Abstract
Synchrotron Fourier transform-infrared (FT-IR) and Raman microspectroscopy were applied to investigate changes in the molecular architecture of mouse oocytes and demonstrate the overall morphology of the maturing oocyte. Here we show that differences were identified between immature mouse oocytes at the germinal vesicle (GV) and mature metaphase II (MII) stage when using this technology, without the introduction of any extrinsic markers, labels, or dyes. GV mouse oocytes were found to have a small, centrally located lipid deposit and another larger polar deposit of similar composition. MII oocytes have very large, centrally located lipid deposits. Each lipid deposit for both cell types contains an inner and outer lipid environment that differs in composition. To assess interoocyte variability, line scans were recorded across the diameter of the oocytes and compared from three independent trials (GV, n = 91; MII, n = 172), and the data were analyzed with principal component analysis (PCA). The average spectra and PCA loading plots show distinct and reproducible changes in the CH stretching region that can be used as molecular maturation markers. The method paves the way for developing an independent assay to assess oocyte status during maturation providing new insights into lipid distribution at the single cell level.
Fourier transform-infrared (FT-IR) microspectroscopy and Raman microspectroscopy (RMS) are powerful techniques for studying the composition of cells and tissues. The spectra obtained provide a unique molecular fingerprint that can be interpreted based on the macromolecular chemistry of the cell or tissue under investigation. Hitherto no FT-IR/Raman imaging or mapping study has been performed on entire oocytes, and there are only two studies that have utilized the FT-IR technique to investigate oocyte components.1,2 These FT-IR studies explored protein secondary structure of the zona pellucida1 and the effect of chilling on the oolemma.2 In an FT-IR/Raman mapping experiment each pixel is essentially a digital element of a hyper-spectral data cube containing absorbance (or counts in the case of Raman), wave-number, and x,y spatial coordinates. Univariate or chemical maps can be plotted based on peak height, integrated areas under specific bands, or band ratios. For example it is possible to generate maps based on protein, lipid, nucleic acid, or carbohydrate concentration. Multivariate imaging techniques including unsupervised hierarchical cluster analysis (UHCA),3-10 K-means clustering,11,12 principal component analysis (PCA),5 linear discriminant analysis,13 fuzzy C-means clustering,12,14 and neural networks5 have proven to be invaluable in the identification and correlation of spectral groups or “clusters” which can be directly compared to stained cells. The achievable spatial resolution of an FT-IR mapping experiment can be improved when the conventional infrared globar source is replaced by a synchrotron source. Synchrotron light is produced from a point source, and the IR component is extracted as a highly collimated beam that is much brighter (100–1000 times that of a globar) enabling wavelength dependent spatial resolution approaching the diffraction limit. The brightness advantage comes from the small size of the source and the fact that light is emitted into a narrow range of angles.15,16 Consequently for a confocal FT-IR microscope coupled to a synchrotron source, the spatial resolution is approximately ∼λ/2.15,17
The oocyte, due to its large diameter, relative flatness, and distinctive maturation states makes it an ideal subject for FT-IR/Raman mapping experiments. Cytoplasmic changes that include cellular processes such as mRNA transcription,18 protein translation,19 post-translational modification of proteins,20 and ultrastructural changes take place as the oocyte grows and matures.21 Other cytochemical changes include alterations in endogenous lipid stores both in terms of abundance and composition.22 Most studies on oocyte lipids concern their accumulation in culture and the damaging effects of cryopreservation as opposed to studying their contribution to oocyte structure, metabolism, and maturation. Given the myriad and unique biochemical changes associated with oocyte growth and maturation, and embryonic development and differentiation, FT-IR would provide a logical alternative to assess developmental changes at the single cell level as opposed to morphological, molecular, and immunohistochemistry techniques or other time-consuming chemical assays, which require large numbers of cells.
In the present study high spatial resolution synchrotron FT-IR and RMS maps that show the macromolecular composition of mouse oocytes at the germinal vesicle (GV) and metaphase II (MII) stage of maturation are presented. The maps provide astonishing insights into the spatial location of lipid deposits within the mouse oocytes, inferring an inherent polarity of the maturing oocyte. Moreover the lipid bands provide potential markers for cytoplasmic and nuclear changes, which in this specific case are associated with oocyte maturation. In addition, spectral maps collected via RMS afford a better spatial resolution and allow a tentative chemical identification of some of the lipid components.
Methods
Sample Preparation
Germinal vesicle (GV) oocytes were obtained from the ovaries of F1 (C57BLxCBA) 6 week old female mice 48 h after a single subcutaneous injection of pregnant mares serum gonadotrophin (PMSG, Folligon, Intervet, Lane Cove, Australia). MII oocytes were obtained from the oviducts of females 12–13 h after subcutaneous injection of human chorionic gonadotrophin (hCG, Chorulon, Intervet) 48 h after the PMSG injection. Animal protocols were approved by the Monash University Animal Ethics Committee and conducted under the Australian National Health and Medical Research Council (NH&MRC) code of practice. MII oocytes were treated with 100 IU/mL hyaluronidase (type IV-S, Sigma Chemicals Co., St. Louis, MO) in phosphate buffer saline (PBS) containing 3 mg/mL bovine serum albumin (BSA; Sigma) for 5 min to allow cumulus cells to separate from the oocytes. GV and MII oocytes were washed in 2 mL of fresh hyaluronidase and BSA free PBS before they were mounted on 2 cm diameter × 0.5 mm thick CaF2 windows, fixed in 100% ethanol, and washed three times in milliQ water. For RMS measurements, oocytes were fixed in formalin and immersed in PBS.
