Summary
We previously demonstrated that the cell-surface lipoprotein MalE contributes to GAS maltose/maltodextrin utilization, but MalE inactivation does not completely abrogate GAS catabolism of maltose or maltotriose. Using a genome-wide approach, we identified the GAS phosphotransferase system (PTS) responsible for non-MalE maltose/maltotriose transport. This PTS is encoded by an open reading frame (M5005_spy1692) previously annotated as ptsG based on homology with the glucose PTS in Bacillus subtilis. Genetic inactivation of M5005_spy1692 significantly reduced transport rates of radiolabeled maltose and maltotriose, but not glucose, leading us to propose its reannotation as malT for maltose transporter. The ΔmalT, ΔmalE, and ΔmalE:malT strains were significantly attenuated in their growth in human saliva and in their ability to catabolize α-glucans digested by purified human salivary α-amylase. Compared to wild-type, the three isogenic mutant strains were significantly impaired in their ability to colonize the mouse oropharynx. Finally, we discovered that the transcript levels of maltodextrin utilization genes are regulated by competitive binding of the maltose repressor MalR and catabolite control protein A. These data provide novel insights into regulation of the GAS maltodextrin genes and their role in GAS host-pathogen interaction, thereby increasing the understanding of links between nutrient acquisition and virulence in common human pathogens.
Keywords: Streptococcus, maltodextrin, transport, amylase, pharyngitis
Introduction
The majority of studies of basic metabolism in bacteria have occurred in two model organisms: Escherichia coli for Gram-negative microbes and Bacillus subtilis for Gram-positives (Deutscher et al., 2006; Hueck and Hillen, 1995; Schonert et al., 2006). However, the proliferation of fully sequenced bacterial genomes has revealed substantial variability between the model organisms and human pathogens in genes putatively devoted to fundamental physiologic processes (Blattner et al., 1997; Holden et al., 2004; Kunst et al., 1997; Parkhill et al., 2001). The sequencing of numerous bacterial genomes also has led to the development of tools for genome-wide investigation of gene transcript levels under various conditions (Rhodius and LaRossa, 2003; Shelburne and Musser, 2004). Transcriptome analyses during bacterial infection in vivo have repeatedly demonstrated highly dynamic transcript levels of genes involved in complex carbohydrate metabolism (Andes et al., 2005; Faucher et al., 2006; Graham et al., 2006; Talaat et al., 2007; Virtaneva et al., 2005). Such findings have led to a recent, dramatic increase in research examining relationships between carbohydrate catabolism and pathogenesis in many microbes (Iyer and Camilli, 2007; Loughman and Caparon, 2006; Munoz-Elias and McKinney, 2005; Tchawa Yimga et al., 2006). For Gram-positive pathogens, a more complete understanding of the relationship between carbohydrate catabolism and bacterial pathogenesis in humans is limited by a relative lack of knowledge of the metabolic pathways in such pathogens, especially in light of the emerging differences with B. subtilis.
Group A Streptococcus is the leading bacterial cause of pharyngitis, with some 15 million cases annually in the United States alone (Brook and Dohar, 2006). The presence of GAS in the human oropharynx is a prerequisite for developing rheumatic fever, which remains endemic in the developing world (Carapetis et al., 2005). Recently, we have been studying the interaction of GAS with human saliva, a major mediator of innate and acquired immunity in the human oropharynx (Shelburne et al., 2005a; Shelburne et al., 2005b). Several of our investigations have focused on GAS catabolism of maltodextrins, which are multiple α-D-glucose units linked by (1→4) glycosidic linkages with two linked glucose called maltose, three maltotriose, four maltotetraose, etc (Shelburne et al., 2006; Shelburne et al., 2007a; Shelburne et al., 2007b). Because of the typical human diet, maltodextrins are present at high quantities in the human oropharynx where they may serve as a key nutrient source for pathogenic and non-pathogenic microbes (Kaczmarek and Rosenmund, 1977; Mormann and Muhlemann, 1981; Scannapieco et al., 1993). Our initial investigation of GAS maltodextrin catabolism demonstrated that MalE, a cell surface maltodextrin-binding lipoprotein that is part of an ATP-binding cassette (ABC) transport system, contributes to GAS colonization of the mouse oropharynx (Shelburne et al., 2006). MalE is important for the growth of GAS in all maltodextrins studied to date, but MalE inactivation did not completely abolish the growth of GAS in a medium where maltose or maltotriose were the sole carbon source (Shelburne et al., 2006; Shelburne et al., 2007a). Thus, there must be a second mechanism in GAS responsible for the remainder of non-MalE maltose and maltotriose transport. However, the identity of this transporter and its role in host-pathogen interaction are unknown.
In this investigation we sought to identify the molecular basis of non-MalE maltose and maltotriose transport in GAS, to define the relative contribution of the GAS maltodextrin transporters to host-pathogen interaction, and to elucidate the regulatory mechanisms of GAS maltodextrin utilization. We have found that relying solely on similarities with B. subtilis in assigning function to a putative GAS carbohydrate transporter obscured the identity of the second GAS maltodextrin transporter. Moreover, we discovered that the GAS maltodextrin transporters participate in the catabolism of polysaccharides digested by human salivary α-amylase, providing a potential mechanism by which maltodextrin transport contributes to the ability of GAS to colonize the oropharynx. Finally, we have elucidated a novel transcriptional regulatory pathway for the GAS maltodextrin utilization genes involving catabolite control protein A (CcpA) and the maltose repressor MalR. The data presented herein add to our understanding of the molecular basis of GAS maltodextrin catabolism and suggest future studies for human genetic susceptibility to GAS pharyngitis.
Results
Transcript level analysis of putative carbohydrate transport systems during growth in various media
In a previous investigation, we showed that inactivation of the maltodextrin binding protein MalE significantly decreased the growth of the serotype M1 strain MGAS5005 in a maltose- or maltotriose-medium (Shelburne et al., 2006; Shelburne et al., 2007a). However, the ability of the ΔmalE isogenic mutant strain to catabolize maltodextrins was not completely abolished (Shelburne et al., 2007a). Bioinformatic analysis of the 12 fully sequenced GAS genomes did not reveal another putative maltodextrin transport system (Banks et al., 2004; Beres et al., 2002; Beres et al., 2006; Ferretti et al., 2001; Green et al., 2005; Holden et al., 2007; Nakagawa et al., 2003; Smoot et al., 2002). To gain insight into what genes in addition to malE might be involved in GAS maltodextrin transport, we determined the transcript levels during growth in various media of genes encoding the 17 known and putative carbohydrate transport systems present in all fully sequenced GAS strains (Table 1). Media tested included a standard laboratory medium (THY), glucose-medium, maltose-medium, and human saliva. We reasoned that the second maltodextrin transport system would have a similar transcript level pattern to malE. As determined by TaqMan real-time QRT-PCR, four main patterns of gene expression emerged: (1) low transcript levels in all media; (2) high transcript levels in all media; (3) low transcript levels in all media except human saliva; and (4) low transcript levels in THY and glucose medium and high transcript levels in maltose-medium and human saliva (Fig. 1). The final transcript level pattern was observed for M5005_spy1058 (malE) and M5005_spy1692, which has been annotated as ptsG due to 34% identity and 54% similarity at the amino acid level with the glucose phosphotransferase system (PTS) of B. subtilis subsp. subtilis strain 168 (Gonzy-Treboul et al., 1991). Given that genes involved in utilization of a particular carbohydrate are often highly expressed in its presence, these data suggest the M5005_spy1692 might participate in GAS maltodextrin transport.
Table 1.
Known and putative carbohydrate transport systems in the serotype M1 strain MGAS5005.
| Gene number(s) in strain MGAS5005 | Putative carbohydrate transported | Type of carbohydrate transport system |
|---|---|---|
| spy0148-spy0150 | 3-keto-L-gulonate | PTSa |
| spy0213-spy0215 | sialic acid | ABCb |
| spy0475 | β-glucoside | PTS |
| spy0519-0521 | N-acetylgalactosamine | PTS |
| spy0662 | fructose | PTS |
| spy0780-spy0783 | mannose/fructose | PTS |
| spy1058-spy1060 | maltodextrinsc | ABC |
| spy1063, spy1064, spy1067d | cyclomaltodextrins | ABC |
| spy1079-spy1083 | cellobiose | PTS |
| spy1308-spy1310 | unknown | ABC |
| spy1399-spy1401 | galactose | PTS |
| spy1479-spy1481 | mannose | PTS |
| spy1542 | sucrose | PTS |
| spy1633-spy1634 | lactose | PTS |
| spy1663-spy1664 | mannitol | PTS |
| spy1692 | glucose | PTS |
| spy1744-spy1746 | cellobiose | PTS |
| spy1784 | trehalose | PTS |
PTS, phosphotransferase system
ABC, ATP-binding cassette
experimentally verified to transport indicated substrate in GAS
orthologs not present in all GAS strains sequenced to date
Fig. 1.
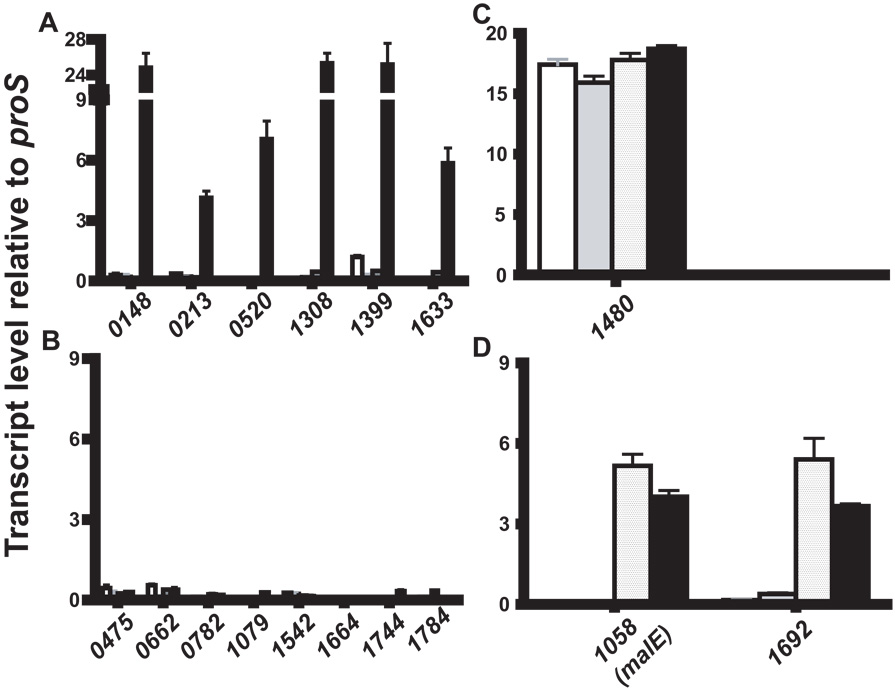
Transcript level analysis of known and putative carbohydrate transport systems in GAS. Strain MGAS5005 was grown to mid-exponential phase in either standard laboratory media (THY, white bars), a chemically defined medium (CDM) containing either 0.5% (wt/vol) glucose (grey bars) or 0.5% maltose (textured bars) or human saliva (black bars). TaqMan real-time QRT-PCR was performed using probe and primers listed in Table S2. The transcript levels of target genes indicated by their M5005_spy gene numbers on the X-axis are shown relative to those of proS, a gene expressed constitutively throughout the GAS cell cycle and whose transcript levels are similar whether grown in THY or saliva (Shelburne et al., 2005a; Virtaneva et al., 2003). (A) Genes whose transcript levels were elevated in human saliva compared to other media; (B) Genes whose transcript levels were low in all media; (C) Genes whose transcript levels were high in all media; (D) Genes whose transcript levels were high in maltose-medium and human saliva. The two genes with the final pattern included malE, which was previously demonstrated to participate in maltodextrin catabolism, and M5005_spy1692 (Shelburne et al., 2007a). Transcript levels are presented as the mean ± standard deviation of four independent experiments done on two separate occasions.
