Abstract
Drug discovery today is impossible without sophisticated modeling and computation. In this review we touch on previous advances in computational biology and by tracing the steps involved in pharmaceutical development, we explore a range of novel, high value opportunities for computational innovation in modeling the biological process of disease and the social process of drug discovery. These opportunities include text mining for new drug leads, modeling molecular pathways and predicting the efficacy of drug cocktails, analyzing genetic overlap between diseases and predicting alternative drug use. Computation can also be used to model research teams and innovative regions and to estimate the value of academy-industry ties for scientific and human benefit. Attention to these opportunities could promise punctuated advance, and will complement the well-established computational work on which drug discovery currently relies.
The identification of chemical agents to enhance the human physiological state—drug discovery—involves coordination of highly complex chemical, biological and social systems and requires staggering capital investment—estimated at between $100 million and $1.7 billion per drug [1, 2].
In the search for new drugs there are numerous sources of error. These root from our limited understanding of the biology of drug action and the sociology of innovation. Biologically, the bottleneck is our poor knowledge of molecular mechanisms underlying complex human phenotypes [3, 4]. Socially, we lack models that accurately capture the link between successful discovery and the dynamic organization of researchers and resources that underpins it.
Computational approaches, if applied wisely, hold the potential to substantially reduce the cost of drug development by broadening the set of viable targets and by identifying novel therapeutic strategies and institutional approaches to drug discovery. Here, we provide an overview of what computational biology and sociology have to offer and what problems need to be solved in their mission to support drug discovery.
Computational biology methods for drug discovery
A number of computational methods have been successfully applied throughout the drug discovery pipeline, from mining textual, experimental and clinical data to building network models of molecular processes, to statistical and causal analysis of promising relationships, as summarized in Figure 1 and Text Box 1.
Figure 1. Role of computational technologies in the drug discovery pipeline.
This figure summarizes how computational biology can impact drug discovery. The various stages of the drug discovery pipeline (See textbox 1 for detailed background on each step) are listed in the left column. We note that the traditional linear process is shifting to become more parallel, simultaneous and cyclical: Red arrows indicate the traditional pipeline while yellow dashed arrows suggest novel workflows that are increasingly adopted by pharmaceutical and biotechnological companies to increase productivity. Biomarkers and analysis of of target molecules’ tissue distribution are the most recently introduced checkpoints and are not required by FDA.
Computational biology methods discussed in the main text* are listed along the top row. Blue lines illustrate how each method is related with others. For example, sequence analysis relies on pattern recognition and classification; Text mining, terminologies and knowledge engineering are entwined, as are pattern recognition and classification. The impact of each computational technique on each stage of drug discovery (dependency between a pair of disciplines) is classified into three categories: actively or heavily used (big black dot), less actively used (small black dot) and our suggestion (small grey dot).
*We do not emphasize chemical informatics in the main text because it relates to issues from chemistry and not biology. Chemical informatics comprises a wide range of approaches from computational and combinatorial chemistry that model lead properties and their interaction with targets. These include chemical structure and property prediction; structure activity relationships; molecular similarity and diversity analysis; compound classification and selection; chemical data collection, analysis and management; virtual drug screening; and the prediction of in vivo compound characteristics.
PK: pharmacokinetics; PD: pharmcodynamics; ADME: absorption, distribution, metabolism, and excretion.
Text Box 1. Drug discovery pipeline.
The traditional drug discovery workflow is shown in Figure 1 (in red). It typically begins with target identification: The target is a human molecule that a drug recognizes and modifies in order to achieve an intended therapeutic effect. Alternatively, the target can be part of a pathogen’s cellular machinery; the role of the drug in this case is to kill the pathogen by interrupting the drug’s target. Most drug targets are proteins, historically drawn from a few families, such as enzymes, receptors and ion channels. Target identification is heavily dependent on 1) analysis of disease mechanisms – to locate the molecular system most likely to incorporate a promising target, 2) genomics – to rank genes with respect to physiological function, and 3) experimental proteomics – to identify candidate proteins and protein interactions that can be inhibited or enhanced by drug.
The next stage is target validation. At this stage researchers use a battery of experimental techniques (genetic engineering, transgenic animal models, antisense DNA/RNA perturbation of pathways and structural biology) to understand the molecular role of the prospective drug target better, and to elucidate whether an agonist or antagonist drug should be designed. It is not uncommon to discover that the initial target is inappropriate for a variety of reasons, in which case target identification must be repeated.
Following target validation, if the targeted molecule appears promising, it is time to identify and optimize a lead or prospective drug. Most frequently, the lead is a small molecule, but it can also be a peptide, antibody or other large substance. To appreciate the difficulty of this stage, consider the number of possible molecules. While finite—molecule size is naturally bounded—the number of potentially relevant compounds is greater than 1024 [56] even if we limit ourselves to small molecules. “Brute force” search approaches – testing all leads exhaustively – are clearly unworkable: Intuition and serendipity are valued highly. There are numerous high-throughput techniques, such as synthetic and combinatorial chemistry, compound screening, design of compound libraries, and a variety of experimental design and sampling approaches (see Figure 1) that enable relatively efficient search through the space of lead compounds.
