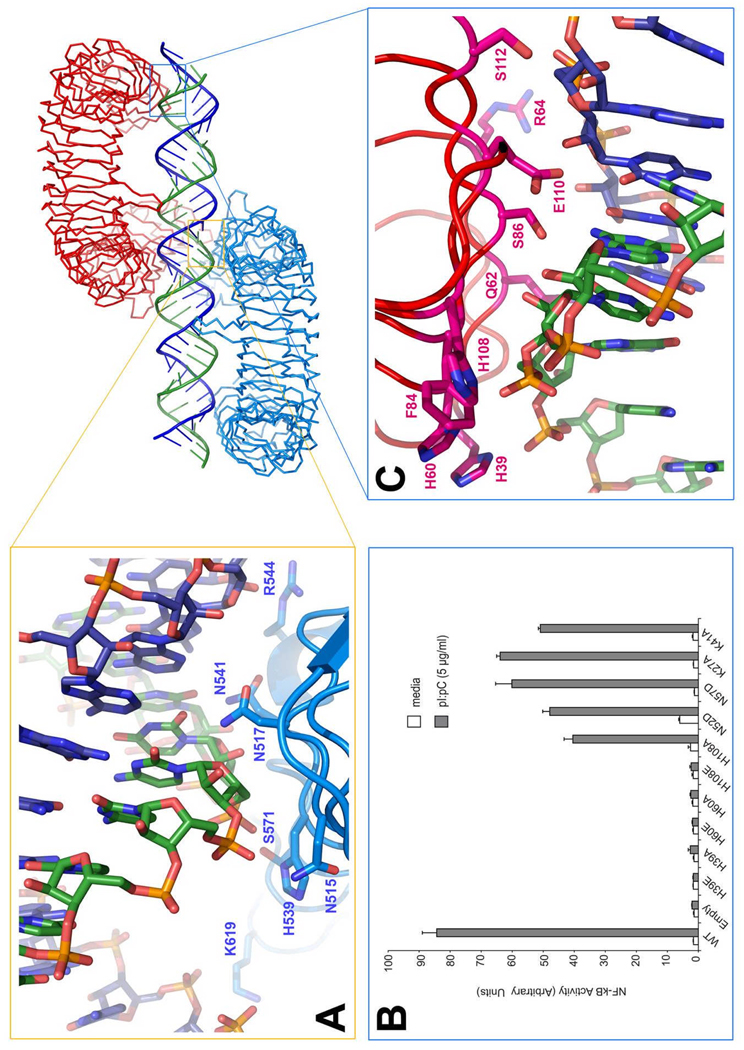
We next solved the structure of the TLR3 signaling complex, consisting of two mTLR3-ECD molecules bound to one 46 bp dsRNA oligonucleotide (Fig 1) by using the apo-mTLR3-ECD structure for molecular replacement. This structure identifies multiple intermolecular contacts that stabilize the complex. The dsRNA interacts with both an N-terminal and a C-terminal site on the glycan-free surface of each mTLR3-ECD, which are on opposite sides of the dsRNA (Fig 1A), with the C-termini in contact (Fig 1B) and the N-termini outstretched at opposing ends of the linear dsRNA molecule. The length of the complex is 141 Å. The overall structure of mTLR3-ECD does not change upon binding to dsRNA, supporting a signaling mechanism in which ligand-induced receptor dimerization brings the two cytoplasmic TIR domains into contact, thus triggering a downstream signaling cascade. The dsRNA in the complex retains a typical A-DNA-like structure, in which the ribose-phosphate backbone and the position of the grooves are the major determinants of binding. The mTLR3-ECD interacts with the sugar-phosphate backbones, but not with individual bases. This explains why TLR3 lacks specificity for any particular nucleotide sequence (4, 10).
Fig 1.
dsRNA:TLR3 signaling complex. Mouse TLR3 ectodomains (green and cyan) form a dimer on the dsRNA (blue and red). The N glycans are shown (light green and light blue). (A) The N- and C-terminal binding sites. (B) Illustration of how the two C-terminal domains are brought together in the complex. Figures generated with PyMol (DeLano Scientific, San Carlos, CA)
The first dsRNA:TLR3 interaction site is located close to the C-terminus, on LRR19-LRR21 (Table S2, Fig 2A). In the complex, these binding sites from two mTLR3-ECD monomers face each other across the dsRNA. Residues within contact distance of the RNA include Asn515, Asn517, His539, Asn541 and Arg544, which are all well-conserved in vertebrates (Fig S3). Mutational analysis previously showed that His539Glu and Asn541Ala were inactive and failed to bind dsRNA, whereas Asn517Ala and Arg544Ala retained activity, indicating that these latter residues are not essential for binding (12).
Fig 2.