FT-IR Synchrotron and RMS Measurements
FT-IR synchrotron measurements were performed on the FT-IR beamline at the Australian Synchrotron. Spectra were collected with a Bruker Hyperion 2000 IR microscope (Bruker Optik Gmbh, Ettlingen, Germany) equipped with a liquid nitrogen cooled HgCdTe (MCT) detector with a 36× IR objective. The Hyperion 2000 microscope is coupled to a Bruker Vertex 80 spectrometer. For FT-IR mapping and line scans, the rectangular aperture was set at 4 μm × 4 μm. Two types of measurements were performed, one set aimed at determining the intracellular variability by recording a map of the entire oocyte (n = 2 for each of the two oocyte stages), while the other measurement assessed the intercellular variability by recording line scans across the diameter of the oocyte (GV oocytes n = 91, MII n = 172). Spectral maps and line scans were collected in transmission by scanning the computer-controlled microscope stage in a raster pattern with a 2 μm step size in both the x and y directions for the maps and a 4 μm step size for the line scans. Interferograms (64 coadded) were collected double sided at a resolution of 6 cm-1 and Fourier transformed using a Blackman-Harris three-term apodization function with a zero-filling factor of 2. Second derivative spectra were calculated using the Savitsky-Golay algorithm with 13 smoothing points. In the calculation of second derivate spectra, the absorbance maxima become minima and intensities are thus positive for absorbance spectra and negative for second derivative spectra. RMS maps were collected at 488 nm excitation using a confocal Raman microscope (CRM200), WITec, Inc. (Ulm Germany), with a water immersion objective (60×/NA = 1). The dwell time per pixel was 500 ms. At this wavelength, the lateral spatial resolution is about 300 nm. The theoretical depth resolution is about twice that value. However, it should be mentioned that the resolution along the z-axis depends on the refractive index of the sample. It usually deviates from the theoretical value and drops with increased penetration depth.23 Spectral maps were obtained from the raw data sets by unsupervised hierarchical cluster analysis.
Data Analysis
Univariate Maps
To examine the intracellular variability, chemical (also known as univariate) maps were generated for individual oocytes from the two groupings. Maps were constructed by integrating the total area between 1800 and 1000 cm−1 (total absorbance map) and the areas underneath the amide I mode (1729-1600 cm-1), CH stretching region (3000−2840 cm−1), and ester carbonyl band (1770−1722 cm−1).
Statistical Analysis
Unsupervised Hierarchical Cluster Analysis
Unsupervised hierarchical cluster analysis also known as agglomerative hierarchical clustering, described in refs 5, 8, and 9, was used to generate false-color maps based on spectral variation. UHCA was performed using Cytospec software.5 The data were quality tested to remove spectra of low absorbance and poor signal-to-noise ratio, and the analysis was performed in the CH stretching region (3100−2800 cm−1) using second derivative spectra. A distance matrix was first calculated so that every spectrum was referenced to every other spectrum and given a relative distance in multivariate space. Ward's algorithm was then used to merge the spectra into groupings, and colors were then assigned to the different clusters.
Principal Component Analysis
The time scale required to record high spatial resolution, high signal-to-noise FT-IR synchrotron maps of entire oocytes (6–8 h) precludes analysis of the interoocyte variability by this means. In order to assess the interoocyte variability, line scans were recorded from GV (n = 91) and MII (n = 172) oocytes collected from three independent trials. For each line scan, approximately 16 spectra were recorded across the diameter of individual oocytes. Each line takes approximately 2 min so it is feasible to record spectra of a large number of oocytes in a reasonable time frame. It is important to note that the line scan represents only a portion of the entire oocyte, and in the case of GV oocytes it is possible to miss the central lipid deposit when performing a line scan. The line scan spectra were processed with principal component analysis which is a proven technique for the analysis of multivariate data.24 A data matrix consisting of 419 spectra that were collected from average spectra from each line scan (∼16 spectra per line scan) for each mouse oocyte in each of the groupings (GV oocytes n = 91, MII n = 172) was decomposed using PCA. The spectra were transformed by performing a second derivative to reduce the influence of baseline variation, and the CH stretching region (3100−2800 cm−1) was selected for the decomposition. The data were mean centered, and a full cross validation PCA was performed using 6 PCs. All data manipulations and the PCA decomposition were performed using the Unscrambler (CAMO, Norway) software package.
Results
GV Oocyte
The total absorbance map from the integrated area between 1800 and 1000 cm−1 of a GV oocyte (Figure 1a) shows very strong absorbance (warm colors) surrounding the periphery of the oocyte that corresponds to the cross-linked glycoproteins of the zona pellucida. The nucleus shows relatively weak absorbance (cool colors) compared to the periphery. The integrated area (Figure 1c) underneath the amide I mode (1729−1600 cm−1) is indicative of the total protein concentration of the egg. The map shows a similar profile to the total absorbance map of the oocyte with strong absorbance around the periphery and weaker absorbance within the nucleus. The northwest pole shows a high protein concentration compared to the rest of the oocyte. Figure 1d shows the integrated area of the CH stretching region (3100−2800 cm−1) associated mainly with the CH bonds from amino acid side chains and lipid moieties for oocytes that have high lipid content. This is similar to the protein map, which indicates that the majority of CH absorbance is from amino acid side chains and not from lipids. This is corroborated in Figure 1e, presenting the integrated area of the ester carbonyl band (1770−1722 cm−1), which shows a very low concentration of lipid throughout the entire oocyte.
