Abstract
The Skillings–Mack statistic (Skillings and Mack, 1981, Technometrics 23: 171–177) is a general Friedman-type statistic that can be used in almost any block design with an arbitrary missing-data structure. The missing data can be either missing by design, for example, an incomplete block design, or missing completely at random. The Skillings–Mack test is equivalent to the Friedman test when there are no missing data in a balanced complete block design, and the Skillings–Mack test is equivalent to the test suggested in Durbin (1951, British Journal of Psychology, Statistical Section 4: 85–90) for a balanced incomplete block design. The Friedman test was implemented in Stata by Goldstein (1991, Stata Technical Bulletin 3: 26–27) and further developed in Goldstein (2005, Stata Journal 5: 285). This article introduces the skilmack command, which performs the Skillings–Mack test.
The skilmack command is also useful when there are many ties or equal ranks (N.B. the Friedman statistic compared with the χ2 distribution will give a conservative result), as well as for small samples; appropriate results can be obtained by simulating the distribution of the test statistic under the null hypothesis.
Keywords: st0167, skilmack, Skillings–Mack, Friedman, block design, nonparametric, unbalanced, missing data, ties
1 Introduction
There are many situations where the assumption of a normally distributed error distribution is not true and hence statisticians need to apply distribution-free methods. A common model for a randomized block design is
where Yij is the response corresponding to the jth treatment in the ith block, μ is the overall mean, τj is the jth treatment effect, and βi is the ith block effect. The errors, εij, are independent and identically distributed with some continuous distribution function. Without loss of generality, let there be k treatments and n blocks, and let nij indicate whether there is an observation for the jth treatment in the ith block. The usual null hypothesis is that the treatment effects are all identical, i.e., τ1 = ⋯ = τk. The Friedman test (Friedman 1937) is the distribution-free test for a completely balanced randomized block design (nij = 1 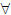 i, j). The Friedman test was implemented in Stata by Goldstein (1991) and further developed in Goldstein (2005). Durbin proposed a Friedman-type test for a balanced incomplete block design (Durbin 1951), and Skillings and Mack (1981) proposed a more general Friedman-type test for an unbalanced incomplete block design with an arbitrary missing-data structure. The missing data can be missing by design or missing completely at random. This article introduces the skilmack command, which calculates the Skillings–Mack (sm) test. skilmack computes the sm test statistic by using the following steps:
i, j). The Friedman test was implemented in Stata by Goldstein (1991) and further developed in Goldstein (2005). Durbin proposed a Friedman-type test for a balanced incomplete block design (Durbin 1951), and Skillings and Mack (1981) proposed a more general Friedman-type test for an unbalanced incomplete block design with an arbitrary missing-data structure. The missing data can be missing by design or missing completely at random. This article introduces the skilmack command, which calculates the Skillings–Mack (sm) test. skilmack computes the sm test statistic by using the following steps:
Remove any block with only one observation.
Within each block, observations are ranked from 1 to ki, where ki is the number of treatments in block i, and when ties occur ranks are averaged.
The rank for Yij will be rij, but when an observation is missing, then the value (ki + 1)/2 is used.
- Compute an adjusted treatment sum, Aj, where
As in the Friedman test, the sm test begins by reducing the data to within-block ranks. In the Friedman test, the ranks are added within each treatment; however, when there are missing data, it is necessary to center the ranks (by subtracting the average rank for that block). The ranks are then weighted by {12/(ki + 1)}1/2. This weight is chosen for two reasons: First, the range of {rij − (ki + 1)/2} is smaller for blocks with missing data, and the weights attempt to address this imbalance. Second, it produces a simple covariance structure on A1,…,Ak. To complete the calculation of the test statistic, the covariance of the treatment sums is required.
Setting A = (A1,…,Ak−1), the covariance matrix for A under the null hypothesis, H0, is given by
where λqt = λtq = (number of blocks in which both treatments q and t are observed). Then the sm statistic is given by
where is any generalized inverse for Σ0. Under H0, for large n.
There are several situations where it would be appropriate to estimate the distribution of sm values under H0, and skilmack simulates this distribution.
The χ2 distribution is inadequate when the sample size is small, and unless n is very large, it is preferable to use simulation to estimate the distribution of sm values under H0, particularly for the tail. The χ2 approximation tends to be conservative. This is especially true for significance levels of 0.01 or less.