The dsRNA binding sites of TLR3. (A) Residues involved in the interaction on the C-terminal site. (B) Illustrates the NF-κB activity of human TLR3 mutants stimulated with pI:pC. (C) Residues involved in binding at the N-terminal site. Note that each interaction involves two strands of the RNA. The dsRNA molecule may undergo a screw rotation of plus or minus one base pair in the crystal.
The second dsRNA:TLR3 interaction site is located on the N-terminal end (LRR-NT-LRR3) of the glycan-free surface and is formed by residues His39, His60, Arg64, Phe84, Ser86, His108, and Glu110 (Table S2, Fig 2B). A striking feature of this site is the presence of three conserved residues: His39, His60, and His108 (Fig S2), which are located in LRR-NT, LRR1, and LRR3, respectively. These residues appear to interact with consecutive phosphate groups on one dsRNA chain. In addition, less well-conserved residues Arg64, Phe84, Ser86 and Glu110 also interact with the ligand (Table S2, Fig 2A). To test the functional importance of the three His residues, we mutated them to Ala or Glu and examined the ability of the mutant proteins to activate NF-κB. As seen in Fig.2B, His39Ala and His60Ala are inactive, indicating that these residues are essential for dsRNA binding. In contrast, His108Ala retains activity, but mutation to glutamate results in loss of function. These findings imply that His108 is not essential for ligand-binding, but due to its close proximity to the negatively-charged phosphate groups in the dsRNA, mutation to a negatively-charged glutamate disrupts ligand binding via electrostatic repulsion. Since His39, His60 and His539 are required for TLR3 function, it is likely that the protonation of the imidazole groups of these residues at mildly acidic pH accounts for the pH dependence of dsRNA binding to TLR3 (9, 10). The N-terminal dsRNA binding site is in the vicinity of a positively charged surface patch that was previously suggested as a potential dsRNA binding site (8). However, the residues from that surface patch are located on the convex face of the N-terminus and do not come into direct contact with the dsRNA.
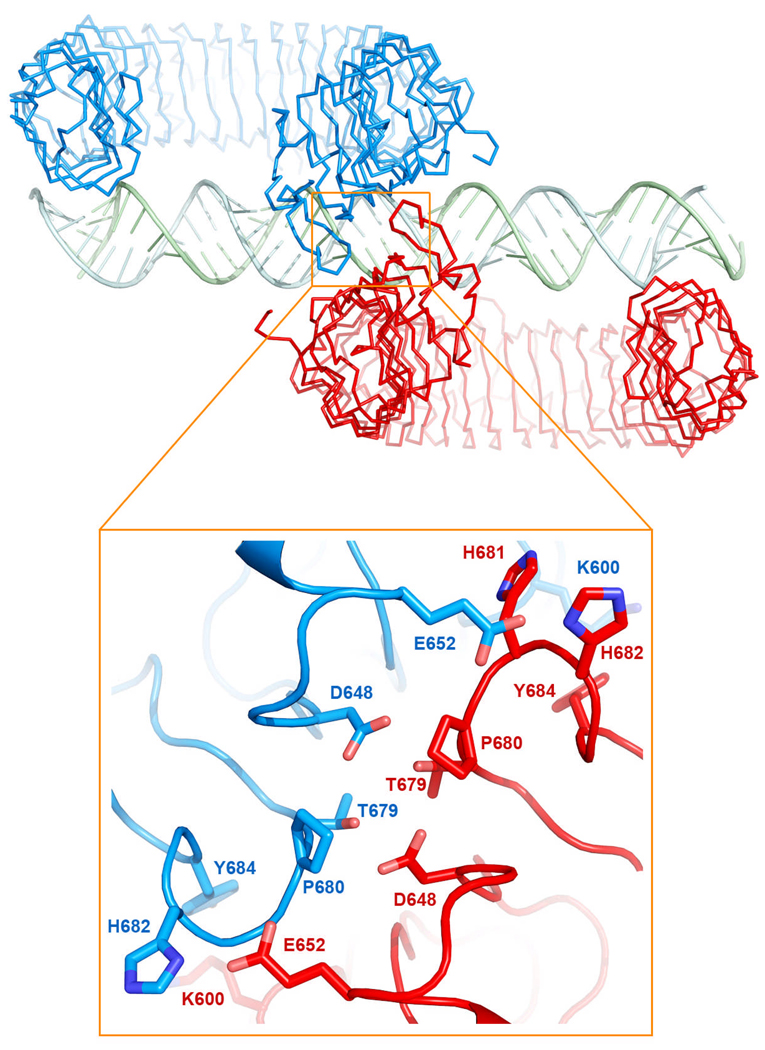
In the mTLR3-dsRNA 2:1 complex, the two LRR-CT domains are brought into close proximity, forming a series of protein-protein interactions (Table S3, Fig 3). Since these are the only interactions between the two mTLR3-ECDs they must be responsible for coordinating the dimer on the dsRNA thus facilitating the dimerization of the cytoplasmic TIR domains as suggested in Fig 4. The distance between the centers of the LRR-CT domains is 26 Å, with approximately 7 Å between the two C-termini. No other interactions exist to cause the two ECDs to dimerize on the dsRNA, and it will be interesting to learn whether similar interactions occur between LRR-CT domains in other TLRs. . This protein-protein interaction also explains why TLR3-ECD was previously observed to bind to dsRNA with a high degree of cooperativity (Leonard et al PNAS); while both the protein-protein interaction and the binding of one TLR3-ECD to dsRNA are relatively weak, the dimeric complex is stabilized by multivalent intermolecular interactions.