Analysis of the M5005_spy1692 genome region and homology in related organisms
Microbial genes encoding enzymes involved in carbohydrate metabolism are often part of an operon of genes involved in both the transport and subsequent catabolism of the carbohydrate of interest (Deutscher et al., 2006). To gain insight into the role of M5005_spy1692, we examined the gene content surrounding M5005_spy1692 in fully sequenced GAS strains as well as the gene content surrounding orthologs of M5005_spy1692 in related organisms. M5005_spy1692 is present in all fully sequenced GAS strains and is predicted to be co-transcribed with a previously unstudied protein, M5005_spy1691, which has been annotated as protein of unknown function (Fig. S1A) (Banks et al., 2004; Beres et al., 2002; Beres et al., 2006; Ferretti et al., 2001; Green et al., 2005; Holden et al., 2007; Nakagawa et al., 2003; Smoot et al., 2002). There are no genes in the immediate vicinity of M5005_spy1692 encoding proteins either known to be or putatively involved in carbohydrate transport or metabolism. M5005_spy1692 has 61% identity and 77% similarity at the amino acid level to a previously unstudied putative PTS in B. anthracis that is not present in the fully sequenced B. subtilis subsp. subtilis strain 168 (Fig. S1B). Orthologs of M5005_spy1692 are present in fully sequenced Streptococcus, Enterococcus, and Clostridium species (Ajdic et al., 2002; Paulsen et al., 2003; Sebaihia et al., 2007). Among available genomes, orthologs of M5005_spy1692 are colocalized with orthologs of M5005_spy1691, whereas none of the other GAS genes in the M5005_spy1692 vicinity are universally present. As noted previously, M5005_spy1692 has similarity to ptsG in B. subtilis. However, none of the genes co-transcribed with ptsG in B. subtilis are found near M5005_spy1692 in GAS (Fig. S1C). Finally, in B. subtilis, maltose uptake occurs via the MalP PTS (Schonert et al., 2006). M5005_spy1692 has 24% identity and 42% similarity at the amino acid level to MalP in B. subtilis subsp. subtilis strain 168, and there is no overlap in the surrounding gene regions (Fig. S1D). We conclude that the gene regions surrounding M5005_spy1692 do not provide insight into its cognate carbohydrate, that M5005_spy1691 and M5005_spy1692 may function in tandem in GAS and other Gram-positive organisms, and that M5005_spy1692 is unlikely to fulfill the same function as ptsG in B. subtilis.
Conservation of M5005_spy1692 among diverse GAS strains
To determine whether M5005_spy1692 is present among assorted GAS strains and to assess its allelic diversity, we performed PCR for M5005_spy1692 followed by sequencing in 73 GAS strains of diverse geographic and temporal origins comprising 24 M serotypes (Table S1). A PCR product was obtained from all tested strains. We identified 257 unique single nucleotide polymorphisms (SNPs) leading to 27 gene alleles with a mean number of 51.8 SNPs in the 2187 bp open reading (Table S1, Fig. S2). We identified no insertions, deletions, or nonsense mutations. The average of one SNP every 42 bps in the M5005_spy1692 ORF is a significantly (P < 0.001) higher rate of nucleotide change compared to the previously identified genome-wide average of one SNP every 115 bp among fully sequenced GAS strains (Beres et al., 2006). The ratio of synonymous to non-synonymous nucleotide substitution in orthologs of M5005_spy1692 was 2.83 ± 0.83 which is not statistically different (P = 0.423) from previous genome-wide ratios for fully-sequenced GAS strains (Beres et al., 2006). These data demonstrate that M5005_spy1692 is present among diverse GAS strains and has a similar genetic selection profile to the remainder of the GAS genome.
M5005_spy1692 participates in maltodextrin catabolism
Our transcript level analysis suggested that M5005_spy1692 may transport maltodextrins. To determine whether M5005_spy1692 participates in maltodextrin catabolism, we replaced M5005_spy1692 with a spectinomycin cassette via non-polar insertional mutagenesis to create strain ΔM5005_spy1692 from the parental serotype M1 strain MGAS5005 (Table 2, confirmatory Southern blot shown in Fig. S3). Given that we previously identified MalE as participating in maltose transport, we also inactivated M5005_spy1692 in strain ΔmalE to create a strain in which both MalE and MalT were inactivated. Finally, we genetically complemented the ΔM5005_spy1692 strain by providing M5005_spy1692 in trans using a plasmid capable of replicating in GAS (see Experimental Procedures). No difference was observed between wild-type, mutant, or complemented mutant strains when grown in THY or glucose-medium (Fig. 2A, B, Table 3). When grown in maltose-medium, the ΔmalE and ΔmalT strains had a prolonged (slower) doubling time and lower final concentration compared to the parental strain, whereas the ΔmalE:malT strain was unable to grow in maltose-medium (Fig. 2C). In a maltotriose-medium, the ΔmalE strain had a significantly lower growth rate in maltotriose-medium whereas the ΔmalE:malT strain again evidenced no growth (Fig. 2D). These data indicate that M5005_SPy1692 participates in the catabolism of maltose and maltotriose, although its role appears to be ancillary to that of MalE and that inactivation of both MalE and M5005_SPy1692 abolishes GAS growth in maltodextrin media.
Table 2.
Strains and plasmids
| Strain or plasmid | Description | Genotype | Source or reference |
|---|---|---|---|
| Strains | |||
| MGAS5005 | Clinical invasive isolate | Wild-type | (Sumby et al., 2005) |
| ΔmalE | malE non-polar mutant | MGAS5005 malE::spc | (Shelburne et al., 2006) |
| compΔmalE | complemented malE mutant | ΔmalE/malE+; Cmr | (Shelburne et al., 2006) |
| ΔmalR | malR non-polar mutant | MGAS5005 malR::spc | (Shelburne et al., 2007b) |
| ΔccpA | ccpA non-polar mutant | MGAS5005 ccpA::spc | (Shelburne et al., 2008) |
| compΔccpA | complemented ccpA mutant | ΔccpA/ccpA+, Cmr | (Shelburne et al., 2008) |
| ΔmalT | malT non-polar mutant | MGAS5005 malT::erm | This study |
| compΔmalT | complemented malT non-polar mutant | ΔmalT/malT+; Cmr | This study |
| pDC123ΔmalT | malT mutant with empty vector | ΔmalT/pDC123+; Cmr | This study |
| ΔmalE:malT | malE/malT double mutant | MGAS5005 malE::spc malT::erm | This study |
| ΔM5005_spy1691 | M5005_spy1691 mutant | MGAS5005 M5005_spy1691:spc | This study |
| compΔM5005_spy1691 | complemented M5005_spy1691 mutant | ΔM5005_spy1691/M5005_spy1691+; Cmr | This study |
| Plasmids | |||
| pDC123 | Chloramphenicol resistance vector | Cmr | (Chaffin and Rubens, 1998) |
| pDCmalE | malE expression vector | Cmr | (Shelburne et al., 2006) |
| pDCccpA | ccpA expression vector | Cmr | (Shelburne et al., 2008) |
| pDCmalT | malT expression vector | Cmr | This study |
| pDCM5005_spy1691 | M5005_spy1691 expression vector | Cmr | This study |
Fig. 2.
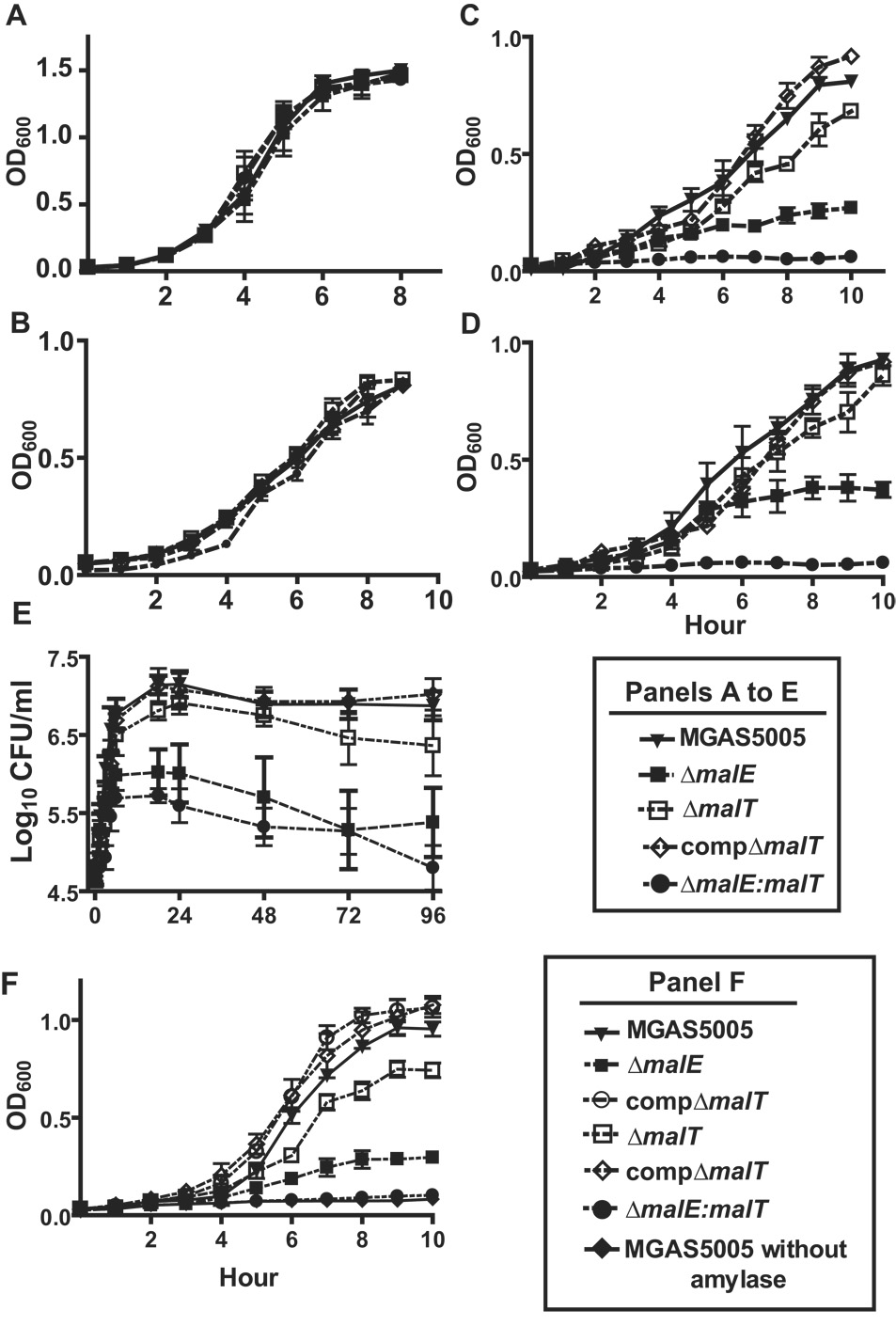
Growth of indicated GAS strains under various conditions. OD600 readings were taken at indicated times for growth in nutrient-rich medium (THY) and chemically defined medium (CDM) with 0.5% (wt/vol) of indicated carbohydrate. Growth in human saliva was monitored using CFU as previously described (Shelburne et al., 2005b). Growth media were: (A) THY; (B) glucose-medium; (C) maltose-medium; (D) maltotriose-medium; (E) human saliva; (F) starch-medium with purified human salivary α-amylase at 200 U/mL. Data graphed are mean values ± standard deviation for five independent experiments done on two separate occasions.