Lead optimization is the process of improving initial “hits” from primary screening. Medicinal chemists draw on principles of organic chemistry to tweak hit molecules and carry out batteries of tests that ensure the modified molecule is more specific to its target, more effective, less toxic, and has a long enough half-life in human tissue to achieve therapeutic effect. In the framework of lead optimization, pharmacodynamics measures the strength of drug effect as a function of drug dosage; pharmacokinetics studies the time course of drug absorption, distribution, metabolism, and excretion; and toxicology deals with any poisonous impact of the new lead.
Clinical trials are the pinnacle of drug development: The general public and patent agencies typically do not learn about the existence of leads until they reach trials. Clinical trials are divided into several stages. First phase trials typically investigate the safety of a drug when administered to healthy humans. Given encouraging phase I results, phase II explores efficiency and safety of the drug in the class of patients expected to benefit from the proposed therapeutic effect. Phase III trials are similar to phase II, but the subject pool is larger to detect subtle influences of the drug and better evaluate its efficiency. There are also optional phase IV trials that may provide improved estimates of drug efficiency and adverse effects: By the time the drug reaches phase IV trials, it has usually reached the market. Clinical trials may be followed by post-market research to discover and validate new applications of existing commercial drugs.
Molecular modeling or structural biology is the most established domain of computational biology. It aims to predict and model properties of biological targets, and rests on the shoulders of chemistry, quantum and biophysics, experimental crystallography and computer science. Alongside established tools of computational chemistry, which model properties of drug leads and their interaction with targets, molecular models form the centerpiece of computational drug discovery and development.
Sequence analysis aims to efficiently compare nucleotide and amino acid sequences, thereby allowing researchers to impute a gene’s function by considering evidence from homologous genes, often from different biological systems. Sequence analysis has now become an indispensable part of target identification in early-stage drug discovery (see Figure 1).
Over the last several years, molecular models and sequence analysis have grown productive and mature. Marginal improvements have diminished in value and there is less room for radical innovation. Thus, we focus this review on emerging computational approaches such as the simulation of molecular networks, probabilistic data integration, and development of drug cocktails. These and related approaches point toward broader biological systems underlying—disease systems that extend beyond single-gene inheritance and pair wise molecular interactions. As such, they promise to extend the repertoire of druggable molecules and suggest novel therapeutic strategies likely to yield big returns in drug discovery.
Text mining (http://people.ischool.berkeley.edu/~hearst/text-mining.html) is one such emerging discipline that aims to extract phrases and statements from electronically stored texts [5, 6], and this information is then assembled and analyzed to generate new knowledge [7]. Text mining techniques draw from computational linguistics, natural language processing, data mining and artificial intelligence. In the biomedical sciences, text mining is currently used to screen articles for biological terms including molecule names and biological statements such as molecular interactions.
The development of terminologies and ontologies consistent—systems of terminologically bounded statements—enables the standardization of text-mined information so that it can be used to model wider biological processes and inspire new drug targets and strategies. Conceptual standardization also facilitates the consistent organization of experimental data. This solves a perennial problem in biomedical research: Data produced with disparate methods from distinct institutions is heterogeneous. For example, drug information provided by Medline Plus (http://www.nlm.nih.gov/medlineplus/) is aimed at the general public and is linguistically incompatible with the drug handbook used by physicians. Standardization enables the merger of diverse resources, increasing the total volume of data for analysis and the number and subtlety of patterns discoverable within it. Together, these steps constitute knowledge engineering. Drawing on philosophy and computer science, knowledge engineering aims to create machine-organized systems of knowledge that enable artificial intelligence applications to make inference that would be difficult or impossible to achieve for human scientists. Knowledge engineering relates to text mining in two distinct ways. Texts are first mined to discover terminologies such as a lexicon of gene names; these terminologies, when organized, enable the more consistent extraction of relations (such as activation of one gene by another) in a subsequent pass through texts. We illustrate this in Figure 2 with an ontology containing the most common concepts in drug discovery and exemplify it in Text Box 2 with the case study for the calibration of warfarin dosage. With this kind of structured knowledge representation, computers can simulate the reasoning process of human scientists on a large scale. For example, the protein BCR-ABL is involved in the apoptosis pathway, which results in cell death, which, in turn, is related to chronic myelogenous leukemia. Reasoning across this chain, we might hypothesize that a BCR-ABL inhibitor could cure chronic myelogenous leukemia, as confirmed by researchers with the discovery of Gleevec [8].