Figure 1.
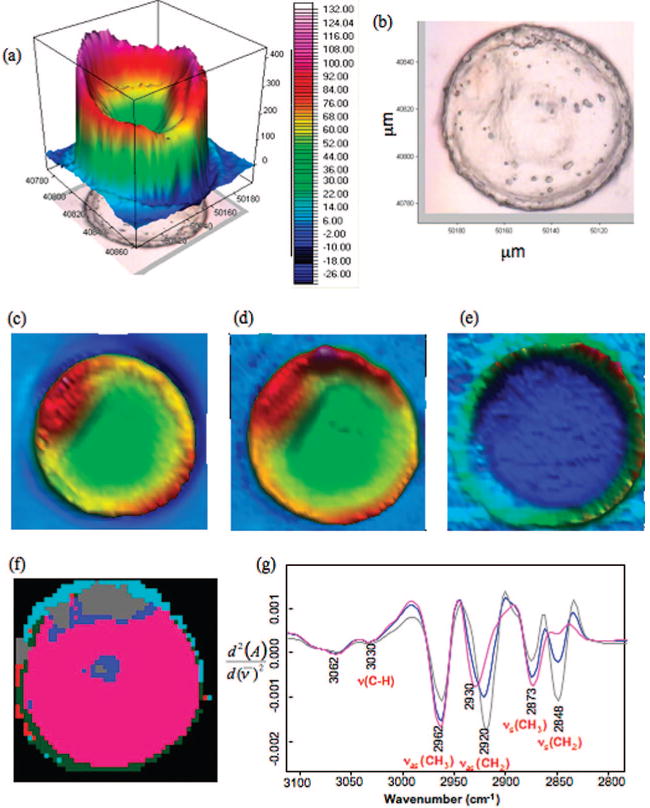
High-resolution synchrotron FT-IR map of a GV cell recorded using a 4 μm × 4 μm aperture: (a) Total absorbance map constructed by integrating the area between 1800 and 1000 cm−1. (b) Photomicrograph of a GV cell showing the clear nucleus and scale bar. (c) Chemical map of the integrated area under the amide I band. (d) Chemical map of the integrated area of the CH2 and CH3 stretching region (3000−2800 cm−1). (e) Chemical map of the integrated area of the ester carbonyl band (1720−1750 cm−1). (f) False color five cluster map generated by performing UHCA on the CH stretching region (3100−2800 cm−1). (g) Mean extracted spectra from the blue, gray, and pink clusters.
Unsupervised hierarchical cluster analysis, which is a proven technique for exploring subtle biochemical differences in FT-IR tissue maps, was applied to further analyze the data.5,8,9 Figure 1f shows a UHCA false cluster map showing five clusters from a UHCA map performed on second derivative spectra in the 3100−2800 cm−1 region of the spectrum. The map provides a detailed insight into lipid distribution within the GV oocyte that is not observable in the univariate maps. It should be noted that the lipid deposit is not observed in the center of the oocyte in Figure 1d because of the high absorbance at the periphery of the egg and the low absorbance from the carbonyl group distorting the range of the color scale. The map in Figure 1f shows three distinct regions: pink, blue, and gray. The small blue and gray clusters, where the average spectra are lipid signatures due to carbonyl absorbance, appear where the nucleolus is expected and toward the pole, while the pink region dominates the rest of the egg. The light blue and dark green clusters are mainly associated with light dispersion, which has previously been described25 and is expected at the edges of tissue sections. The mean extracted spectra from each cluster in Figure 1f is presented in Figure 1g, which show significant differences in the CH stretching region. The full range spectra are shown in Figure 1S of the Supporting Information where the gray spectrum is indicative of saturated long chain fatty acids. The increase in the ratio of the intensity of bands assigned to the asymmetric CH2 stretching vibration (νas(CH2)) and the symmetric CH2 stretching vibration (νs(CH2)) relative to the νas(CH3) along with the lack of change in the CH stretching vibration ν(CH)), associated with CH vibration of unsaturated fatty acids, proves that the gray region is comprised of predominately long chain saturated fatty acids. The blue spectrum in this figure, which is representative of the outer cluster on both the central and polar lipid deposits, shows a distinctly different spectrum where the νas(CH2) band has red-shifted 5 cm−1 compared to the corresponding band in the gray spectrum. In addition, the ratio of νs(CH3)/νs(CH2) is significantly greater (p < 0.05 for one-tail ANOVA) compared to the gray spectrum than the blue spectrum, indicating a higher percentage of branched chain fatty acids in the outer lipid deposit compared to the inner deposit. The pink spectrum is consistent with CH2 and CH3 stretching vibrations from amino acid side chains of proteins as can be gauged by the weak intensity of the νs(CH2) bands relative to the νas(CH3) band and the total lack of carbonyl signature. The νas(CH3) band is enhanced in the pink spectrum because there is a high proportion of methyl groups associated with amino acid side chains.