Table 3.
Growth of GAS strains in various media.
| Doubling time (min)a | ||||||
|---|---|---|---|---|---|---|
| Strain | THYb | Glucosec | Maltosec | Maltotriosec | Maltotetraose | Human saliva |
| MGAS5005 | 47.2 ± 4.2 | 57.3 ± 5.1 | 72.5 ± 3.7 | 58.5 ± 4.7 | 61.3 ± 4.7 | 67.2 ± 5.7 |
| ΔmalE | 46.2 ± 4.6 | 55.3 ± 3.7 | 92.1 ± 4.9d | 81.9 ± 5.1d | N.D.e | 92.1 ± 8.7 d |
| compΔmalE | 48.3 ± 5.9 | 60.3 ± 6.1 | 74.5 ± 4.6 | 61.2 ± 6.1 | 64.1 ± 5.7 | 73.1 ± 7.2 |
| ΔmalT | 44.2 ± 5.1 | 56.2 ± 5.2 | 78.5 ± 4.1d | 63.3 ± 3.8 | 62.5 ± 5.1 | 76.6 ± 6.5d |
| compΔmalT | 49.6 ± 5.2 | 61.2 ± 5.4 | 71.2 ± 4.9 | 61.6 ± 4.2 | 64.3 ± 4.7 | 71.2 ± 4.5 |
| ΔmalE:malT | 45.1 ± 4.9 | 54.3 ± 3.9 | N.D.e | N.D.e | N.D.e | 102.3 ± 5.2d |
| ΔM5005_spy1691 | 46.3 ± 3.9 | 58.1 ± 4.9 | 73.1 ± 5.2 | 83.7 ± 5.9d | 78.3 ± 5.2d | 95.6 ± 6.2 |
| compΔM5005_spy1691 | 49.1 ± 2.7 | 59.9 ± 3.2 | 74.9 ± 4.1 | 62.2 ± 5.7 | 65.7 ± 7.2 | 72.5 ± 5.9 |
Doubling-time determined by non-linear regression during exponential growth phase.
Todd-Hewitt broth with 0.2% yeast extract.
Chemically-defined medium with indicated carbohydrate added at 0.5% concentration.
P < 0.05 compared to parental strain MGAS5005
Doubling time could not be determined because of inadequate growth.
M5005_SPy1692 mediates transports of maltose and maltotriose
Our growth data suggested that M5005_SPy1692 participates in the catabolism of maltose, but not glucose. We tested the hypothesis that M5005_SPy1692 participates in the transport of maltodextrins, but not glucose, by determining the uptake of radiolabeled carbohydrates among the various GAS strains. We observed no difference in the uptake of glucose for the wild-type, ΔmalE, ΔmalT, ΔmalE:malT, or complemented mutant strains (Table 4). However, the Km was higher and vmax lower for maltose and maltotriose in the ΔmalE and ΔmalT strains compared to wild-type indicating less solute transport in the isogenic mutant strains (Table 4). There were no significant differences between the complemented mutant strains and wild-type in the uptake of either maltose or maltotriose. There was no detectable uptake of either maltose or maltotriose in the ΔmalE:malT strain so Km and vmax of maltodextrins could not be calculated for this strain. These findings support the hypothesis that M5005_SPy1692 participates in the uptake of maltose and maltotriose, leading us to hereafter refer to M5005_SPy1692 as MalT for maltose transporter.
Table 4.
Transport of 14C labeled sugars by group A Streptococcus
| Apparent Km(µM)a/vmaxb | |||
|---|---|---|---|
| Strain | Glucose | Maltose | Maltotriose |
| MGAS5005 | 4.3/131 | 9.7/83 | 3.3/119 |
| ΔmalE | 4.1/136 | 13.9c/62c | 16.5c/31c |
| compΔmalE | 5.1/142 | 8.5/72 | 2.5/129 |
| ΔmalT | 4.7/122 | 26.3c/37c | 6.7c/95c |
| compΔmalT | 4.8/118 | 8.4/91 | 3.1/129 |
| ΔmalE:malT | 5.1/114 | N.D.d | N.D.d |
| ΔM5005_spy1692 | 4.8/126 | 10.3/91 | 4.1/110 |
| compΔM5005_spy1692 | 3.9/115 | 9.8/87 | 4.7/105 |
Apparent Km were determined assuming that transport follows Michaelis-Menton kinetics.
vmax = nmoles/min/1010 CFU
P < 0.05 compared with parental strain MGAS5005
N.D. = Not done as transport of maltose or maltotriose was not sufficient in strain ΔmalE:malT to calculate Km or vmax
MalT and MalE are needed for optimal growth of GAS in human saliva and the catabolism of α-glucans digested by human salivary α-amylase
To begin to study the role of MalT in host-pathogen interaction, we tested the ability of the wild-type, ΔmalE, ΔmalT, and ΔmalE:malT strains to grow in human saliva. Consistent with previously published data, we observed a significant decrease in the ability of strain ΔmalE to proliferate in human saliva (Fig. 2E). Similarly, inactivation of MalT significantly decreased the ability of GAS to grow in human saliva, whereas the ΔmalE:malT strain had severely attenuated growth. The complemented mutant strains showed no significant growth difference compared to wild-type (Fig. 2E, Table 3).
GAS is unable to catabolize α-glucans, such as starch and glycogen, which are present at high concentrations in the human oropharynx (unpublished data). Given that human salivary α-amylase degrades α-glucans mainly to maltodextrins, we hypothesized that MalE and MalT would be needed for optimal catabolism of α-glucans digested by human salivary α-amylase. Similar to our findings in human saliva, the ΔmalE:malT strain showed a marked growth defect in the starch/amylase growth medium (Fig. 2F). The ΔmalE strain grew significantly more slowly and to a lower final density compared to the ΔmalT strain whereas both strains grew significantly less compared to the wild-type parental strain. Complementation of the mutant strains restored growth to wild-type levels. There was no growth in the starch medium when human salivary α-amylase was not added. We found similar growth patterns in a glycogen/amylase medium compared to the starch/amylase medium (data not shown). Therefore, we conclude that MalT and MalE are needed for optimal growth of GAS in human saliva and for GAS to catabolize α-glucans degraded by human salivary α-amylase but that MalE is the predominant contributor to growth in both situations.
MalT and MalE participate in GAS colonization of the mouse oropharynx
We previously demonstrated that MalE contributed to GAS colonization of the mouse oropharynx (Shelburne et al., 2006). Therefore, we next sought to determine whether MalT participates in GAS colonization of the mouse oropharynx and to determine the relative contribution of MalE and MalT to GAS oropharyngeal colonization. To this end, groups of 25 adult outbred CD-1 mice were challenged intranasally with ~ 1.0 × 107 CFU GAS in a total of 100 µl (50 µl per nostril). The oropharynx of each mouse was then swabbed daily using a fine tip, sterile cotton applicator (Fisher), which was then used to inoculate a BSA plate. Over time, significantly more mice were colonized with strain MGAS5005 and the compΔmalT strain compared to strain ΔmalT (Fig. 3A). Similar to our previous findings, mice infected with the ΔmalE strain were also colonized at a lower level compared to the parental strain (Shelburne et al., 2006). Of the three isogenic mutant strains, the ΔmalE:malT strain was the most significantly attenuated in its ability to colonize mice. Moreover, at each time point tested, the number of GAS CFU was significantly higher in the animals infected with the wild-type and complemented strains compared to the isogenic mutant strains (Fig. 3B). These data support our hypothesis that MalT and MalE are involved in colonization of the mouse oropharynx, and that when both are inactivated, GAS colonization is severely affected.
Fig. 3.
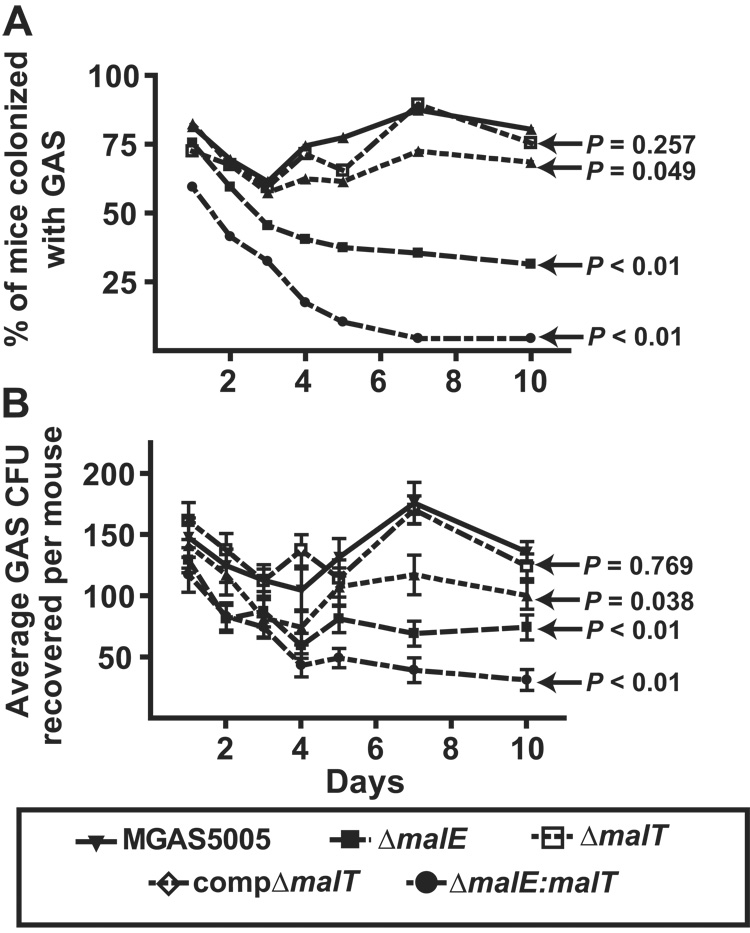
Colonization and colony forming unit (CFU) recovery rates among mice infected with GAS. Adult outbred CD-1 mice (25 per group) were inoculated with 1 × 107 CFU of indicated GAS strains. Mice oropharynx were swabbed daily onto BSA. Plates were incubated for 24 hrs, and β-hemolytic colonies were counted and tested for GAS carbohydrate antigen using latex agglutination. (A) % of mice with GAS isolated from oropharynx by day; (B) average number of GAS CFU per mouse isolated by day. Data graphed are mean values ± standard deviation. P values refer to repeated measures analysis of indicated strains compared to parental wild-type strain MGAS5005.
malT and malE are expressed during human pharyngitis
Inasmuch as malT and malE contribute to the growth of GAS in human saliva ex vivo and in the ability of GAS to colonize the mouse oropharynx, we hypothesized that malT and malE are transcribed and expressed in humans with GAS pharyngitis. We tested this hypothesis by using TaqMan real-time PCR to assay malT and malE gene transcript levels in RNA isolated from the throat swabs of 6 patients with GAS pharyngitis (Virtaneva et al., 2003). Transcripts of malT and malE were present in all 6 specimens at levels ~ 1.5 fold greater than the control gene proS (Fig. 4). Therefore, we conclude that malT and malE are actively transcribed during GAS pharyngitis in humans.
Fig. 4.