Fig 3.
Closeup of the C-terminal domain interacting residues. Some of these residues (678–681) are located on a conserved loop observed in other TLR structures.
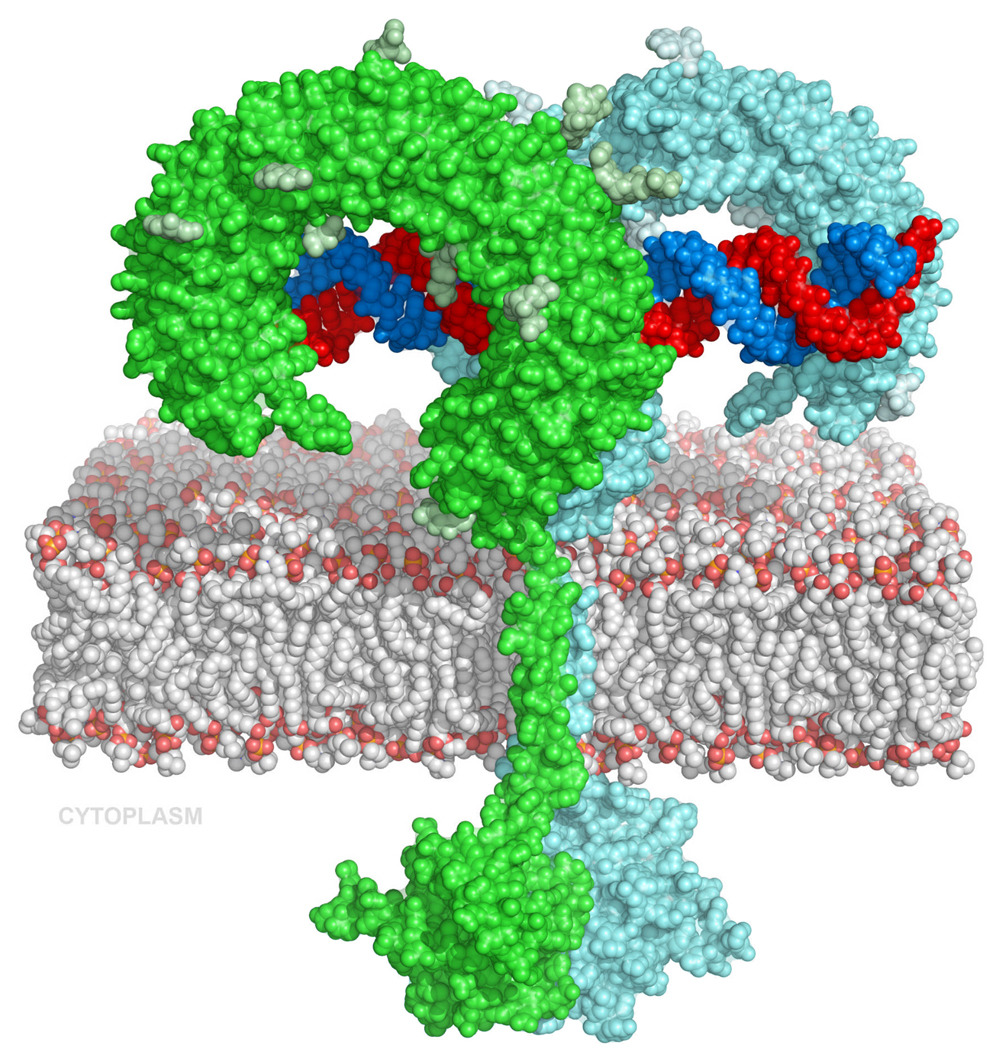
Fig 4.
Model of the full-length dsRNA:TLR3 signaling complex. The proximity of the two C-termini permits association of the trans-membrane helices and the dimerization of the cytoplasmic TIR domains. The TIR domains were homology modeled from the structure of the TLR10 TIR domains (pdb code 2J67).