For ties, or equal ranks, we will also want to simulate because there is less variation between the weighted sum of centered ranks, Aj, or the sum of ranks in the Friedman test, and so the χ2 distribution will give conservative p-values.
Unlike with the friedman command (Goldstein 2005), for skilmack the data are required to be in the more usual long format, i.e., one column for the outcome measure, one for the block identifier or ID, and one for the treatment or within-block repeated variable.
2 The skilmack command
2.1 Syntax
skilmack varname [if] [in], id (varname) repeated (varname) [covariance forcesims(on | off) reps(#) seed(#) notable(noties | tiescov | both)]
2.2 Options
id(varname) is required and specifies the factor variable containing the block identifiers.
repeated(varname) is required and specifies the factor variable containing the treatment identifiers.
covariance specifies that the estimated covariance matrix is used in place of the no-ties covariance matrix. The estimated covariance matrix is the sample covariance matrix of the weighted sum of centered ranks from the simulations.
forcesims(on | off) forces whether simulations are used. Simulations will be run if and only if there are ties, unless overridden by this option.
reps(#) sets the number of simulations. The default is reps(1000).
seed(#) specifies the random-number seed; time is used as the default seed. This option allows an exact replication of the Monte Carlo simulations.
notable(noties | tiescov | both) suppresses the output table produced in the no-ties section or in the case of ties when the covariance option is used (or both).
3 Example
The following example was used in Hollander and Wolfe's book on nonparametric statistical methods (Hollander and Wolfe 1999), which in turn is taken from Brady (1969). The skilmack command will be demonstrated using this dataset. (Numbers in parentheses are within-subject ranks.)
| Dysfluencies under each condition | |||
|---|---|---|---|
| ID | R | A | N |
| 1 | 3 (1) | 5 (2) | 15 (3) |
| 2 | 1 (1) | 3 (2) | 18 (3) |
| 3 | 5 (2) | 4 (1) | 21 (3) |
| 4 | 2 (1) | 6 (2) | |
| 5 | 0 (1) | 2 (2) | 17 (3) |
| 6 | 0 (1) | 2 (2) | 10 (3) |
| 7 | 0 (1) | 3 (2) | 8 (3) |
| 8 | 0 (1) | 2 (2) | 13 (3) |
The sm results from the above data are
. skilmack score, id(id) repeated(cond)
Weighted Sum of Centered Ranks
| cond | N | WSumCRank | SE | WSum/SE |
|---|---|---|---|---|
| A | 7 | −1.73 | 3.74 | −0.46 |
| N | 8 | 13.12 | 3.87 | 3.39 |
| R | 8 | −11.39 | 3.87 | −2.94 |
| Total | 0 | |||
Skillings Mack = 13.281
P-value (No ties) = 0.0013
N.B. As P-value <0.02, it is likely to be conservative (unless n large) Consider obtaining a p-value from a simulated null distribution of SM - see options.
As cautioned in the output, the p-value did turn out to be conservative, because when we typed skilmack score, id(id) repeated(cond) forcesims(on) seed(1) reps(100000), we got an empirical p-value of 0.
N.B. A large, negative WSumCRank (or WSum/SE) means a low ranking (e.g., 1) because of typically low scores. This was the case for condition R, which had the fewest dysfluencies.
An informal examination of differences in the repeated measures can be made by comparing the values of WSum/SE.
4 Simulation
A dataset is simulated by sorting on random numbers, for each individual, to randomly shuffle which data point belongs to which repeat. The sorting on random numbers is not applied where there are missing data to preserve the missing-data structure. A value of the test statistic is then calculated. This process is then repeated (default 1,000 times). The p-value is the proportion of times that randomly generated values of sm are at least as large as the value of sm from the actual data.
If there are ties (equal ranks), average ranks are assigned, e.g., 1.5, 1.5, 3. Assigning average ranks is perhaps the most common way of dealing with ties. However, one may prefer to force ranks to be randomly assigned when they are tied. (This can effectively be done by adding a small random amount to each score.)
The sm statistic can be calculated when there are ties; however, the p-value calculated from the assumed χ2 null distribution becomes more and more conservative the more ties there are. To provide a more accurate p-value, simulations are used to approximate the distribution of sm values under the null hypothesis, conditional on the particular missing-data structure and tied rankings.
With the covariance option, the sm statistic can be redefined by estimating the covariance matrix of the weighted sums of centered ranks and using this in place of the covariance matrix (which is accurate when there are no ties, but not when there are many ties). A new table is produced with different standard errors, and a new sm statistic and p-value are calculated. The tables can be suppressed by using the notable() option.