MII Oocyte
Figure 2a depicts the total absorbance map (1800-1000 cm−1) of the MII oocyte while Figure 2b shows the corresponding photomicrograph. The total absorbance map shows strong absorbance associated with the oocyte cortex (pink) and strong to moderate absorbance toward the center of the oocyte (red). The absorbance toward the center of the egg is much greater than that observed in the nucleus of the GV oocyte. Figure 2c shows the integrated area of the amide I mode, which shows a high concentration of protein toward the periphery of the oocyte (pink) and moderate absorbance toward the center (red). Figure 2d shows the integrated area of the CH stretching region (3100-2800 cm−1), which unlike the corresponding maps in Figure 1c,d, is vastly different compared to the amide I map. This indicates that unlike in GV oocytes, lipid contributions rather than amino acid side chain vibrations dominate the CH stretching region. This conclusion is supported by the map of the integrated area of the ester carbonyl band (1770-1722 cm−1) in Figure 2e, which shows a very similar pattern to the CH2-CH3 stretching map in Figure 2d. The lipid deposits are far larger and in higher concentration in MII oocytes compared to GV oocytes. This is further corroborated by the UHCA false-color map of the CH stretching region (3100-2800 cm−1) presented in Figure 2f. The map shows a similar pattern to the maps presented in parts d and e of Figure 2 with the gray cluster indicative of the large lipid deposits, while the pink, which is identical to the pink cluster for GV oocytes, is mainly dominated by protein. The gray cluster also appears around the periphery of the oocyte in the region of the zona pellucida and indicates that lipids become distributed around the periphery of the oocyte at this later stage of maturation. The midblue cluster, located around the gray cluster, is predominantly lipid but of a different composition to the gray. The stronger contribution of the νs(CH3) to the midblue spectrum distinguishes it from the gray cluster and indicates a higher concentration of branched chain fatty acids in the outer region of the lipid deposit. The mean extracted spectra associated with the gray clusters from both the GV and MII oocytes reveal a similar profile (Figure 2g). The entire spectra are presented in Figure 1S of the Supporting Information. The spectra show strong intensity contributions from the νas(CH2) and νs(CH2) bands relative to the νas(CH3) indicating a high concentration of long chain saturated fatty acids. However, there is a shift of the νas(CH2) from 2920 cm−1 in the GV oocyte to 2927 cm−1 in the MII oocyte indicating distinctly different lipid environments between the two eggs. Other differences include a change in the νs(CH2)/νas(CH2) ratio between the lipid environments of the two oocyte types. The MII oocyte has a significantly lower νas(CH3)/νs(CH2) ratio compared to the GV oocyte (p < 0.05, for one-tail ANOVA). This correlates with a decrease in the concentration of branched chain fatty acids compared to the GV oocyte lipid deposits. The light blue and green cluster spectra are affected by dispersion, and consequently the spectra are not shown.
Figure 2.
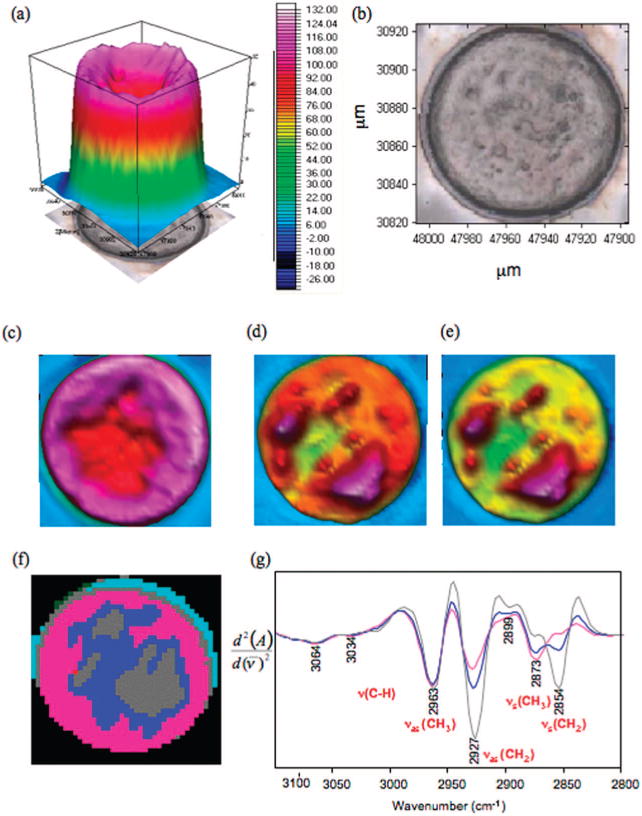
FT-IR synchrotron map of MII cell: (a) Total absorbance map, 1800−1000 cm−1. (b) Photomicrograph of an MII cell. (c) Integrated areas of amide I mode. (d) Integrated area of CH stretching region. (e) Integrated area of ester carbonyl band. (f) False color five cluster map generated by performing (UHCA on the CH stretching region (3100−2800 cm−1). (g) Mean extracted spectra from each cluster color coded to match the UHCA map.
Similar conclusions are reached from the analysis of the Raman maps shown in Figure 3. Figure 3a shows a bright field image of a MII oocyte, and Figure 3b shows the corresponding Raman image, obtained via UHCA. The oocyte presented in this Raman image was obtained, analyzed, and processed at Northeastern University and serves as a totally independent control. Large lipid deposits are also observed in this oocyte confirming the lipid enrichment observed in the FT-IR image of the MII oocyte. In this image, the gray and blue areas represent high cytoplasmic protein content. The light blue regions are likely to be rich in mitochondria. Mitochondria are very abundant in oocytes to support nourishing processes, and their quantity is known to be related to oocyte health and an indicator for viability prior to implantation. The distribution pattern of the light blue areas agrees well with the distribution of mitochondria typically found in oocytes.26 The possibility to image mitochondrial allocation employing UHCA has been demonstrated on cultured cells.27 The pink areas represent regions of very high lipid concentrations. The Raman mean cluster spectrum of the pink areas is shown in Figure 3C (pink trace) along with a reference spectrum of pure 1,2-dioleoyl-3-trimethylammonium-propane (black trace), a common membrane lipid, indicating that the pink areas of the oocyte are mostly composed of lipids or phospholipids, with small amounts of protein (as indicated by the broadening of the peak at 1660 cm−1 due to the presence of the amide I vibration). Spectra with significant lipid or phospholipid contributions presumably reveal complexes of smooth-surfaced endoplasmatic reticulum as commonly found in these cells.28
Figure 3.