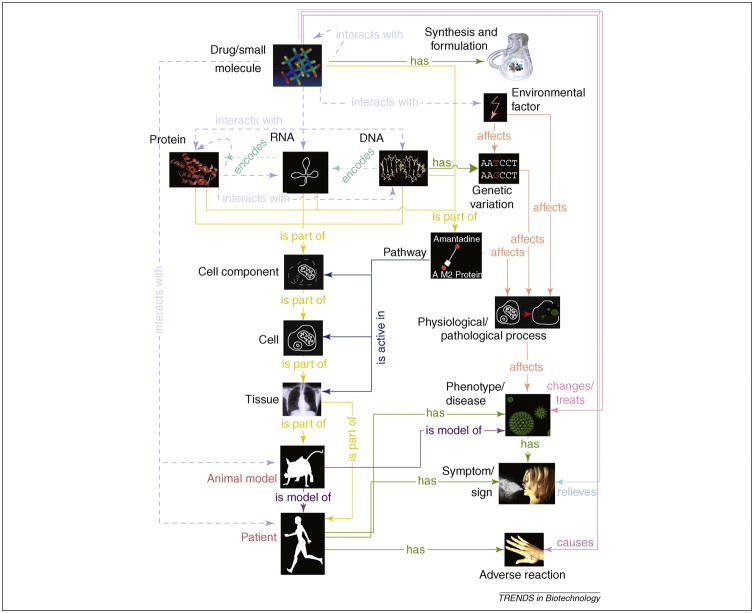
Figure 2. A provisional ontology for drug discovery.
The ontology features a concept for drugs or small chemical molecules and another for drug synthesis and formulation. Further, a drug is known to target a specific molecule: protein, RNA, or DNA. A drug’s target is biologically active in the context of a pathway, cellular component, cell, and tissue. The drug is typically tested using an animal model for a specific human phenotype, usually a disease. A patient or human with the disease phenotype has a set of symptoms/signs, some of which can be unrelated to the disease. Drugs treat patients, but can cause adverse reactions. Finally, we link the patient’s drug response to the genetic variation in her genome and environmental factors encountered. These concepts are linked with directed or undirected relations, such as encodes (DNA encodes RNA), interacts with (drug-drug interactions), is part of (a cell is part of a tissue), affects (environmental factor affects genetic variation), and several others. We illustrate our ontology with the drug amantadine, which acts as a prophylactic agent against several RNA virus-induced influenzas that afflict the respiratory system and result in coughing and sneezing. Amantadine can cause a skin rash as a side effect.
Text Box 2. Successful applications of computational biology in drug discovery: two case studies.
Case study I: Determining patient-specific warfarin dosage based on clinical and genetic data
Warfarin is the most widely prescribed anticoagulant in North America. Delivering the right amount is critical – if the dose is too high, patients could bleed profusely; if it is too low, they could develop life-threatening clots. This task is difficult for doctors because ideal dosage varies from patient to patient.
In a recent study [57], a consortium of researchers pooled data about 5,052 patients who underwent warfarin treatment across several organizations. Information describing 4,043 patients was used to develop and fit a statistical model, while data about the remaining 1,009 patients was used to validate it. Parameters included traditional clinical variables such as demographic characteristics, primary justification for warfarin treatment, concomitant medications and genetic variables, primarily single nucleotide polymorphisms in genes VKORC1 (the therapeutic target of warfarin) and CYP2C9 (the warfarin metabolism-related gene) for each patient.
The problem was then framed as a statistical regression: the outcome variable was the stable therapeutic dose of warfarin, and input variables included all clinical and genetic variables mentioned above. The researchers used a battery of regression methods: support vector regression, regression trees, model trees, multivariate adaptive regression splines, least-angle regression, Lasso, and ordinary linear regression. Their results show that for the 46% of warfarin recipients who are difficult to dose because they require abnormal quantities of warfarin, the ordinary linear regression model with two biomarkers performed best, and beat models with clinical data alone. For these patients, model predictions could be life saving.
The work drew on PharmGKB (Pharmacogenomics Knowledge Base), which required knowledge engineering to structure its database schema and content [58]. Patient-specific amounts of known biomarkers were used as input variables for the analysis.
Extensions to this work could include text- and data-mining efforts [11, 59] to identify additional markers for analysis (e.g., genetic loci correlating with the phenotype) as well as interactions between warfarin and food additives or other drugs. In turn, this would require the redesign of regression models to include additional variables and likely second-order interactions between them. Analysis would then require a larger sample of patient records. To obtain such a sample, still larger multi-institutional collaborations would be required, the design of which would, in turn, benefit from dynamic models of research collaboration.
Case study 2:Fragile Xsyndrome, FMR1 gene, and fenobam
Fragile X syndrome [60] is an X-linked dominant genetic disorder [61] most frequently caused by transcriptional inactivation of the FMR1 gene, either due to expansion of a CGG repeat found in the 5′-untranslated region or deletion of the gene [62]. Fragile X syndrome is associated with intellectual and emotional disabilities, ranging from mild impairment in cognition to mental retardation and autism.