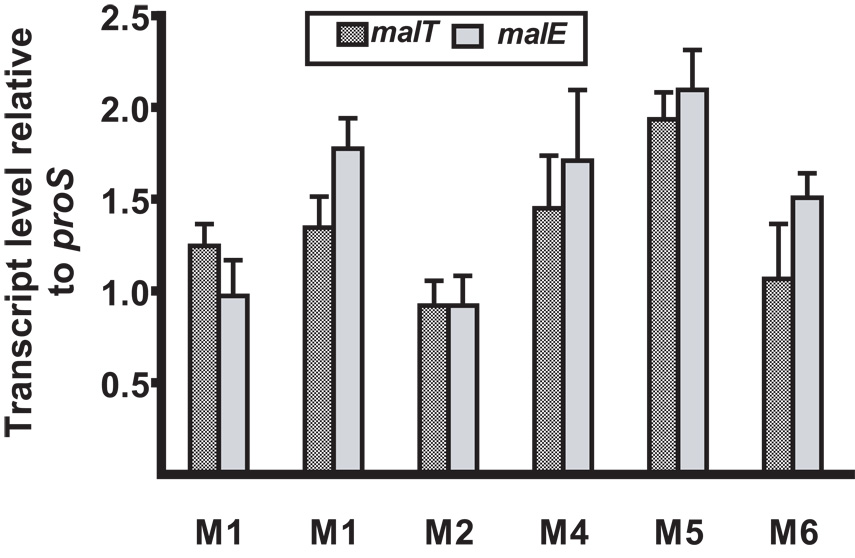
Presence of malT and malE transcripts in vivo during pharyngitis in humans. malT and malE transcript levels for 6 patients with GAS pharyngitis were determined by TaqMan real-time PCR. X-axis label indicate the M serotype of the infecting GAS strains. Column height indicates the median level of the indicated gene compared to the endogenous GAS control gene proS (43). Transcript levels are presented as the mean ± standard deviation of four independent experiments done on two separate occasions with samples analyzed in triplicate.
M5005_spy1691 contributes to maltodextrin catabolism
As indicated earlier, in fully sequenced bacterial genomes orthologs of GAS MalT are found adjacent to an unstudied open reading frame that encodes a 272 amino-acid protein (M5005_spy1691) in strain MGAS5005. BLASTn and BLASTp searches of M5005_spy1691 revealed no significant homology with a gene or protein of known function, whereas analysis of predicted protein function suggested that M5005_spy1691 belongs to the endonuclease/exonuclease/phosphatase protein family. M5005_spy1691 does not contain a predicted transmembrane domain or an N-terminal secretion signal sequence suggesting that it likely has a cytoplasmic location. Given that M5005_spy1691 appears to be co-transcribed with malT, we tested the hypothesis that it participates in maltodextrin metabolism by insertionally inactivating M5005_spy1691 to create the ΔM5005_spy1691 strain. There was no significant difference in the growth of strain ΔM5005_spy1691 versus strain MGAS5005 in either THY or a glucose medium (Table 3). We also observed no difference in the ability of the ΔM5005_spy1691 strain to grow in a maltose medium compared to its parental wild-type strain. However, compared to wild-type, the ΔM5005_spy1691 strain had an increased (slower) doubling-time and reached a lower final density in maltotriose and maltotetraose media. The uptake of radiolabeled glucose, maltose, or maltotriose was not significantly different in the ΔM5005_spy1691 strain compared to strain MGAS5005 (Table 4). Therefore, we conclude that M5005_spy1691 likely participates in the intracellular processing of maltotriose and longer maltodextrins to utilizable energy sources.
malT and M5005_spy1691 transcript levels are influenced by MalR
In a previous study we demonstrated that the putative transcriptional regulator MalR negatively influences the transcript level of genes involved in maltose/maltodextrin catabolism (Shelburne et al., 2007b). Therefore, we hypothesized that MalR similarly influences the transcript levels of MalT and M5005_spy1691. Indeed, the transcript levels of malT and M5005_spy1691 were negatively influenced by MalR, being higher in the ΔmalR strain compared to its parental wild-type strain MGAS5005 (Fig. 5A). There was no statistically significant difference in the transcript level of malT compared with M5005_spy1691, consistent with the hypothesis that the two genes are co-transcribed. Similar to previous findings regarding genes influenced by MalR, we observed significant increases in the transcript level of malT and M5005_spy1691 in the ΔmalR strain during the late-exponential phase of growth in saliva compared to THY or maltotriose-medium (Fig. 5B) (Shelburne et al., 2007b). These data indicate that malT and M5005_spy1691 are influenced by MalR in a similar fashion to other GAS genes involved in maltodextrin catabolism.
Fig. 5.
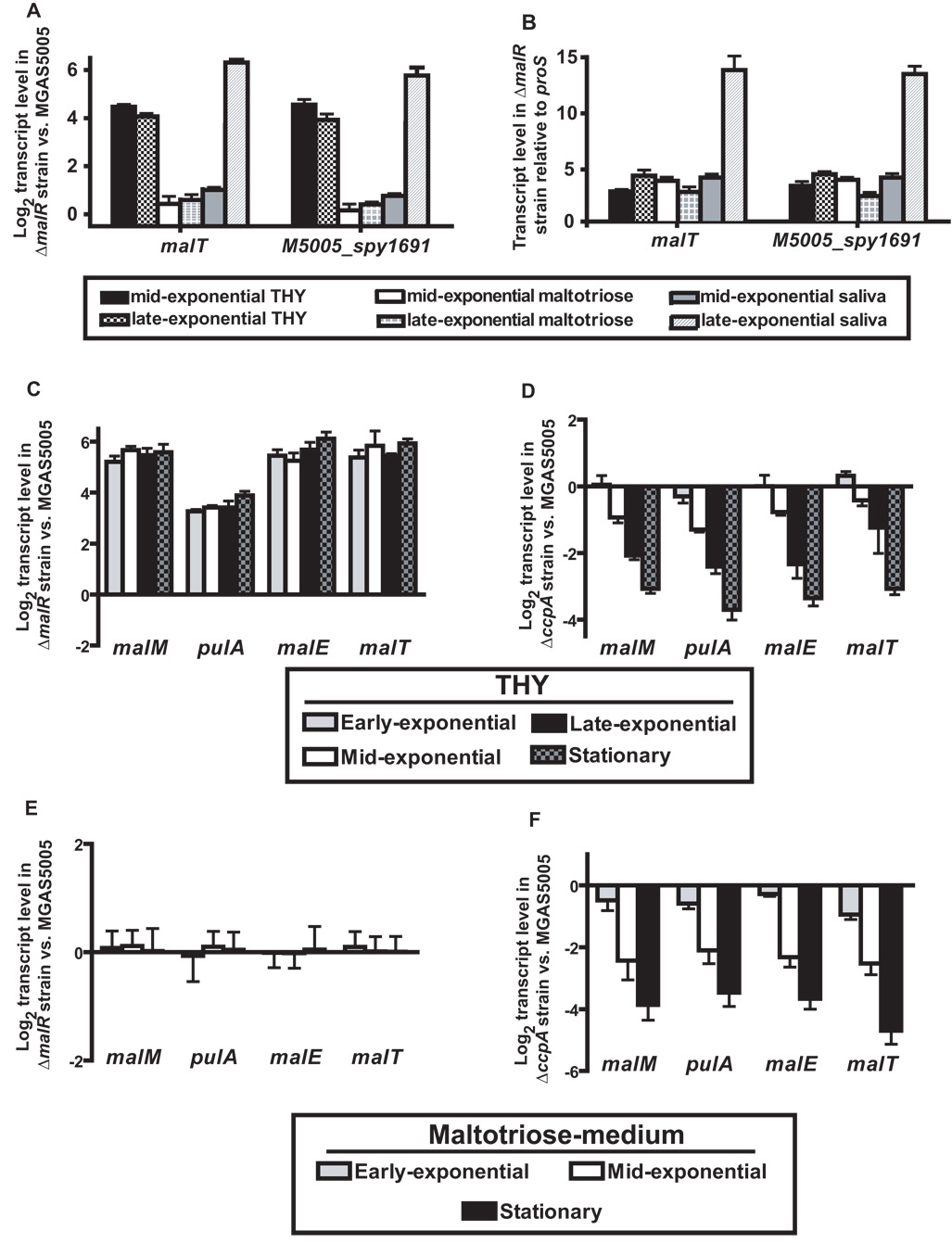
Influence of MalR and CcpA on maltodextrin utilization genes. Strain MGAS5005 and its isogenic ΔmalR or ΔccpA derivatives were grown to indicated growth phases in standard laboratory media (THY), a maltotriose-medium, or human saliva. TaqMan real-time QRT-PCR was performed using probe and primers listed in Table S2. The transcript levels of target genes were normalized to those of proS, a gene expressed constitutively throughout the GAS cell cycle and whose transcript levels are similar during growth in THY or saliva (Shelburne et al., 2005a; Virtaneva et al., 2003). (A) malT and M5005_spy1691 transcript levels are expressed as log2 of the fold difference in the ΔmalR versus wild-type strain (ΔΔCT method), therefore positive values indicates higher transcript levels in the ΔmalR strain. (B) Gene transcript levels in the ΔmalR strain relative to proS (ΔCT method). (C) and (D) Log2 fold difference of maltodextrin utilization genes for ΔmalR (C) and ΔccpA (D) strains compared to wild-type during growth in THY. (E) and (F) Log2 fold difference of maltodextrin utilization genes for ΔmalR (E) and ΔccpA (F) strains compared to wild-type during growth in maltotriose-medium. Data are presented as the mean ± standard deviation of quadruplicate measurements done on two separate occasions with samples analyzed in triplicate.
Putative catabolite response elements are present in the promoter regions of GAS maltodextrin utilization genes
MalR is a member of the LacI-GalR family of transcriptional regulators (Puyet et al., 1993). The LacI-GalR family also includes catabolite control protein A (CcpA), a key regulator of carbohydrate utilization in GAS and other Gram-positive organisms (Abranches et al., 2008; Almengor et al., 2007; Shelburne et al., 2008; Sonenshein, 2007). CcpA affects transcription by binding to cis-acting catabolite response element (cre) sites for which consensus sequences have been determined in Bacillus species (WTGNAANCGNWNNCWW where W = A or T and N = any base; Miwa et al., 2000; Weickert and Chambliss, 1990). The crystal structure of CcpA shows that particular amino acids contribute to the base specificity of the cre binding sequence (Schumacher et al., 2004). The amino acid sequences of the DNA binding domains of GAS MalR and GAS CcpA are highly conserved, including the key DNA interacting residues R21 and L55, suggesting that MalR might interact with the same cre sites as CcpA (Fig. S4). Potential cre sites are present in the promoters of the GAS maltodextrin utilization genes malM (encodes a putative glucanotransferase), malE, pulA (encodes a pullulanase), and malT (Fig. S5). To determine the location of the putative cre sites relative to transcription initiation, we determined the transcription start sites for the four genes using 5’rapid amplification of cDNA ends (RACE, see Experimental Methods). We found that the putative cre sites are located upstream of the transcription initiation sites for each of the four genes (Fig. S6). Thus, we conclude that there are putative cre sites in the promoter regions of the maltodextrin utilization genes and that these DNA sequences may serve as a binding site for CcpA and MalR.