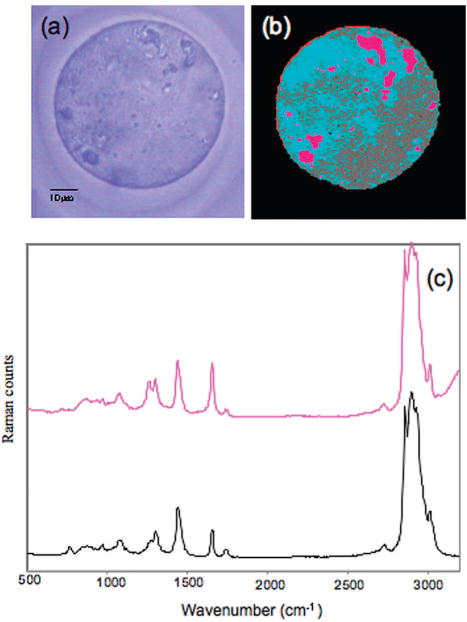
Microscopic (a) and false color map reconstructed from hierarchical cluster analysis of a Raman image collected of MII oocyte at 0.5 μm spatial resolution (b). (c) The spectra show a Raman spectrum from lipid or phospholipid rich regions (pink) in comparison with a spectrum of 1,2-dioleoyl-3-trimethylammonium-propane a common membrane lipid (black spectrum).
Comparison of Averaged Spectra
The raw average FT-IR spectra (Figure 4a) of the MII and GV oocytes are markedly different across the entire spectral range. The MII have a greater total absorbance compared to the GV oocyte and a much higher concentration of protein. On average MII have a higher lipid content across the diameter of the oocyte compared to the GV as evinced by the intensity of the ester carbonyl band at 1740 cm−1. In the GV oocytes, the ester carbonyl is barely discernible and the profile in the CH stretching region is characteristic of absorbance from amino acid side chains. It should be noted that in averaging the spectral information, the signals from small lipid deposits in the center and the periphery of the GV oocyte are overwhelmed and hence there is only a hint of the carbonyl band in Figure 4a. The inset in Figure 4b shows the averaged spectrum of the CH stretching region for MII and GV oocytes. There are clear differences between the intensity ratios for the CH2 and CH3 stretching vibrations between oocyte types. MII oocytes have a significantly higher vas(CH2)/vas(CH3) ratio (p < 0.05 one-tail ANOVA) compared to the GV oocytes indicating a higher concentration of predominantly longer chain fatty acids. On the other hand, GV oocytes have a significantly higher vs(CH3)/vs(CH2) ratio (p < 0.05 one-tail ANOVA) compared to MII oocytes indicating a larger concentration of methyl groups associated with amino acid side chains.
Figure 4.
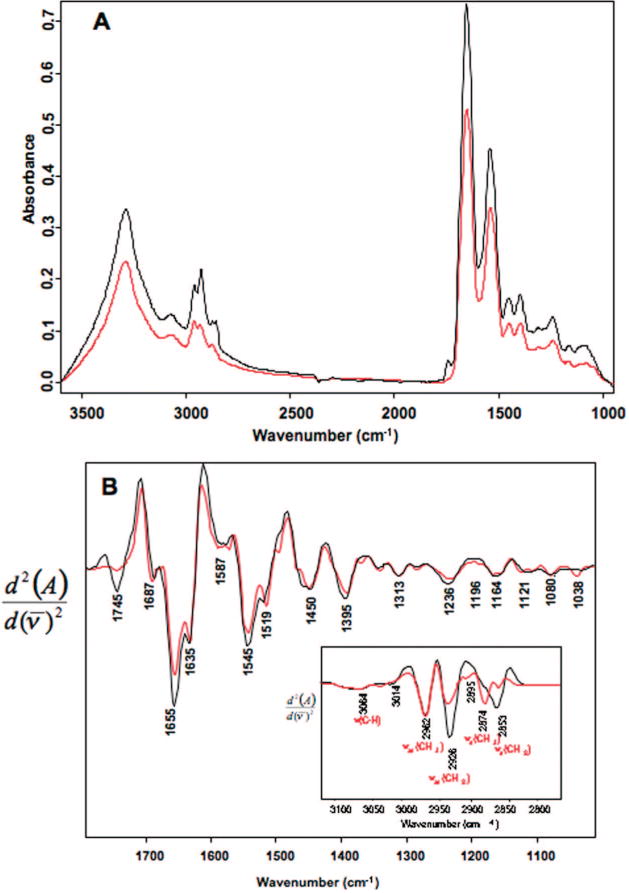
(A) Mean FT-IR synchrotron spectra calculated from line scans recorded of 90 GV (red) and 93 MII (black) cells showing major band assignments for the CH stretching region. (B) Second derivative spectra calculated from the mean spectra presented in part A.