Experiments with transgenic mice [63] having the Fmr1 gene knocked-out demonstrated that in the absence of functional FMR1 protein, neuronal synapses are altered and dendritic protrusions result are structurally malformed [64]. It was shown [64] that two different antagonists of the mGluR5 receptor can rescue Fragile X-related neuronal protrusion morphology.
The unexpected turn in this story was the discovery that researchers could compensate for the absence of FMR1 protein by tuning the state of molecules in its interacting neighborhood, namely, by inhibiting the mGluR5 receptor.
Fenobam is a relatively old anti-anxiety drug [65] and a potent antagonist of mGluR5 receptor [66]. It was therefore natural to suggest that fenobam might rescue Fragile X-related abnormalities. Preliminary results have been encouraging and a clinical trial of an oral drug therapy option for adult patients with Fragile X syndrome is currently underway [67].
Data integration deals with the challenges arising from fusing multiple data types, such as relationships established from literature-mining, gene expression data from microarrays, protein-protein interactions from yeast two-hybrid experiments, and even clinical patient records. Data integration involves not only an accurate ontological representation of relevant factors, but also an estimate of uncertainty and bias in the data from each source. As a field, data integration is still in its infancy, a state of “productive anarchy” where approaches abound but no universal standards exist. We believe data integration will become increasingly important for drug discovery as available data for biosystems that are relevant to human disease grows (see Figure 3).
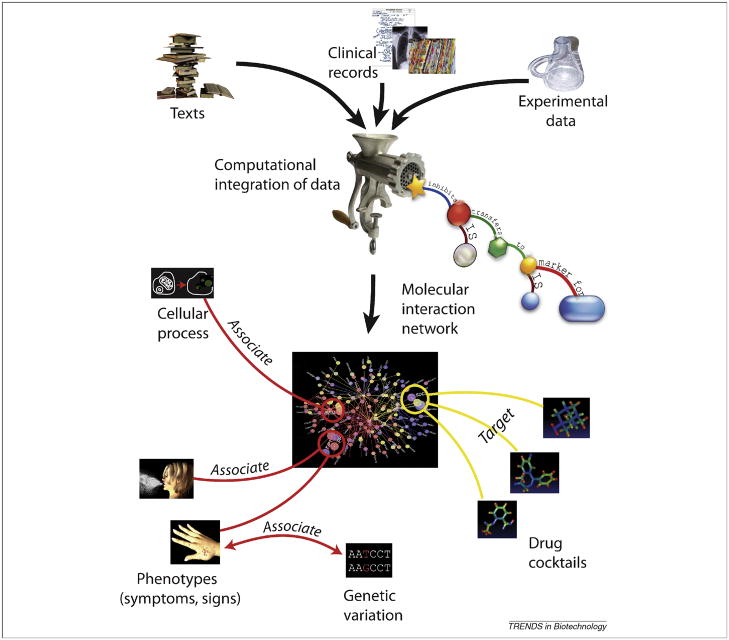
Figure 3. Overview of promising computational opportunities in drug discovery.
Text-mining enables the extraction of information from publications and clinical records. Mathematical modeling helps to assess experimental data in context of previously collected facts, while computational data integration distills multiple raw data types into a collection of computable biological statements. The resulting network of semantic relations can serve as a scaffold for modeling biological processes, design and optimization of therapeutic drug cocktails, and linking complex phenotypes to genotypes. The figure incorporates ontological concepts outlined in figure 2: cellular process (such as tissue necrosis), symptoms (in this case, sneezing, allergic rash), genetic variation (depicted as a single nucleotide polymorphism), and drugs (amantadine, valium, and aspirin, listed here top to down.
Data mining takes these large, complex arrays of data as input, to which a range of statistical and mathematical techniques are then employed to discover unanticipated regularities. Data mining has become important in diverse commercial applications including the mining of movie review databases, credit card purchase data, and stock exchange transactions. In biomedicine, the mining of databases of molecular interactions has already proven successful in suggesting novel disease-related processes [9, 10] (note the hypothetical association between cellular process and molecular pathway in Figure 3).
Data mining has also been used to identify patterns in clinical patient records that point toward novel therapeutic interventions (see Figure 3). In combination with patient-specific genomic data, such as single nucleotide polymorphisms and copy number variation, clinical data mining could lead to better understanding of drug action—the mechanisms underlying drug successes and failures. This could ultimately increase the match between patients and drugs by inspiring development of customized drugs. There are, however, many challenges associated with the use of clinical data. Physicians are not consistent in entering data and, for billing purposes, it is acceptable to encode diseases with their “equivalents,” such that diabetes mellitus I is often coded diabetes mellitus II and male breast cancer as its female counterpart. Moreover, patient records are often incomplete as patients move across hospitals and regions. Even a perfect data series will often not contain other relevant patient data, such as diet and over-the-counter drug use. Concerns about patient privacy also limit access to clinical data. Nevertheless, we believe that mining medical records will become increasingly important for drug discovery and development.