Inactivation of CcpA and MalR produce opposing effects on maltodextrin utilization gene transcript levels
To begin to investigate the role of MalR and CcpA in the regulation of GAS maltodextrin utilization genes, we measured the transcript levels of malM, malE, pulA, and malT in the wild-type and ΔmalR strains at four stages of growth in THY. At each time point, the transcript levels of all four genes were significantly increased in the ΔmalR strain consistent with the notion that MalR represses transcription during growth in a nutrient-rich medium (Fig. 5C). Conversely, inactivation of CcpA led to a decrease in the transcript level of the maltodextrin utilization genes indicating that CcpA positively influences the transcript level of the maltodextrin utilization genes under nutrient-rich conditions (Fig. 5D). When grown in a maltotriose-medium, there were no significant differences in maltodextrin gene transcript levels between the wild-type and ΔmalR strains, in accordance with the hypothesis that maltodextrins induce release of MalR from its DNA binding site (Fig. 8E). Compared to wild-type, the ΔccpA strain had significantly lower maltodextrin gene transcript levels in the maltotriose-medium (Fig. 8F). Therefore, we conclude that MalR negatively influences maltodextrin gene transcript levels in the absence of inducing maltodextrins and that CcpA is needed to activate maltodextrin gene transcription under the conditions tested.
Purified CcpA and MalR bind specifically to cre sites in maltodextrin utilization gene promoter regions
In light of the bioinformatic and gene transcript level data presented above, we next tested the hypothesis that CcpA and MalR bind to the putative maltodextrin utilization cre sites. To this end, we overexpressed and purified GAS CcpA and MalR along with the phosphorylated form of the histidine-containing phosphocarrier protein of the phosphotransferase system (HPr-Ser46-P), a key cofactor for CcpA binding (Experimental Methods, Fig. S7) (Deutscher et al., 2005). As determined by fluorescence anisotropy, recombinant GAS CcpA-Ser46-P and MalR bound to the four maltodextrin cre sites with relatively high affinity (apparent Kd between 10–50 nM, Fig. 6, Table 5). As expected, specific binding of MalR and CcpA-Ser46-P was not observed for DNA from the promoter region of ftsX, a control gene whose transcript level is not influenced by CcpA or MalR and whose promoter does not contain a putative cre site (i.e. a negative control, Fig. 6). The CcpA-HPr-Ser46-P complex bound all tested cre sites with similarly high affinity that was some 50–100 fold higher than that observed for CcpA in the absence of HPr-Ser46-P (Fig. 6B, Table 5). MalR also bound these cre sites with high affinity, although with 2 to 2.7-fold higher equilibrium dissociation constants (Kd) than CcpA (Table 5). This small difference likely reflects minor variations in the amino acid sequences of the DNA binding domains of CcpA and MalR (Figure S4). Interestingly, MalR displayed the highest affinity, i.e., the lowest Kd, for the malT cre, consistent with the observation that this cre site has a perfect match with the consensus CcpA binding sequence (Table 5). The presence of maltotriose markedly increased the Kd of MalR for all cre sites (Fig. 6D), which is in accord with the idea that the presence of maltotriose induces MalR release from cognate DNA (Nieto et al., 1997). The Kd of CcpA was unaffected by the presence of maltodextrins. The Kd of MalR-cre binding remained unchanged in the presence of non-maltodextrin carbohydrates (glucose, lactose, sucrose), indicating a specific effect of maltodextrins on MalR. In toto, these binding data demonstrate that CcpA and MalR can bind specifically and with affinity to the same cre sites in the promoter regions of the maltodextrin utilization genes, providing a direct mechanism for the influence of CcpA and MalR on maltodextrin utilization gene transcript levels.
Fig. 6.
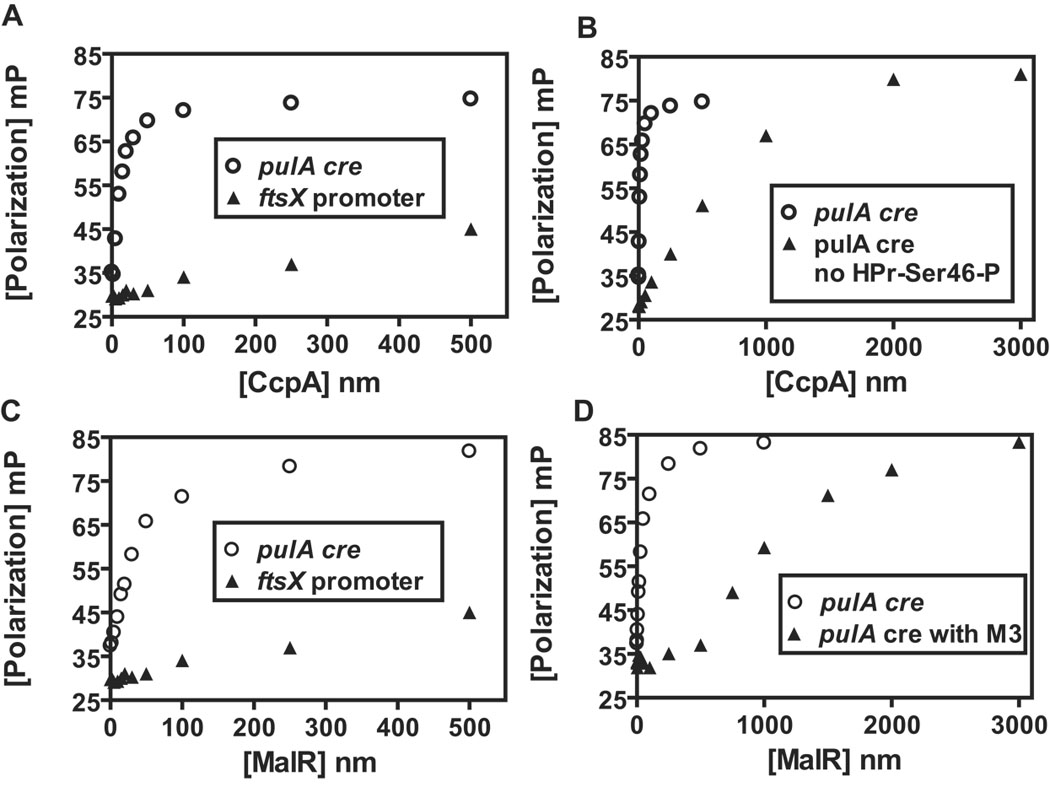
Binding of GAS CcpA and MalR to a representative maltodextrin utilization gene cre site. (A) Purified GAS CcpA was titrated into a binding buffer containing 1 nM fluorescein-labeled pulA cre (open circle) or 1 nM fluorescein labelled ftsX promoter DNA (negative control, closed triangle) in the presence of 50µM HPr-Ser46-P, a co-effector necessary for high affinity CcpA binding to cre DNA. Millipolarization units (mP) are plotted against the CcpA concentration (nM). (B) Comparison of CcpA pulA-cre biding in the presence (open circle) and absence (closed triangle) of 50 µM HPr-Ser46-P. (C) Purified GAS MalR was titrated as for CcpA except that Hpr-Ser46-P was not included in the binding buffer. (D) Comparison of MalR pulA-cre binding in the presence (closed triangle) and absence (open circle) of 10 mM maltotriose. The hyperbolic nature of the CcpA-HPr-Ser46-P and MalR interaction with the pulA cre indicates specific binding to this dideoxynucleotide.
Table 5.
Equilibrium dissociation binding constants (Kd) for complexes of purified GAS CcpA and MalR and cre sites involved in maltodextrin utilization.
| Kda(nm) | |||
|---|---|---|---|
| Gene | CcpA-Hpr-Ser46-P | CcpA | MalR |
| malM | 15.4 ± 1.0 | 1150 ± 80 | 41.7 ± 1.7 |
| malE | 14.0 ± 1.2 | 1047 ± 70 | 37.5 ± 2.2 |
| pulA | 13.2 ± 1.0 | 1125 ± 69 | 33.5 ± 1.9 |
| malt | 11.2 ± 0.9 | 935 ± 54 | 22.7 ± 2.1 |
Kd was determined in triplicate using the equation Y= (Bmax * X)/(Kd + X) where Y = minipolarization units, Bmax = maximum polarization value, and X = protein concentration.
Discussion
Given that we had previously established a role for the maltodextrin-binding lipoprotein MalE in GAS colonization of the mouse oropharynx, herein we sought to more completely define the molecular basis of maltodextrin catabolism in GAS and its role in host-pathogen interaction (Shelburne et al., 2006). By using an unbiased, genome-wide strategy we determined that the GAS gene (M5005_spy1692) annotated as encoding the glucose PTS (PtsG) actually contributes to maltose and maltotriose transport in GAS. The PTS responsible for maltose transport in B. subtilis is annotated as MalP for maltose PTS, but malP in streptococcal organisms is already used to refer to maltose phosphorylase (Schonert et al., 2006). Moreover, as noted earlier, MalT in GAS shares limited homology with MalP in B. subtilis. Thus re-annotating M5005_spy1692 as MalT, rather than MalP, is more accurate and will reduce confusion. We note that a recent investigation in Streptococcus mutans found that a gene previously annotated as ptsG (SMU.2047) was the principal transporter of maltose and maltotriose (Webb et al., 2007). These investigators also recommended annotating SMU.2047, which has 71% identity and 83% similarity at the amino acid level to M5005_spy1692, as malT.
Our data demonstrating a central role of MalE in GAS maltodextrin transport provides rationale for our earlier finding, replicated here, that MalE is needed for optimum GAS colonization of the mouse oropharynx (Shelburne et al., 2006). Whereas we discovered that the maltodextrin transport systems encoded by MalE and MalT are needed for GAS utilization of polysaccharides digested by human salivary α-amylase, we again found that MalE appears to play a central role in this process. Therefore we hypothesize that GAS relies on human salivary α-amylase in vivo to initiate the digestion of ingested polysaccharides which can subsequently be transported, mainly by a MalE-mediated mechanism, and utilized for the proliferation of organisms which then results in pharyngitis (Fig. 7). It was recently noted that the salivary amylase locus is the genetic region with the highest degree of large scale copy number variation in humans, which accords well with the observation that human salivary α-amylase levels are highly variable (Bellavia et al., 1979; Iafrate et al., 2004). Thus, our data raise the possibility that the activity of human salivary α-amylase in combination with diet may underlie the predisposition of certain persons to develop GAS pharyngitis. Such a proposition is supported by the observation that GAS pharyngitis rarely occurs in children less than 3 years old, an age at which salivary α-amylase activity is relatively low (Dezan et al., 2002; Woods et al., 1999).
Fig. 7.