Principal Component Analysis
To further investigate spectral changes between the oocyte groupings, principal component analysis was applied to the line scan data to identify potential oocyte maturation markers. PCA enables the decomposition of original data into a model consisting of a signal part and a noise part. It is ideally suited to spectroscopic data where there are thousands of variables (wavenumber values) per spectrum. Tools such as score plots enable the data to be presented in a form where the spectra can be represented as single points in multivariate space. The scores indicate which objects are responsible for most of the variance in the data. Each score represents a whole spectrum plotted in 1, 2, or 3 spatial dimensions. A loading plot is used to assign coefficients to relevant principal components (PCs) from a variable data set. Loadings express relations between variables (i.e., wavenumber values) and enable one to tell which variables are dominating or influencing the model and how they are associated with one another. In a PCA scores plot, each point represents an entire spectrum based on its contribution to each PC or eigenvector. A PCA decomposition performed on the second derivative averaged line scan spectra for each oocyte investigated over the entire spectral region (3500−1000 cm−1) revealed no clear separation between the GV and MII groupings (data not shown). However, a PCA decomposition performed solely on the CH stretching region (3100−2800 cm−1) showed an excellent separation for the two oocyte groups (Figure 5a) when plotted on a principal component 1 (PC1) versus PC2 scores plot. The separation between the clusters of GV and MII oocytes occurs primarily along PC1. The GV oocytes form a much tighter cluster compared to the MII oocytes. The spread of the MII oocyte spectra across PC1 is indicative of different lipid concentrations associated with individual MII eggs. The most lipid rich oocytes have more negative PC1 scores, while those lower in lipid have more positive PC1 scores.
Figure 5.
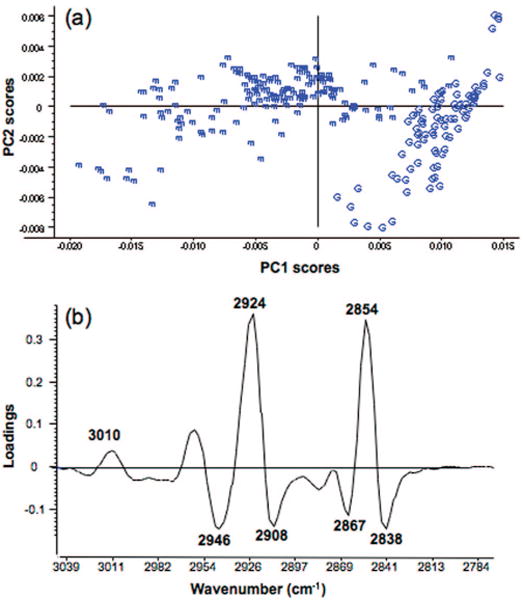
Principal component analysis PC1 and PC2 scores plot showing a clear separation between GV (G) and MII (m) cells along PC1. The PCA decomposition was performed on 265 spectra which included 173 averaged spectra extracted from the line scans of 90 MII cells and a further 92 averaged spectra collected from the line scans of 92 GV cells from 4 independent trials. The PCA was performed in the CH stretching region (3050−2780 cm−1) on second derivative spectra. (b) Corresponding PC1 loadings plot showing strong positive loadings for the vas(CH2) and vs(CH2) modes at 2924 and 2854 cm−1 associated with the MII cells and strong negative loadings for vas(CH3) and vs(CH3) at 2946 and 2867 cm−1 associated with the GV cells.
The PC1 loadings plot (Figure 5b) highlights the bands that are important in the separation observed along PC1 in the PC1 versus PC2 scores plot. Strong positive loadings are associated with negative scores, while strong negative loadings are associated with positive scores. The apparent opposite correlation is because the PCA decomposition is performed on second derivatives where the positive bands in the raw spectra become minima in the second derivative. It is apparent that the bands at 2924 and 2854 cm−1 (assigned to the vas(CH2) and vas(CH2) characteristic of long chain saturated fatty acids) have strong loadings contributing to the separation of MII oocytes, while GV oocytes are correlated with the negative loadings at 2946, 2908, 2867, and 2838 cm−1 associated with CH vibrations from amino acid side chains. The band at 3010 cm−1 assigned to the v(CH) from unsaturated fatty acids appears as a small but notable positive loading that is associated with the MII oocytes. This indicates that there is an increase in unsaturated fatty acids in the MII oocytes compared to the GV. The increase in intensity of the 3010 cm−1 band for the MII oocytes was not as evident in the averaged spectrum demonstrating the power of the PCA technique in resolving “hidden patterns” in the data. The distinct separation achieved with the PCA analysis indicates that the CH stretching region could provide a way to fingerprint oocytes at different stages of maturation.
Discussion
Polarity and oocyte asymmetry has long been studied, yet established solely in invertebrates such as the Drosophila.29,30 Even though research is ongoing in documenting the same for mammals,29,31 the search has not yielded many confirming conclusions. Thus, we have been employing two techniques which are based on biochemical mapping of the functional groups from all the subcellular constituents, which can offer a novel assessment and insight into the overall morphology and potential polarity of the growing and maturing oocyte. High spatially resolved synchrotron FT-IR and Raman maps of mammalian cells provide a way to visualize the macromolecular changes that accompany physiological and biological transformations in in vivo or in vitro systems. The present study utilized immature and mature mouse oocytes to demonstrate this powerful technology. Current morphological techniques to assess cell differentiation in general and more specifically oocyte maturation are hampered by the lack of suitable standards and the inherent variability associated with subjective grading systems. FT-IR/Raman mapping/imaging spectroscopy can overcome these limitations through identifying unique spectral features of functional groups that can be spatially located. These can be associated with cell morphology in relation to differentiation and maturation.