There are several approaches to identify patterns within the data-mining toolbox. Pattern recognition and classification are methods that mimic human perception of real-world phenomena, such as recognizing faces and handwritten letters, detecting contours of objects and threatening sounds, and separating relevant from irrelevant text in search queries. These methods have been used in biomedicine to discover multi-molecular processes in complex databases containing biological and clinical information [11]. Information theory and signal processing are other approaches to pattern-identification. They originated in response to practical problems of communication [12] in which the analyst attempts to separate a communicated signal from the “noise” that clouds it. For example, a sender may emit an encoded message over a noisy channel such as radio waves in the open air. A receiver records the signal, separates it from noise and decodes the message. The corresponding mathematical toolbox allows estimation of mutual information from several variables to detect non-linear dependencies among them. These approaches are commonly used in computational biology and drug discovery [10,13-15] to identify pathways involving multiple genes, proteins and other molecules.
Uncovering relationships through data mining naturally turns up spurious associations that derive from noise and artifacts of the method or data source. Statistical models are required to rigorously test these associations as hypotheses. Perhaps the most common family of statistical models used in drug discovery and development is regression analysis (see the warfarin example in Text Box 2). Regressions characterize the stochastic relationship between a response variable, such as drug toxicity, and input or causally independent variables, such as the presence of certain structural motifs in the drug, its molecular weight or solubility. Although regression is one of the oldest approaches to statistical modeling, it permeates virtually every stage of drug discovery, from ranking prospective targets and leads to analyzing clinical trial data.
As statistical modeling has evolved, increased care has been taken to isolate empirical measurements that allow the separation of causal from coincidental associations. Analysts have also worked to systematically compare competing causal models. Causality analysis is a paradigm within statistics and hypothesis testing devoted to these concerns. Its methods have been employed in biomedicine to determine causal relations between drugs and dangerous side effects [16], link genetic variation and phenotype [18] (see Figure 3 and Text Box 2), and separate genetic regulatory relationships from the co-occurrence of molecular events [17].
Promising directions in computational biology for drug discovery
Molecular network-based models
If we conceive the molecules inside a human cell—genes, proteins, RNAs, and exogenous small molecules—as vertices of a graph, and interactions between pairs of molecules as directed edges of this graph, we can represent the cell as a molecular network as illustrated in the centre scheme of Figure 3. Drugs are external substances that enter and interrupt this complex network of molecular processes. A robust, dynamic model of human cellular machinery could explain clinical outcomes for existing drugs and predict the effect of new leads. This is, of course, the Holy Grail of computational biology—a dream rather than reality. Nevertheless, recent advances in data integration have enabled the construction of complex ‘hairball’ networks based on text-mining and high-throughput experiments [19] ( Figure 3). Most of these networks are static, but as they become more precise and incorporate the temporality of biomolecular processes, they will likely enable identification and systematic screening of novel drug targets. Even current, static networks suggest design principles and metrics that could accelerate the discovery of successful drugs. For example, population variability in the genes underlying molecular networks, especially in the region of the drug target, could pinpoint patient subgroups for which a drug might be most (and least) effective. The occurrence of rare variation in patient subgroups might even warrant undertaking clinical trials for a personalized drug. On the other hand, molecules that are highly connected in the network of cellular processes or that are present in a number of human tissues are likely to be poor drug targets: Their interruption will have broad and often unintended consequences. (See Fragile X case study in Text Box 2, in which the druggable protein was distinct from the molecular cause of the condition). High-quality targets will instead have the property of high “betweenness centrality”, i.e. they will ideally link normal and pathological pathways in such a way that interrupting their function will eliminate only disease-related processes while leaving healthy ones intact [20, 21].
It is difficult, however, to isolate exclusive links between vicious and virtuous molecular processes. More often, multiple targets exist whose concerted interruption would exert a greater therapeutic effect than the disturbance of any one in isolation. This is the goal of multi-drug cocktails, a growing strategy that applies compounds jointly to interact with one or more molecular targets as illustrated in Figure 3. One successful drug of this type is Advair, a cocktail of fluticasone and salmeterol used to manage asthma and chronic obstructive pulmonary disease. Most available cocktails have been discovered serendipitously leaving much room for research into the action of multiple drugs and their systematic identification [22]. The potential of this direction is highlighted by the deep link between the genetic basis of disease and the biology of drug action. Many common pathological phenotypes, such as diabetes and neurodegenerative disorders, appear to have multifactorial genetic bases that are confounded with environmental risk factors. For example, patients with very similar disease phenotypes may have distinct genetic variants (genetic heterogeneity). Alternately, multiple genetic aberrations may act synergistically to contribute to a specific disease phenotype (genetic epistasis). For these disorders, multiple genes or proteins would need to be targeted at once to achieve the desired outcome with minimal side effects. Current approaches to identify cocktails range from high-throughput combinatory screening for drug leads [23] to large-scale modeling of molecular systems [24, 25]. Because the combinatorial search space for drug cocktails grows exponentially with the number of ingredients, experimental design methodologies will be critical to efficiently sample that space.