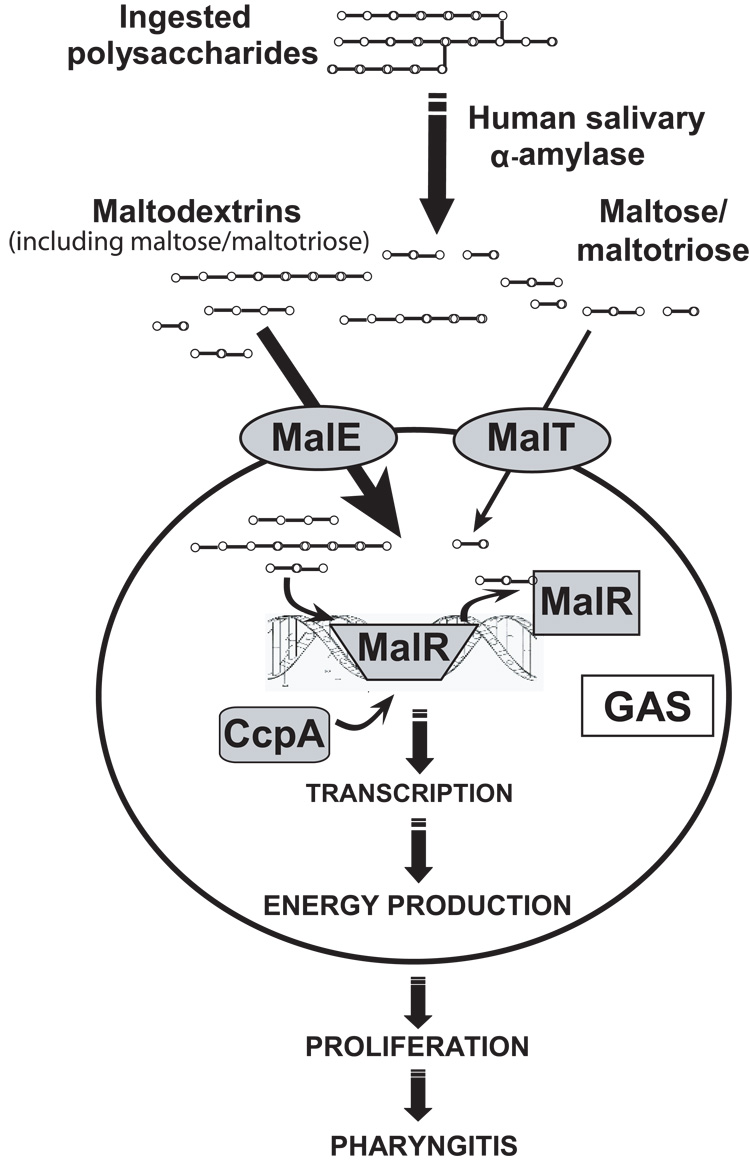
Schematic for potential contribution of GAS maltodextrin utilization to pharyngitis including regulatory mechanisms. Host-ingested polysaccharides are broken down by salivary α-amylase to maltodextrins that are mainly transported by MalE with some transport of maltose and maltotriose by MalT. The entry of maltodextrins leads to release of MalR from the promoter regions of the maltodextrin utilization genes with subsequent binding by CcpA. CcpA binding induces transcription of maltodextrin utilization genes allowing for further transport and subsequent catabolism of maltodextrins. The transported maltodextrins eventually enter into glycolytic pathways providing energy needed for proliferation and subsequent pharyngitis.
The data presented herein also provide new insights into the function of the previously uninvestigated gene M5005_spy1691 and into the coordinated regulation of maltose/maltodextrin genes with diverse chromosomal locations. Given that PTS phosphorylate transported carbohydrates and that orthologs of MalT and M5005_spy1691 are colocalized, it seemed likely that M5005_spy1691 would be a maltose phosphorylase or hydrolase that contributes to maltose catabolism. However, we found that the ΔM5005_spy1691 strain had no growth defect in a maltose-medium but did have decreased growth in a maltotriose or maltotetraose medium. Thus, it appears that M5005_spy1691 participates in the catabolism of maltodextrins longer than maltose. In this fashion it would be analogous to the maltose phosphorylase of E. coli that is involved in breaking down maltopentaose and longer maltodextrins (Boos and Shuman, 1998). Our findings raise the question of why M5005_spy1691 would be colocalized with MalT if MalT does not transport the longer maltodextrins apparently processed by M5005_spy1691. One possible answer is that all of the maltose/maltodextrin utilization genes so far identified in GAS are influenced by MalR, including MalE (Shelburne et al., 2007b). Our data show that MalR also influences MalT and M5005_spy1691, likely accounting for the similar transcriptional profiles of MalE and MalT under various growth conditions and in the human oropharynx.
MalR is classified as a member of the LacI-GalR family of transcriptional regulators, the best studied of which in Gram-positive organisms is the catabolite control protein A (CcpA). Bioinformatic analysis suggested that MalR and CcpA share common DNA binding sites (Fig. S3), and our purified protein binding data confirmed this hypothesis. Competition for binding of LacI-GalR family members to the same DNA sequence makes physiologic sense and may be more widespread than is currently appreciated given the presence of multiple LacI-GalR members in many Gram-positive pathogens (Muller et al., 2006). A major purpose of CcpA is to ensure that the organism preferentially utilizes high energy carbohydrates when available (Sonenshein, 2007). Given that maltodextrins enter into energy producing pathways with minimal modifications, GAS apparently uses CcpA to activate transcription of the maltodextrin utilization genes. However, as the in vivo supply of maltodextrins for GAS is inconsistent, MalR represses transcription of these genes in the absence of inducing agents. Thus, the ability of GAS to transport and catabolize maltodextrins depends on the coordinated depression of gene transcription by MalR along with transcriptional activation by CcpA acting through the same DNA binding motifs (Fig. 7). Moreover, these data provide a mechanism explaining the previous observations that elimination of cre sites, but not CcpA inactivation, leads to release of catabolite repression for certain carbohydrate utilization genes (Wen and Burne, 2002).
The relationship between complex carbohydrate utilization and virulence in human microbial pathogens has recently become an area of intensive investigation (Iyer and Camilli, 2007; Loughman and Caparon, 2006; Moyrand et al., 2007; Munoz-Elias and McKinney, 2005; Shelburne et al., 2006; Tanzer et al., 2006; Tchawa Yimga et al., 2006). Elucidating the mechanisms by which microbes obtain specific key nutrients in different host environments may provide a rational basis for the development of novel antimicrobial strategies. Our findings regarding the relative contribution of two maltodextrin transporters to the ability of GAS to colonize the mouse oropharynx significantly advance understanding of the molecular basis of GAS pathogenesis.
Experimental Procedures
Strains and culture conditions
The genome of serotype M1 strain MGAS5005 has been sequenced (Sumby et al., 2005). Strain MGAS5005 has been studied extensively in models of GAS infection and in vitro, including transcriptome analysis during growth in human saliva (Shelburne et al., 2005a; Shelburne et al., 2006; Sumby et al., 2006; Virtaneva et al., 2005). Additional GAS strains used in this study are listed in Table S1. M serotype was inferred by sequencing the emm gene (i.e. emm typed www.cdc.gov/ncidod/biotech/strep/strepindex.htm). GAS was grown on Trypticase soy agar containing 5% sheep blood (BSA; Becton Dickinson) or in Todd-Hewitt broth containing 0.2% (wt/vol) yeast extract (THY; Difco). Spectinomycin, erythromycin, or chloramphenicol (Sigma) was added to THY agar or broth at a concentration of 150 µg/mL, 1 µg/mL, or 4 µg/mL, respectively, when appropriate. Ultra-pure carbohydrates (Sigma) were added at a concentration of 0.5% (wt/vol) to a carbohydrate-free preparation of a commercially available chemically-defined medium (CDM; SAFC Biosciences) (Vise et al., 2003). For the purposes of this manuscript, we will use the terms glucose-medium, maltose-medium, etc., to refer to the carbohydrate-free CDM supplemented with the specified carbohydrate. We have previously demonstrated that GAS grows in the CDM only when exogenous carbohydrates are added (Shelburne et al., 2006). When indicated, purified human salivary amylase (Sigma) was added to CDM at physiologic concentration (200 U/mL) (Soderling et al., 1993; Takai et al., 2004).
RNA isolation and transcript level analysis
For analysis of carbohydrate-transport system gene expression under diverse growth conditions, various GAS strains were grown to mid-logarithmic phase in indicated media. RNA was isolated and purified using an RNeasy kit (Qiagen) (Shelburne et al., 2005a). The quality and the concentration of RNA were assessed with an Agilent 2100 Bioanalyzer and by analysis of the A260/A280 ratio. cDNA was reverse transcribed from RNA using Superscript III (Invitrogen) following the manufacturer’s instructions. TaqMan quantitative real-time PCR (QRT-PCR) was performed in quadruplicate with an ABI Thermocycler 7700 (Applied Biosystems) using the ΔCT method of analysis with proS used as the internal control gene (Chaussee et al., 2001). To determine malE and malT gene transcript levels in vivo, GAS RNA was isolated from throat swabs obtained from six patients with GAS pharyngitis and analyzed as described above (Virtaneva et al., 2003). To compare gene transcript levels between the parental, ΔmalR, and ΔccpA isogenic mutant strains, the ΔΔCT method was employed (User Bulletin no. 2, ABI PRISM 7700 Sequence Detection System; Applied Biosystems). TaqMan primers and probes for genes of interest and the internal control gene proS are listed in Table S2. To ensure accuracy, all QRT-PCR primers and probes were tested to confirm similar amplification efficiencies using GAS genomic DNA as template. All transcript level experiments were performed in quadruplicate on at least two separate occasions with samples analyzed in triplicate.
Growth of GAS strains in various media
All growth experiments were done at least in triplicate on a minimum of three occasions. For comparison growth studies in THY or CDM, GAS strains were grown overnight in THY. Overnight culture was added to fresh medium to achieve a uniform starting OD600 of 0.05, generally about a 1:100 dilution. Spectrophotometric density readings were then taken hourly until completion of the experiment. GAS was grown in human saliva as described (Shelburne et al., 2005b). Saliva was collected on ice from healthy volunteers as described under a Baylor College of Medicine Institutional Review Board human subjects protocol (Shelburne et al., 2005b). Pooled saliva collected from at least 4 donors was used to minimize effects of donor variation on study results.
Assessment of malT allelic diversity
The presence of malT and its allelic diversity in the genomes of the strains studied was determined by PCR amplification. Chromosomal DNA was isolated using a DNeasy kit (Qiagen). PCR primers used for malT amplification and sequencing were designed on the basis of the serotype M1 strain MGAS5005 genome (Table S2) (Sumby et al., 2005). A separate 3’ primer was used for M3 strains due to sequence diversity at the primer annealing site. The malT nucleotide sequence was determined from data obtained from both DNA strands using an Applied Biosystems 3730XL DNA Analyzer. Sequences were assembled and edited using Sequencher v4.5, and the inferred amino acid sequences were aligned using ClustalW. All sequence polymorphisms identified relative to strain MGAS5005 were confirmed by a second independent iteration of the entire DNA sequencing determination process.
Isogenic mutant strain construction
Creation of the ΔmalE, ΔmalR, and ΔccpA strains using non-polar insertional mutagenesis and spectinomycin selection was previously described (Shelburne et al., 2006; Shelburne et al., 2007b; Shelburne et al., 2008). The ΔmalE:malT strain was created by non-polar insertional mutagenesis from parental serotype M1 strain MGAS5005 using the PCR splicing overlap extension method described by Kuwayama et al. (Kuwayama et al., 2002). Primers were designed to amplify the 5’ and 3’ ends of the malT gene region along with nucleotide sequences that were complementary to the 5’ or 3’ portion of the erythromycin (erm) resistance cassette from plasmid pJRS233 (Perez-Casal et al., 1993) (Table S2). A third set of primers containing nucleotide sequences complementary to the 5’ and 3’ ends of the malT gene region was used to amplify the erm cassette. Fusion PCR was then used to link the 5’ and 3’ malT gene region PCR products to the erm cassette via the overlapping nucleotide sequence regions (Dalton and Scott, 2004; Kuwayama et al., 2002). This resulted in a PCR product in which the erm cassette, flanked by the 5’ and 3’ malT gene regions, was inserted in the place of nearly the entire malT open reading frame. The gene-disruption PCR construct was used to transform competent GAS cells, with the double recombination event selected via erythromycin resistance. The same gene disruption PCR construct was used to transform previously described ΔmalE competent cells with resultant selection for the ΔmalE:malT mutant strain. A similar process was used to the create the ΔM5005_spy1691 strain, except that the spectinomycin cassette (spc) from plasmid pSL60 instead of the erm cassette was used to create the gene disruption construct as previously described (Lukomski et al., 2000). The isogenic mutant strains were analyzed by Southern hybridization and DNA sequencing to confirm that the proper genetic construct was obtained (Fig. S3).