While most dsRNA contacts occur via residues on the glycan-free surface of mTLR3-ECD, the N-glycosyl moiety of Asn413, which is located on the concave surface of mTLR3-ECD at LRR15, extends towards the dsRNA and directly contacts it. This contact may be more extensive in vivo, since mammalian glycan is typically larger than the insect glycan observed in our protein expression system. Although the Asn413 glycosylation site is well conserved (Fig S2), mutational analysis indicates that deletion of this glycan does not completely abrogate activity (12, 13).
Our results also help to explain some observations from a previous study by Takada et al. (14) in which single LRRs were sequentially deleted from hTLR3. Deletion of most LRRs located between the two dsRNA binding sites resulted in loss of function, which might be expected since such deletions would perturb the relative position of the two dsRNA binding sites on each TLR3-ECD. However, deletion LRRs 4, 11 and 17 did not abolish TLR3 function. The retention of function in these mutants may indicate that the TLR3 solenoid possesses sufficient flexibility such that it may compensate for the deletion and allow proper binding.
The dsRNA:TLR3 signaling complex features both similarities and differences with the structure of a complex containing TLR1 and TLR2 ECDs bridged by the ligand Pam3CSK4 (15). Two lipid chains of the ligand bind to a pocket in TLR2 and the third chain binds to a channel located in approximately the same region on TLR1. In both complexes, the ligand bridges two TLR molecules by the same glycan-free surface, resulting in the formation of a dimer in which the two TLR-ECDs are related by an approximate two-fold symmetry axis. TLR3 binds its ligand exclusively via surface contacts (mainly hydrogen bonding and electrostatic interactions), and although approximately the same surface area is buried in the TLR1:TLR2 interaction as in the TLR3 dimer, the intermolecular interactions involved differ substantially (Table S4). The protein-protein interactions occur only at the LRR-CT in the TLR3:dsRNA complex, while the extensive interactions between TLR1 and TLR2 occur near the binding pockets. In the dsRNA:TLR3-ECD complex the two C-terminal residues are ∼7 Å apart, whereas in the TLR1:TLR2 complex they are 42 Å apart. In the latter case however, the native LRR-CT domains were replaced with a hagfish VLR LRR-CT. In both these complexes the glycan-free surfaces are brought together by interaction with the ligands. For some TLRs, at least, a pattern may be emerging in which pathogen-associated ligands bind to TLRs by different mechanisms, but in each case, binding bridges two TLRs on the same glycan-free surface, and forms dimers with similar overall architecture.
Supplementary Material
Reference List
- 1.Kaisho T, Akira S. J. Allergy Clin. Immunol. 2006;117:979. doi: 10.1016/j.jaci.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 2.Iwasaki A, Medzhitov R. Nat. Immunol. 2004;5:987. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 3.Gay NJ, Gangloff M. Annu. Rev. Biochem. 2007;76:141. doi: 10.1146/annurev.biochem.76.060305.151318. [DOI] [PubMed] [Google Scholar]
- 4.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Nature. 2001;413:732. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 5.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. Nat. Immunol. 2003;4:161. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 6.Zhang SY, et al. Science. 2007;317:1522. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 7.Bell JK, et al. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10976. doi: 10.1073/pnas.0505077102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choe J, Kelker MS, Wilson IA. Science. 2005;581:309. doi: 10.1126/science.1115253. [DOI] [PubMed] [Google Scholar]
- 9.de Bouteiller O, et al. J. Biol. Chem. 2005;280:38133. doi: 10.1074/jbc.M507163200. [DOI] [PubMed] [Google Scholar]
- 10.Leonard JN, et al. Proc. Natl. Acad. Sci. U. S. A. 2008;105:258. doi: 10.1073/pnas.0710779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto M, et al. J. Immunol. 2003;171:3154. doi: 10.4049/jimmunol.171.6.3154. [DOI] [PubMed] [Google Scholar]
- 12.Bell JK, Askins J, Hall PR, Davies DR, Segal DM. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8792. doi: 10.1073/pnas.0603245103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun J, et al. J. Biol. Chem. 2006;281:11144. doi: 10.1074/jbc.M510442200. [DOI] [PubMed] [Google Scholar]
- 14.Takada E, et al. Mol. Immunol. 2007;44:3633. doi: 10.1016/j.molimm.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 15.Jin MS, et al. Cell. 2007;130:1071. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 16.We thank Di Xia for assistance with data collection and Fred Dyda for valuable discussions. This work was supported by the intramural Research Program of the NIDDK and the NCI and by an NIH/FDA Intramural Biodefense Award from NIAID. We acknowledge the valuable assistance of the Protein Expression laboratory, Science Applications International Corp., Frederick MD, in developing protein expression systems. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38. The coordinates were deposited in the Protein Data Bank, accession codes 3CIG and 3CIY.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.