As with single drug leads, computational methods can be harnessed to rank combinations of drug leads. As a first step, historical publications could be mined for associations between currently under investigated molecules and their impact on human biological processes. Subsequently, clinical databases can be screened to detect serendipitous connections between approved drugs and harmful or beneficial interactions when taken by the same patient [26]. Finally, predictive models can be designed to relate the molecular structure of cocktail components with the clinical phenotype of treated patients. These models would incorporate information about cellular networks and patient genetic variability.
Even when associations within a molecular network do not prove causal, the persistent association of molecules with pathological pathways could be exploited as biomarkers (see Figure 3). Biomarkers [20, 21] are molecules present in human tissues, such as proteins and lipids, which can change state in response to fluctuations of normal or pathogenic cellular processes or even therapeutic intervention. Biomarkers are extremely useful for disease diagnosis [19] and prognosis, calibration of appropriate drug dosage [27], evaluation of drug efficacy and toxicity [28], as well as patient stratification in clinical trials.
Biomarkers might also help us understand the genetic heterogeneity associated with pathological phenotypes [29]. This could suggest patient-specific treatment recommendations [30] so that different drugs are administered to people with distinct genetic variation underlying the same phenotype. Despite this promise, the area of biomarker development has recently been shrouded by controversy [31] as a number of published associations between putative biomarkers and target phenotypes that were based on microarray and other genomic data now appear irreproducible, possibly due to flaws in statistical analysis and interpretation.
We close this section with discussion of clinical data mining, an area emerging with the rise of information technology in health care. There are many sources of clinical observation, from hospital billing records, patient discharge summaries and insurance archives to clinical trial statistics. All of these resources have been implemented in response to specific practical needs—usually related to billing for care. The richest of these resources, electronic health records, store information about tens of millions of patients and include demographic data, drug prescription history and side effects linked to the time series of diagnosed phenotypes and laboratory tests (see Text Box 2). Despite limitations described earlier, analyzing these records promises to be extremely useful. We anticipate using clinical databases for evaluating drug safety and the discovery of subtle adverse effects. Moreover, analogous to the rationale for using drug cocktails when multiple pathways lead to disease, knowledge of multiple diseases that result from the same pathway suggests opportunities to repurpose existing drugs, a considerably less expensive discovery strategy.
Modeling the social context of drug discovery
Drug discovery and development require expansive research teams that have grown more complex over the past 25 years, spanning academy and industry and incorporating diverse expertise. Poorly orchestrated, disconnected or duplicative research programs can cause significant losses in research productivity and funds, leading to the inflation of health care costs. The systematic study of innovation patterns in academia and industry has led to detailed models of the knowledge discovery process. We believe that improving these models will elucidate successful paths to drug discovery and highlight opportunities for institutional innovation that could increase the efficiency and public benefit of drug-related expenditures.
Based on analysis of research articles and patent bylines, social scientists have developed inventories of authors and inventors, as well as of their evolving collaboration networks. [32, 33] Social networks, like idealized molecular networks, provide a coarse view of complex social processes. They are nevertheless useful in capturing social patterns and trends. Recent findings point to the increasing importance of “big science” as multi-author or team research has become more frequent and their publications have the highest citation impact [34]. When team composition has been analyzed, those bridging different fields were more likely to achieve breakthroughs [35, 36], and those mixing newcomers and incumbents resulted in the most referenced research [37]. As we join author and inventor information with the text-mined research topic data described earlier, it will become possible to build predictive models that guide scientists and organizations in assembling more successful teams for drug discovery.
Recent analyses of research articles and patents have revealed innovation patterns that vary considerably across the institutions in which biomedical research takes place [38, 39]. Analysis of regional biotechnology clusters suggests that as the number of institutional neighbors that fund research increases, the success of an institution’s own research also increases. These so-called ‘spillovers’ have been measured in the form of successful patent applications, [40, 41] described innovations, [39, 42] and community recognition of innovations (e.g., article and patent citations) [32]. This suggests clear benefits to working in dense, well-funded research environments. The mobility of local labor is one likely factor contributing to spillovers, as is the ease of informal data and idea sharing. Some ideas that spread locally through informal means never become widely disseminated because the review process for publications and patents systematically censors negative results, such as drug leads that do not inhibit certain targets. This highlights an opportunity for biomedical institutions to foster interpersonal and Internet venues where hypotheses, findings and negative results can spread.