Construction of complemented isogenic mutant strains
For complementation purposes, we used plasmid pDC123 which encodes the chloramphenicol acetyl transferase (cat) gene for antibiotic selection and the phoZ gene for blue-white screening via alkaline phosphatase activity (27). pDC123 is a low-copy plasmid capable of replicating in GAS (Chaffin and Rubens, 1998). The complete malT gene along with ~ 250 bps upstream, which includes the promoter region, and ~15 bps downstream was amplified from strain MGAS5005 chromosomal DNA using primers that introduced EcoR1 and BamH1 cut sites at the 5’ and 3’ ends, respectively (Table S2). The resulting PCR product was digested with EcoR1 and BamH1 (Invitrogen) and directionally cloned into the EcoR1 and BamH1 sites of plasmid pDC123. The resulting plasmid, named pDCmalT, was used to transform competent ΔmalT cells to create the compΔmalT strain. Chloramphenicol resistance and blue/white screening were used to choose transformed strains for further analysis. Presence of the desired pDCmalT construct was confirmed using PCR and DNA sequencing. pDC123 lacking the malT gene was used to transform strain MGAS5005 and the ΔmalT isogenic mutant strain as controls. There was no statistical difference in the growth or transport assays between the ΔmalT strain and the pDC123ΔmalT strain (data not shown). The same procedure was previously employed to create the compΔmalE strain and was also used to create the compΔM5005_spy1691 strain (Shelburne et al., 2006).
Carbohydrate transport assays
All transport assay experiments were performed in quadruplicate on two separate occasions. GAS strains were grown to mid-exponential phase in CDM with the carbohydrate of interest added at 0.5% concentration. The bacteria were collected by centrifugation, washed, and suspended to an OD600 = 0.5 in 160 µl of CDM lacking carbohydrates. Under these conditions a 1 mL suspension contained 1.75 × 108 GAS colony forming units (CFU). [14C]-glucose (300 mCi/mmol; 200 µCi/ml), [14C]-maltose (600 mCi/mmol; 200 µCi/ml), or [14C]-maltotriose (900 mCi/mmol; 100 µCi/ml) were purchased from American Radiolabeled Chemicals, St. Louis, MO. [14C]-maltose was judged to be >98% pure by thin-layer chromatography whereas [14C]-maltotriose was found to be ~90% pure (personal communication from ARC). To determine Km and vmax, the final 14C labeled sugar concentration was varied from 10 nM to 50 µM in 40 µl of carbohydrate-free CDM to provide a total of 200 µl of bacteria plus labeled carbohydrate. Following addition of the labeled carbohydrate, 40 µl samples were taken every 30 sec for the first 120 sec, a time period during which carbohydrate transport rates were found to be linear, and filtered on a 0.45 µm nitrocellulose membrane (Millipore) which was then rinsed twice with CDM lacking carbohydrates. Liquid scintillation counting was used to determine the retained radioactivity on each filter (Beckman Model LS7500).
Mouse colonization experiment
All experiments in mice were performed according to a protocol approved by the Baylor College of Medicine Institutional Animal Care and Use Committee. Mouse throat colonization studies were conducted in adult (18- to 20-g) female outbred CD-1 Swiss mice (Harlan Sprague-Dawley Inc.) (Lukomski et al., 2000; Shelburne et al., 2006). Indicated GAS strains were grown in THY and harvested at an OD600 of approximately 0.5. The cells were washed once with sterile phosphate buffered saline (PBS) and suspended in PBS at 1 × 108 colony forming units (CFU)/ml. Both nares of each mouse were inoculated with 50 µL of the GAS suspension (total inoculation 100 µl, 1 × 107 CFU) with 25 mice in each group. The throat of each mouse was swabbed before inoculation and then at indicated time periods thereafter. The throat swabs were streaked onto BSA, grown overnight, and β-hemolytic colonies were counted. A minimum of five of the resultant β-hemolytic colonies per plate were then tested for the presence of GAS carbohydrate antigen via latex agglutination (BD Biosciences) before all β-hemolytic colonies were presumed to be GAS. No non-GAS β-hemolytic colonies were identified during the course of the experiment.
Detection of transcription start sites for maltodextrin utilization genes
The transcription start sites for GAS maltodextrin utilization genes were determined using 5’ rapid amplification of cDNA ends (RACE) according to the manufacturer’s protocol (Invitrogen). In brief, gene specific cDNA was created from total RNA using indicated primers (Table S2). Following TdT tailing of the cDNA, a second primer was used along with a conserved anchor primer to amplify the cDNA which was subsequently cloned into a TOPO TA vector (Invitrogen). The vector was chemically incorporated into Escherichia coli TOP10 cells, isolated using a MiniPrep Kit (Qiagen), and sequenced.
Overexpression and purification of GAS MalR and CcpA
Recombinant GAS CcpA was overexpressed in E. coli and purified as previously described (Shelburne et al., 2008). For purification of recombinant MalR, PCR was used to introduce NcoI and EcoR1 restriction sites (Table S2) onto the 5’ and 3’ ends of the malR gene lacking the initial methionine codon. The PCR product obtained from strain MGAS5005 was digested with NcoI and EcoR1 and cloned into the pET-His2 plasmid previously digested with the same enzymes (Lei et al., 2004). The resultant plasmid was cloned into Escherichia coli strain NovaBlue (Novagen) and then into BL21 (DE3) (Novagen) for protein expression. The resultant recombinant MalR protein had 12 amino acids MHHHHHHLETMG fused to the second amino acid residue at the aminoterminus. For expression of the recombinant protein, E. coli BL21 (DE3) cells were grown in 4L of Luria-Burtani broth supplemented with 100 mg/L of ampicillin to an OD600 of 1.0. The cells were induced with 0.5 mmol/L of isopropyl β-D-thiogalactopyranoside (IPTG, Sigma), allowed to grow for 6 hrs, and harvested by centrifugation. The cells were chemically and mechanically lysed, centrifuged, and the resultant supernatant loaded onto a nickel His-trap column (Amersham). Protein was eluted with increasing concentrations of imidazole and pooled fractions loaded onto a gel filtration column and eluted with 20 mm Tris-HCl pH 7.5, 200 mm NaCl. Pooled fractions were concentrated via centrifugation using a 10,000 molecular weight cut off filter (Millipore). The resultant proteins were >95% pure (Fig. S6).
Fluorescence Polarization
The DNA binding activities of CcpA and MalR were measured by fluorescence polarization on a Beacon 2000 fluorescence polarization instrument (Pavera, Madison, WI) (Kliegman et al., 2006; Lundblad et al., 1996). 42-bp 5′-fluoresceinated oligodeoxynucleotides (Sigma) were constructed of complementary 16-bp oligodeoxynucleotide sequences linked by 5 cytosines (Table S1). CcpA was titrated into 1 nM labelled DNA in 20 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA, and 10 µg/ml poly(dIdC) in the presence or absence of 50 µM HPr-Ser46-P (Shelburne et al., 2008). MalR was titrated into 1 nM labelled DNA in the same binding buffer but without the addition of HPr-Ser46-P. When indicated carbohydrates were added to the MalR binding buffer at a final concentration of 10 mM. Equilibrium binding constants were determined in triplicate as described with all experiments carried out at 25 °C (Newberry and Brennan, 2004).
Statistical analysis
Growth and RNA transcript levels were compared using Student’s 2-sided t-test. A repeated measures analysis was used to calculate the difference in the relatively ability of the GAS strains to colonize the mouse oropharynx. Statistical significance was assigned a 2-sided P value of 0.05 using Bonferroni’s adjustment for multiple comparisons when appropriate. Statistical calculations were performed using NCSS software version 2004.
Acknowledgements
This work was supported by American Heart Association grants 0565133Y/ 0765115Y and National Institute Allergy and Infectious Diseases K08 Career Development Award AI-064564 to Samuel A. Shelburne and grant G-0040 from the Robert A. Welch Foundation to Richard G. Brennan. Portions of this work were presented at the 2007 American Society of Clinical Investigation/American Academy of Physicians annual meeting in Chicago, IL.
References
- Abranches J, Nascimento MM, Zeng L, Browngardt CM, Wen ZT, Rivera MF, Burne RA. CcpA regulates central metabolism and virulence gene expression in Streptococcus mutans. J Bacteriol. 2008;190:2340–2349. doi: 10.1128/JB.01237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci USA. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almengor AC, Kinkel TL, Day SJ, McIver KS. The catabolite control protein CcpA binds to Pmga and influences expression of the virulence regulator mga in the group A Streptococcus. J Bacteriol. 2007;189:8405–8416. doi: 10.1128/JB.01038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andes D, Lepak A, Pitula A, Marchillo K, Clark J. A simple approach for estimating gene expression in Candida albicans directly from a systemic infection site. J Infect Dis. 2005;192:893–900. doi: 10.1086/432104. [DOI] [PubMed] [Google Scholar]
- Banks DJ, Porcella SF, Barbian KD, Beres SB, Philips LE, Voyich JM, et al. Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. J Infect Dis. 2004;190:727–738. doi: 10.1086/422697. [DOI] [PubMed] [Google Scholar]
- Bellavia SL, Moreno J, Sanz E, Picas EI, Blanco A. alpha-Amylase activity of human neonate and adult saliva. Arch Oral Biol. 1979;24:117–121. doi: 10.1016/0003-9969(79)90059-1. [DOI] [PubMed] [Google Scholar]
- Beres SB, Sylva GL, Barbian KD, Lei B, Hoff JS, Mammarella ND, et al. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc Natl Acad Sci USA. 2002;99:10078–10083. doi: 10.1073/pnas.152298499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Richter EW, Nagiec MJ, Sumby P, Porcella SF, DeLeo FR, Musser JM. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc Natl Acad Sci USA. 2006;103:7059–7064. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Boos W, Shuman H. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol Mol Biol Rev. 1998;62:204–229. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook I, Dohar JE. Management of group A beta-hemolytic streptococcal pharyngotonsillitis in children. J Fam Pract. 2006;55:S1–S11. quiz S12. [PubMed] [Google Scholar]
- Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- Chaffin DO, Rubens CE. Blue/white screening of recombinant plasmids in Gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene. 1998;219:91–99. doi: 10.1016/s0378-1119(98)00396-5. [DOI] [PubMed] [Google Scholar]
- Chaussee MS, Watson RO, Smoot JC, Musser JM. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes. Infect Immun. 2001;69:822–831. doi: 10.1128/IAI.69.2.822-831.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton TL, Scott JR. CovS inactivates CovR and is required for growth under conditions of general stress in Streptococcus pyogenes. J Bacteriol. 2004;186:3928–3937. doi: 10.1128/JB.186.12.3928-3937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Herro R, Bourand A, Mijakovic I, Poncet S. P-Ser-HPr--a link between carbon metabolism and the virulence of some pathogenic bacteria. Biochim Biophys Acta. 2005;1754:118–125. doi: 10.1016/j.bbapap.2005.07.029. [DOI] [PubMed] [Google Scholar]
- Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezan CC, Nicolau J, Souza DN, Walter LR. Flow rate, amylase activity, and protein and sialic acid concentrations of saliva from children aged 18, 30 and 42 months attending a baby clinic. Arch Oral Biol. 2002;47:423–427. doi: 10.1016/s0003-9969(02)00032-8. [DOI] [PubMed] [Google Scholar]
- Faucher SP, Porwollik S, Dozois CM, McClelland M, Daigle F. Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc Natl Acad Sci USA. 2006;103:1906–1911. doi: 10.1073/pnas.0509183103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, et al. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci USA. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzy-Treboul G, de Waard JH, Zagorec M, Postma PW. The glucose permease of the phosphotransferase system of Bacillus subtilis: evidence for IIGlc and IIIGlc domains. Mol Microbiol. 1991;5:1241–1249. doi: 10.1111/j.1365-2958.1991.tb01898.x. [DOI] [PubMed] [Google Scholar]