We have identified unique spectra of lipid deposit for each of the oocyte groups analyzed with the most interesting findings being the small central and polar lipid deposits in GV oocytes which are differently distributed in mature MII oocytes. Both lipid deposits in GV oocytes are similar in composition as evinced by the UHCA and show an inner and an outer region, which vary dramatically in composition. In MII oocytes a much larger lipid deposit is centrally located, which also consists of two main regions that appear gray (inner) and blue (outer) in the UHCA map.
While the line scans presented herein were performed using a synchrotron light source, initial spectra were obtained with more conventional focal plane array and linear array detectors yielding similar results for the PCA analysis. The main advantage of the synchrotron light source in these experiments was to generate the high spatial resolution maps of the entire cell. The theoretical spatial resolution achieved by a focal plane array detector is dependent on the pixel size (5.5 μm2 for Varian “Stingray” FT-IR imaging spectrometers). However, due to “pixel overlap”, the actual spatial resolution is closer to 15 μm.15 At the IR beamline at the Australian Synchrotron, a 4 μm resolution can be achieved due to the confocality of the microscope, the extremely bright photon source, and the numerical aperture of the Schwarzschild objective (NA of ∼0.6). Maps collected with the focal plane array detector could not resolve the difference in inner and outer lipid deposits observed in the GV oocyte. However, to determine oocyte maturation routinely, a standard focal plane array, linear array, or conventional FT-IR mapping instrument would suffice.
Variation in the sizes and numbers of lipid droplets is a well-established feature of mammalian oocytes.32–34 “Fat globules” in the center of oocytes were first reported by Subramaniam and Aiyar in 1935.35 As confirmed by our results, the central distribution of lipid droplets was shown to be a characteristic of immature oocytes, while in mature oocytes the droplets are peripherally distributed.36 The importance of triglycerides as compact energy reserves is well documented and it is known that phospholipids and cholesterol are prerequisites for the formation of membranes required for rapid cell division after fertilization.22 The PCA analysis indicated an increase in unsaturated fatty acids in MII stage oocytes compared to GV stage oocytes, which is dominated mainly by protein. This is consistent with an increase in membrane fluidity, which is a requirement for rapid division.37
The phosphoinositide–phospholipase C pathway is strongly implicated in the control of mouse oocyte meiosis.38,39 The pathway becomes progressively functional as oocyte growth advances, and it appears to play a role in the G2/M transition when meiosis resumes in in vitro maturation models. Phospholipase C β1 (PLC β1) is first exclusively localized to the nucleus and then migrates to the cytoplasm when the oocyte is fully grown. During oocyte maturation, PLC β1 is active in the cytoplasm before it migrates and becomes active in the nucleus just prior to germinal vesicle breakdown. This indicates the important role of the nucleus phosphoinositide cycle during oocyte meiosis. It appears also that this chronology is crucial and that defects leading to an inappropriate intracellular localization can have dramatic consequences in the production of competent oocytes and lead to fertility problems.
The examination of the CH stretching region in combination with PCA analysis confirms that distinct vibrational markers for maturation are related to the composition of lipids within the oocyte. The FT-IR spectral signature of the CH stretching region could in the future be used to predict oocyte developmental competence specifically and to interrogate cell variability and change during embryo development and cell differentiation through development.
Supplementary Material
Acknowledgments
This work is supported by an Australian Research Council Discovery Grant, the Australian Synchrotron, and the NICHD Cooperative Program on Female Health and Egg Quality Grant 1U01 HD044778-01. Especially, we would like to thank Judith A. Newmark and Carrol M. Warner for providing the mouse oocytes for the Raman experiments and advice on experimental procedures.
Footnotes
Supporting Information Available: Mean extracted spectra from cluster analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Nara M, Yonezawa N, Shimada T, Takahashi K, Tanokura M, Yumoto F, Nakagawa H, Ohashi K, Hamano S, Nakano M. Exp Biol Med. 2006;231:166–171. doi: 10.1177/153537020623100206. [DOI] [PubMed] [Google Scholar]
- 2.Ghetler Y, Yavin S, Shalgi R, Arav A. Hum Reprod. 2005;12:3385–3389. doi: 10.1093/humrep/dei236. [DOI] [PubMed] [Google Scholar]