Most notable on the landscape of drug discovery is the clear distinction between institutional sectors. Companies develop drugs for an anticipated market of patients, leaving fundamental investigation into treatments for small or impoverished populations underfunded. Governments and philanthropies aim to counteract these so-called market failures by selectively supporting such work in universities, research institutes and small companies. Fundamental research, however, has become more relevant for drug discovery since the detection of ‘disease genes.’ As a result, more scientists from academy and industry are converging to investigate the same diseases, leads and targets. Even though distinctions blur at the boundaries where researchers circulate between sectors, [43] scientists in academy and industry have different incentives and resources that continue to inspire distinct activities and research cultures. In universities and research institutes, reputational credit is awarded scientists for priority in publishing [44] the most general scientific discoveries [40]. In companies, monetary rewards are achieved by controlling and appropriating benefits from the most powerful technological innovations [41]. As a result, university research is rapidly published in order to maximize dissemination, while company research is patented to maximize the firm’s ability to benefit from exclusive rights to the market [42]. Pharmaceutical companies surveyed in 1994 reported that secrecy was even more effective protection for innovations than patenting, especially in manufacturing drugs [42].
Other institutional differences exist: Because universities have a higher proportion of independent researchers per capital investment, at any given moment academic research will appear less focused than industry research, while companies channel money toward the exploitation of singular opportunities, such as an extensive molecular screen. Over time, however, industry shifts quickly between different research areas to identify and appropriate value from product-relevant discoveries, while academic researchers focus and swarm [38, 39]. As a result, industry leaves behind many expensive draft genome sequences, partial screening experiments, and incomplete collections of biomaterials that could be valuable for academic research.
These differences suggest unexploited complementarities between academic and industrial drug discovery efforts. Recent data [45] on the demography of illness, obtained from epidemiologists and researchers in the emerging field of population health, might enable us to improve models of existing and potential markets for drugs. This will, in turn, facilitate estimation of the potential market size for different treatments and answer important questions like whether the afflicted are rich or poor; how many there are; and how serious is the affliction’s debilitating effect? Models built with these data have recently been used to estimate the value that G8 states might be willing to prepay pharmaceutical and biotech companies for developing successful treatments afflicting impoverished populations unable to justify the commercial expense on their own [46].
Modeling potential drug markets, research institutions and investment alongside the biological systems described above will allow us to predict those research areas for which cast-off information from industry could be most valuable. Some research suggests that cross-organization collaborations host the most innovation in drug discovery [47]. Analyzing academy-industry complementarities explicitly will suggest where productive relationships could yet be consummated and where governments could facilitate them by paying companies for: (i) leads or targets relevant for noncommercial diseases; (ii) rights to redirect drugs for noncommercial diseases that have a genetic overlap with commercially relevant diseases; (iii) industrial ‘draft’ research made available for academic investigation and discovery.
Governments have already attempted to shape other aspects of the innovation landscape, which should now be reevaluated with available data. Because a rising proportion of biological research is medically relevant, the U.S. National Institutes of Health has begun to increase its sponsorship of ‘translational’ science to further encourages the application of fundamental research (see http://nihroadmap.nih.gov/clinicalresearch/overview-translational.asp). Governments have also expanded opportunities to obtain intellectual property (IP) for drug-relevant discoveries. Thirty years ago, patents were unavailable for many discoveries in basic science, but now it is possible to patent genes and altered organisms, as well as basic scientific ‘research tools’ [48]. In 1976, after 9 years of trial and error, University of Chicago’s Eugene Goldwasser isolated erythropoietin [49], the human protein that stimulates red blood cell proliferation and soon after identified the producing gene, rHuEpo, publishing both discoveries[50]. These publications enabled Amgen and Johnson & Johnson to produce and to patent two synthetic erythropoietins, Epogen and Procrit, which have gone on to become blockbuster drugs prescribed for a variety of blood scarcity conditions. Today, this process would have been much different, likely involving academic patents with the help of a technological transfer office, and pharmaceutical licensing or the formation of a ‘spin-off’ company.
The choice governments make to stimulate translational versus fundamental research might have major implications for the sustainability of pharmaceutical innovation. Translational research builds on existing biological knowledge and is most productive in short-term drug identification and optimization. Fundamental research into biological systems, however, will likely be critical for the long-term success of drug discovery inasmuch as it proposes entirely new target families and therapeutic strategies. Researchers have begun to model this process and quantify the costs and benefits of research investment [41]. The importance of this approach is suggested by recent work that demonstrates ‘the burden of knowledge’ in modern biomedical science [51]. Much knowledge has already accumulated and the diminishing success of established techniques suggests that ‘low hanging fruit’ on the tree of knowledge has been harvested.
Finally, many have observed the creation of patent ‘fences’ or ‘thickets’ surrounding fundamental biological techniques and resources [52]. Some have expressed concern that proliferating legal rights could result in an ‘anti-commons’ where fewer and fewer resources are commonly available for researchers to build upon [53], slowing drug discovery and stifling life-saving innovations [54]. Computational methods can estimate the impact intellectual property (IP) rights exert on the accumulation of pharmaceutical knowledge and the speed of drug discovery. A recent analysis of paper-patent pairs covering identical biotechnologies suggested that once patents are granted, the dissemination of ideas slows slightly [55]. We believe the increasing resolution of data on published biomedical ideas and IP will enable realistic models of pharmaceutical innovation and legal protection that could become a powerful tool to advise economic and government policy.