5 Friedman test dealing with ties
Because the sm test is equivalent to the Friedman test in a completely balanced design with no missing data (Skillings and Mack 1981) even when there are ties, skilmack can be used to perform the Friedman test when there are ties.
. recode score .=2 /* to create a full dataset with a tie */(score: 1 changes made)
. skilmack score, id(id) repeated(cond) seed(1) reps(10000)
Weighted Sum of Centered Ranks
| cond | N | WSumCRank | SE | WSum/SE |
|---|---|---|---|---|
| A | 8 | −2.60 | 4.00 | −0.65 |
| N | 8 | 13.86 | 4.00 | 3.46 |
| R | 8 | −11.26 | 4.00 | −2.81 |
| Total | 0 | |||
Skillings Mack = 13.562
P-value (No ties) = 0.0011
N.B. As P-value <0.02, it is likely to be conservative (unless n large). Consider obtaining a p-value from a simulated null distribution of SM - see options.
Ties exist. Above SEs and P-value approximate, if not too many ties; 24 rows of [id, score]; 23 different combinations; n(id) = 8
Consider using the p-value below, (which is found from a simulated conditional null distribution of SM - see options - simulating ...........)
Empirical P-value (Ties) ~ 0.0000
Equivalence with the Friedman test is illustrated below.
. reshape wide score, i(cond) j(id)
(note: j = 1 2 3 4 5 6 7 8)
| Data | long | −> | wide |
|---|---|---|---|
| Number of obs. | 24 | −> | 3 |
| Number of variables | 3 | −> | 9 |
| j variable (8 values) | id | −> | (dropped) |
| xij variables: | |||
| score | −> | score1 score2 ... score8 |
. friedman score*
Friedman = 13.5625
Kendall = 0.8477
P-value = 0.0011
There are several things to notice here. First, the sm statistic is the Friedman statistic, and because they have the same approximate χ2 distribution under the null hypothesis, the p-values (ignoring the issue of ties) are the same. Second, the skilmack command gives information on how many ties there are. There were 23 different combinations of (id, score) (maximum possible 24 and minimum possible 8). Because there were few ties, one could argue that there is little need to get a better approximate null distribution since it will differ only slightly from the χ2 distribution assumed in the no-ties case. However, a simulated null distribution is desirable here because the dataset and the p-value are small. The p-value conditional on the particular ties, and coming from comparison of the test statistic with the simulated null distribution, should be, and was, lower than the p-value ignoring ties and coming from the χ2 distribution. The approximate null distribution will differ each time unless the seed is set and will be more accurate with more simulations leading to a more reliable p-value.
6 Summary
The sm test does not seem to be readily available in main statistical packages. skilmack is the first Stata implementation of the sm test, which is a generalization of the Friedman test. It is a distribution-free method and can be applied to a variety of situations; for example, it can be used to analyze an incomplete block design. The sm test is equivalent to the Friedman test when there are no missing data. The simulations option allows a more theoretically correct p-value to be estimated when ties exist in the dataset, and this can be used regardless of whether there are missing data.
References
- Brady JP. Studies on the metronome effect on stuttering. Behaviour Research and Therapy. 1969;7:197–204. doi: 10.1016/0005-7967(69)90033-3. [DOI] [PubMed] [Google Scholar]
- Durbin J. Incomplete blocks in ranking experiments. British Journal of Psychology, Statistical Section. 1951;4:85–90. [Google Scholar]
- Friedman M. The use of ranks to avoid the assumption of normality implicit in the analysis of variance. Journal of the American Statistical Association. 1937;32:675–701. [Google Scholar]
- Goldstein R. snp2: Friedman's ANOVA & Kendall's coefficient of concordance. Stata Technical Bulletin. 1991;3:26–27. Reprinted in Stata Technical Bulletin Reprints, vol. 1, pp. 157–158. College Station, TX: Stata Press. [Google Scholar]
- Goldstein R. Software update: snp2 1: Friedman's ANOVA test and Kendall's coefficient of concordance. Stata Journal. 2005;5:285. [Google Scholar]
- Hollander M, Wolfe DA. Nonparametric Statistical Methods. 2nd ed. Wiley; New York: 1999. [Google Scholar]
- Skillings JH, Mack GA. On the use of a Friedman-type statistic in balanced and unbalanced block designs. Technometrics. 1981;23:171–177. [Google Scholar]