- 3.Bhargava R, Levin L. Spectrochemical Analysis Using Multichannel Detectors. Blackwell; Oxford, U.K: 2005. [Google Scholar]
- 4.Lasch P, Haensch W, Lewis EN, Kidder LH, Naumann D. Appl Spectrosc. 2002;56(1):1–9. [Google Scholar]
- 5.Lasch P, Naumann D. Cell Mol Biol. 1998;44:189–202. [PubMed] [Google Scholar]
- 6.Chew SF, Wood BR, Kanaan C, Browning J, MacGregor D, Davis ID, Cebon J, Tait BD, McNaughton D. Tissue Antigens. 2007;69(s1):252–258. doi: 10.1111/j.1399-0039.2006.00775.x. [DOI] [PubMed] [Google Scholar]
- 7.Wood BR, Bambery K, Quinn MA, McNaughton D. Proc SPIE Smart Mater, Nano-, and Micro-Smart Syst. 2005;5651:78–84. [Google Scholar]
- 8.Wood BR, Bambery KR, Evans CJ, Quinn MA, McNaughton D. BMC Med Imaging. 2006;6 doi: 10.1186/1471-2342-1186-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wood BR, McNaughton D, Chiriboga L, Yee H, Diem M. Gynecol Oncol. 2003;93:59–68. doi: 10.1016/j.ygyno.2003.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bambery K, Wood BR, Quinn MA, McNaughton D. Aust J Chem. 2004;57:1139–1143. [Google Scholar]
- 11.Zhang L, Small GW, Haka AS, Kidder LH, Lewis EN. Appl Spectrosc. 2003;57:14–22. doi: 10.1366/000370203321165151. [DOI] [PubMed] [Google Scholar]
- 12.Lasch P, Haensch W, Naumann D, Diem M. Biochim Biophys Acta. 2004;1688:176–186. doi: 10.1016/j.bbadis.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Mansfield JR, McIntosh K, Crowson AN, Mantsch HH, Jackson M. Appl Spectrosc. 1999;53:1323–1330. [Google Scholar]
- 14.Mansfield JR, Sowa MG, Scarth GB, Somorjai RL, Mantsch HH. Anal Chem. 1997;69:3370–3374. doi: 10.1016/s0895-6111(97)00018-9. [DOI] [PubMed] [Google Scholar]
- 15.Miller LM, Smith RJ. Vib Spectrosc. 2005;38:237–240. [Google Scholar]
- 16.Carr GL. Vib Spectrosc. 1999;19:53–60. [Google Scholar]
- 17.Carr GL. Rev Sci Instrum. 2001;72:1613–1619. [Google Scholar]
- 18.Hunter AG, Moore RM. J Dairy Sci. 1987;70:1646–1651. doi: 10.3168/jds.S0022-0302(87)80192-3. [DOI] [PubMed] [Google Scholar]
- 19.Sirard MA, Florman HM, Leibfried-Rutledge ML, Barnes FL, Sims ML, First NL. Biol Reprod. 1989;40:1257–1263. doi: 10.1095/biolreprod40.6.1257. [DOI] [PubMed] [Google Scholar]
- 20.Levesque JT, Sirard MA. Mol Reprod Dev. 1995;42:114–121. doi: 10.1002/mrd.1080420115. [DOI] [PubMed] [Google Scholar]
- 21.Kruip TAM, Cran DG, van Beneden TH, Dieleman SJ. Gamete Res. 19:23–31. [Google Scholar]
- 22.McEvoy TG, Coull GD, Broadbent PJ, Hutchinson JS, Speake BK. J Reprod Fertil. 2000;118:163–170. [PubMed] [Google Scholar]
- 23.Everall N. Spectroscopy. 2004;19:22–27. [Google Scholar]
- 24.Wold S. Pattern Recogn. 1976;8:127–139. [Google Scholar]
- 25.Mohlenhoff B, Romeo M, Diem M, Wood BR. Biophys J. 2005;88:3635–3640. doi: 10.1529/biophysj.104.057950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warner CM, Newmark JA, Comiskey M, De Fazio SR, O'Malley DM, Rajadhyaksha M, Townsend DJ, McKnight S, Roysam B, Dwyer PJ, DiMarzio CA. Reprod Fertil Dev. 2004;16:729–741. doi: 10.1071/rd04088. [DOI] [PubMed] [Google Scholar]
- 27.Matthäus C, Chernenko T, Newmark JA, Warner CM, Diem M. Biophys J. 2007;93:668–673. doi: 10.1529/biophysj.106.102061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Blerkom J. Reproduction. 2004;128:269–280. doi: 10.1530/rep.1.00240. [DOI] [PubMed] [Google Scholar]
- 29.Albertini DF, Barrett SL. Semin Cell Dev Biol. 2004;15:599–606. doi: 10.1016/j.semcdb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Edwards RG, Beard HK. Mol Hum Reprod. 1997;3:863–905. doi: 10.1093/molehr/3.10.863. [DOI] [PubMed] [Google Scholar]
- 31.Edwards RG. Reprod BioMed Online. 2001;3:138–160. doi: 10.1016/s1472-6483(10)61983-9. [DOI] [PubMed] [Google Scholar]
- 32.Zamboni L, Mastroianni L. J Ultrastruct Res. 1966;14:95–117. [Google Scholar]
- 33.Wassarman PM, Josefowicz WJ. J Morph. 1978;156:209–236. doi: 10.1002/jmor.1051560206. [DOI] [PubMed] [Google Scholar]
- 34.Assey RJ, Hyttel P, Kanuya N. Anat Embryol. 1994;190:461–468. doi: 10.1007/BF00235493. [DOI] [PubMed] [Google Scholar]
- 35.Subramaniam MK, Aiyar RG. J R Microsc Soc. 1935;55:174. [Google Scholar]
- 36.Nili H, Mesbah F, Kafi M, Esfahant MHN. Anat Histol Embryol. 2004;33:196–199. doi: 10.1111/j.1439-0264.2004.00536.x. [DOI] [PubMed] [Google Scholar]
- 37.Schaefter BE. J Cell Sci. 1977;26:47–55. doi: 10.1242/jcs.26.1.47. [DOI] [PubMed] [Google Scholar]
- 38.Komatsu J, Yamano S, Kuwahara A, Tokumura A, Irahara M. Life Sci. 2006:506–511. doi: 10.1016/j.lfs.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 39.Lefèvre B, Pesty A, Courtot AM, Martins CV, Broca O, Denys A, Arnault E, Poirot C, Avazeri N. Crit Rev Eukaryotic Gene Expression. 2007;17:259–269. doi: 10.1615/critreveukargeneexpr.v17.i4.10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