- Graham MR, Virtaneva K, Porcella SF, Gardner DJ, Long RD, Welty DM, et al. Analysis of the transcriptome of group A Streptococcus in mouse soft tissue infection. Am J Pathol. 2006;169:927–942. doi: 10.2353/ajpath.2006.060112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green NM, Zhang S, Porcella SF, Nagiec MJ, Barbian KD, Beres SB, et al. Genome sequence of a serotype M28 strain of group A Streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J Infect Dis. 2005;192:760–770. doi: 10.1086/430618. [DOI] [PubMed] [Google Scholar]
- Holden MT, Scott A, Cherevach I, Chillingworth T, Churcher C, Cronin A, et al. Complete genome of acute rheumatic fever-associated serotype M5 Streptococcus pyogenes strain Manfredo. J Bacteriol. 2007;189:1473–1477. doi: 10.1128/JB.01227-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden MTG, Feil EJ, Lindsay JA, Peacock SJ, Day NPJ, Enright MC, et al. Complete genomes of two clinical Staphylococcus aureus strains: Evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci USA. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueck CJ, Hillen W. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the gram-positive bacteria? Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Iyer R, Camilli A. Sucrose metabolism contributes to in vivo fitness of Streptococcus pneumoniae. Mol Microbiol. 2007;66:1–13. doi: 10.1111/j.1365-2958.2007.05878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarek MJ, Rosenmund H. The action of human pancreatic and salivary isoamylases on starch and glycogen. Clin Chim Acta. 1977;79:69–73. doi: 10.1016/0009-8981(77)90462-4. [DOI] [PubMed] [Google Scholar]
- Kliegman JI, Griner SL, Helmann JD, Brennan RG, Glasfeld A. Structural basis for the metal-selective activation of the manganese transport regulator of Bacillus subtilis. Biochemistry (Mosc) 2006;45:3493–3505. doi: 10.1021/bi0524215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- Kuwayama H, Obara S, Morio T, Katoh M, Urushihara H, Tanaka Y. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 2002;30:E2. doi: 10.1093/nar/30.2.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei B, Liu M, Chesney GL, Musser JM. Identification of new candidate vaccine antigens made by Streptococcus pyogenes: purification and characterization of 16 putative extracellular lipoproteins. J Infect Dis. 2004;189:79–89. doi: 10.1086/380491. [DOI] [PubMed] [Google Scholar]
- Loughman JA, Caparon MG. A novel adaptation of aldolase regulates virulence in Streptococcus pyogenes. EMBO J. 2006;25:5414–5422. doi: 10.1038/sj.emboj.7601393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S, Hoe NP, Abdi I, Rurangirwa J, Kordari P, Liu M, et al. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect Immun. 2000;68:535–542. doi: 10.1128/iai.68.2.535-542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad JR, Laurance M, Goodman RH. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol Endocrinol. 1996;10:607–612. doi: 10.1210/mend.10.6.8776720. [DOI] [PubMed] [Google Scholar]
- Miwa Y, Nakata A, Ogiwara A, Yamamoto M, Fujita Y. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res. 2000;28:1206–1210. doi: 10.1093/nar/28.5.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mormann JE, Muhlemann HR. Oral starch degradation and its influence on acid production in human dental plaque. Caries Res. 1981;15:166–175. doi: 10.1159/000260514. [DOI] [PubMed] [Google Scholar]
- Moyrand F, Fontaine T, Janbon G. Systematic capsule gene disruption reveals the central role of galactose metabolism on Cryptococcus neoformans virulence. Mol Microbiol. 2007;64:771–781. doi: 10.1111/j.1365-2958.2007.05695.x. [DOI] [PubMed] [Google Scholar]
- Muller W, Horstmann N, Hillen W, Sticht H. The transcription regulator RbsR represents a novel interaction partner of the phosphoprotein HPr-Ser46-P in Bacillus subtilis. Febs J. 2006;273:1251–1261. doi: 10.1111/j.1742-4658.2006.05148.x. [DOI] [PubMed] [Google Scholar]
- Munoz-Elias EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa I, Kurokawa K, Yamashita A, Nakata M, Tomiyasu Y, Okahashi N, et al. Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome Res. 2003;13:1042–1055. doi: 10.1101/gr.1096703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newberry KJ, Brennan RG. The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus. J Biol Chem. 2004;279:20356–20362. doi: 10.1074/jbc.M400960200. [DOI] [PubMed] [Google Scholar]
- Nieto C, Espinosa M, Puyet A. The maltose/maltodextrin regulon of Streptococcus pneumoniae. Differential promoter regulation by the transcriptional repressor MalR. J Biol Chem. 1997;272:30860–30865. doi: 10.1074/jbc.272.49.30860. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- Perez-Casal J, Price JA, Maguin E, Scott JR. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol Microbiol. 1993;8:809–819. doi: 10.1111/j.1365-2958.1993.tb01628.x. [DOI] [PubMed] [Google Scholar]
- Puyet A, Ibanez AM, Espinosa M. Characterization of the Streptococcus pneumoniae maltosaccharide regulator MalR, a member of the LacI-GalR family of repressors displaying distinctive genetic features. J Biol Chem. 1993;268:25402–25408. [PubMed] [Google Scholar]
- Rhodius VA, LaRossa RA. Uses and pitfalls of microarrays for studying transcriptional regulation. Curr Opin Microbiol. 2003;6:114–119. doi: 10.1016/s1369-5274(03)00034-1. [DOI] [PubMed] [Google Scholar]
- Scannapieco FA, Torres G, Levine MJ. Salivary alpha-amylase: role in dental plaque and caries formation. Crit Rev Oral Biol Med. 1993;4:301–307. doi: 10.1177/10454411930040030701. [DOI] [PubMed] [Google Scholar]
- Schonert S, Seitz S, Krafft H, Feuerbaum EA, Andernach I, Witz G, Dahl MK. Maltose and maltodextrin utilization by Bacillus subtilis. J Bacteriol. 2006;188:3911–3922. doi: 10.1128/JB.00213-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Allen GS, Diel M, Seidel G, Hillen W, Brennan RG. Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P. Cell. 2004;118:731–741. doi: 10.1016/j.cell.2004.08.027. [DOI] [PubMed] [Google Scholar]
- Sebaihia M, Peck MW, Minton NP, Thomson NR, Holden MT, Mitchell WJ, et al. Genome sequence of a proteolytic (Group I) Clostridium botulinum strain Hall A and comparative analysis of the clostridial genomes. Genome Res. 2007;17:1082–1092. doi: 10.1101/gr.6282807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, Musser JM. Virulence gene expression in vivo. Curr Opin Microbiol. 2004;7:283–289. doi: 10.1016/j.mib.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Shelburne SA, Sumby P, Sitkiewicz I, Granville CN, DeLeo FR, Musser JM. Central role of a two-component gene regulatory system of previously unknown function in pathogen persistence in human saliva. Proc Natl Acad Sci USA. 2005a;102:16037–16042. doi: 10.1073/pnas.0505839102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, 3rd, Granville C, Tokuyama M, Sitkiewicz I, Patel P, Musser JM. Growth characteristics of and virulence factor production by group A Streptococcus during cultivation in human saliva. Infect Immun. 2005b;73:4723–4731. doi: 10.1128/IAI.73.8.4723-4731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, 3rd, Sumby P, Sitkiewicz I, Okorafor N, Granville C, Patel P, et al. Maltodextrin utilization plays a key role in the ability of group A Streptococcus to colonize the oropharynx. Infect Immun. 2006;74:4605–4614. doi: 10.1128/IAI.00477-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, 3rd, Fang H, Okorafor N, Sumby P, Sitkiewicz I, Keith D, et al. MalE of group A Streptococcus participates in the rapid transport of maltotriose and longer maltodextrins. J Bacteriol. 2007a;189:2610–2617. doi: 10.1128/JB.01539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, 3rd, Okorafor N, Sitkiewicz I, Sumby P, Keith D, Patel P, et al. Regulation of polysaccharide utilization contributes to the persistence of group A Streptococcus in the oropharynx. Infect Immun. 2007b;75:2981–2990. doi: 10.1128/IAI.00081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, 3rd, Keith D, Horstmann N, Sumby P, Davenport MT, Graviss EA, et al. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc Natl Acad Sci U S A. 2008;105:1698–1703. doi: 10.1073/pnas.0711767105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot JC, Barbian KD, Van Gompel JJ, Smoot LM, Chaussee MS, Sylva GL, et al. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc Natl Acad Sci USA. 2002;99:4668–4673. doi: 10.1073/pnas.062526099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderling E, Pienihakkinen K, Alanen ML, Hietaoja M, Alanen P. Salivary flow rate, buffer effect, sodium, and amylase in adolescents: a longitudinal study. Scand J Dent Res. 1993;101:98–102. doi: 10.1111/j.1600-0722.1993.tb01096.x. [DOI] [PubMed] [Google Scholar]
- Sonenshein AL. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 2007 doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, Ricklefs SM, et al. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192:771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2006;2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai N, Yamaguchi M, Aragaki T, Eto K, Uchihashi K, Nishikawa Y. Effect of psychological stress on the salivary cortisol and amylase levels in healthy young adults. Arch Oral Biol. 2004;49:963–968. doi: 10.1016/j.archoralbio.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Talaat AM, Ward SK, Wu CW, Rondon E, Tavano C, Bannantine JP, et al. Mycobacterial bacilli are metabolically active during chronic tuberculosis in murine lungs: insights from genome-wide transcriptional profiling. J Bacteriol. 2007;189:4265–4274. doi: 10.1128/JB.00011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer JM, Thompson A, Wen ZT, Burne RA. Streptococcus mutans: fructose transport, xylitol resistance, and virulence. J Dent Res. 2006;85:369–373. doi: 10.1177/154405910608500417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchawa Yimga M, Leatham MP, Allen JH, Laux DC, Conway T, Cohen PS. Role of gluconeogenesis and the tricarboxylic acid cycle in the virulence of Salmonella enterica serovar Typhimurium in BALB/c mice. Infect Immun. 2006;74:1130–1140. doi: 10.1128/IAI.74.2.1130-1140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtaneva K, Graham MR, Porcella SF, Hoe NP, Su H, Graviss EA, et al. Group A Streptococcus gene expression in humans and cynomolgus macaques with acute pharyngitis. Infect Immun. 2003;71:2199–2207. doi: 10.1128/IAI.71.4.2199-2207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtaneva K, Porcella SF, Graham MR, Ireland RM, Johnson CA, Ricklefs SM, et al. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc Natl Acad Sci USA. 2005;102:9014–9019. doi: 10.1073/pnas.0503671102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vise PD, Kodali K, Hoe N, Paszczynski A, Musser JM, Daughdrill GW. Stable isotope labeling of a group A Streptococcus virulence factor using a chemically defined growth medium. Protein Expr Purif. 2003;32:232–238. doi: 10.1016/S1046-5928(03)00235-3. [DOI] [PubMed] [Google Scholar]
- Webb AJ, Homer KA, Hosie AH. A phosphoenolpyruvate-dependent phosphotransferase system is the principal maltose transporter in Streptococcus mutans. J Bacteriol. 2007;189:3322–3327. doi: 10.1128/JB.01633-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert MJ, Chambliss GH. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen ZT, Burne RA. Analysis of cis- and trans-acting factors involved in regulation of the Streptococcus mutans fructanase gene (fruA) J Bacteriol. 2002;184:126–133. doi: 10.1128/JB.184.1.126-133.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods WA, Carter CT, Schlager TA. Detection of group A streptococci in children under 3 years of age with pharyngitis. Pediatr Emerg Care. 1999;15:338–340. doi: 10.1097/00006565-199910000-00011. [DOI] [PubMed] [Google Scholar]