Conclusion
We have discussed emerging computational approaches that enable the modeling of entire biological pathways relevant to health and disease. These hold great promise for drug discovery, especially building molecular network models of disease, mining biomedical literature and patient records, and harnessing computation to drive discovery of new targets, multi-drug cocktails and novel purposes for existing drugs. In parallel, electronic publication and patent data have facilitated the development of detailed models of the social process underlying biomedical innovation. We anticipate that advances in modeling the networks of scientists, inventors, institutions, resources and rights that drive drug discovery will illuminate successful strategies and opportunities for institutional innovation that ensure the rate and social relevance of drug development.
Computation has traditionally been used intensively in biological and social science—to solve difficult existing problems. Recent developments suggest that utilizing computation extensively to link disparate data and support integrative models can broaden our vision of biological and social processes. We believe this new view will fundamentally expand the possibilities of drug action, reorganize and reignite innovation in drug discovery.
Acknowledgments
We are grateful to Rajeev Aurora, Carolyn Cho, Murat Cokol, Preston Hensley, Ivan Iossifov, Aaron J. Mackey, and Nathan Siemers, for insightful discussions and comments on earlier versions of the manuscript. This work was supported by the U.S. National Institutes of Health (GM61372 and U54 CA121852-01A1) and the National Science Foundation (0242971).
References
- 1.Oberholzer-Gee F, Inamdar SN. Merck’s recall of rofecoxib--a strategic perspective. N Engl J Med. 2004;351:2147–2149. doi: 10.1056/NEJMp048284. [DOI] [PubMed] [Google Scholar]
- 2.Citizen P. Rx R&D Myths: The Case Against The Drug Industry’s R&D “Scare Card”. Public Citizen Congress Watch 2001 [Google Scholar]
- 3.Tobert JA. Lovastatin and beyond: the history of the HMG-CoA reductase inhibitors. Nat Rev Drug Discov. 2003;2:517–526. doi: 10.1038/nrd1112. [DOI] [PubMed] [Google Scholar]
- 4.Apic G, et al. Illuminating drug discovery with biological pathways. FEBS Lett. 2005;579:1872–1877. doi: 10.1016/j.febslet.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 5.Ananiadou S, et al. Text mining and its potential applications in systems biology. Trends Biotechnol. 2006;24:571–579. doi: 10.1016/j.tibtech.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Rzhetsky A, et al. Seeking a new biology through text mining. Cell. 2008;134:9–13. doi: 10.1016/j.cell.2008.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garten Y, Altman RB. Pharmspresso: a text mining tool for extraction of pharmacogenomic concepts and relationships from full text. BMC Bioinformatics. 2009;10(Suppl 2):S6. doi: 10.1186/1471-2105-10-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Druker BJ, Lydon NB. Lessons learned from the development of an abl tyrosine kinase inhibitor for chronic myelogenous leukemia. J Clin Invest. 2000;105:3–7. doi: 10.1172/JCI9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anastassiou D. Genomic signal processing. Ieee Signal Processing Magazine. 2001;18:8–20. [Google Scholar]
- 10.Anastassiou D. Computational analysis of the synergy among multiple interacting genes. Mol Syst Biol. 2007;3:83. doi: 10.1038/msb4100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campillos M, et al. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 12.Shannon CE, Weaver W. The mathematical theory of communication. University of Illinois Press; 1949. [Google Scholar]
- 13.Butte AJ, Kohane IS. Mutual information relevance networks: functional genomic clustering using pairwise entropy measurements. Pac Symp Biocomput. 2000:418–429. doi: 10.1142/9789814447331_0040. [DOI] [PubMed] [Google Scholar]
- 14.Basso K, et al. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 15.Watkinson J, et al. Identification of gene interactions associated with disease from gene expression data using synergy networks. BMC Syst Biol. 2008;2:10. doi: 10.1186/1752-0509-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Almenoff JS, et al. Novel statistical tools for monitoring the safety of marketed drugs. Clin Pharmacol Ther. 2007;82:157–166. doi: 10.1038/sj.clpt.6100258. [DOI] [PubMed] [Google Scholar]
- 17.Chen LS, et al. Harnessing naturally randomized transcription to infer regulatory relationships among genes. Genome Biol. 2007;8:R219. doi: 10.1186/gb-2007-8-10-r219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, et al. Analysis of Drosophila segmentation network identifies a JNK pathway factor overexpressed in kidney cancer. Science. 2009;323:1218–1222. doi: 10.1126/science.1157669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marrer E, Dieterle F. Promises of biomarkers in drug development--a reality check. Chem Biol Drug Des. 2007;69:381–394. doi: 10.1111/j.1747-0285.2007.00522.x. [DOI] [PubMed] [Google Scholar]
- 21.Marrer E, Dieterle F. Biomarkers in oncology drug development: rescuers or troublemakers? Expert Opin Drug Metab Toxicol. 2008;4:1391–1402. doi: 10.1517/17425255.4.11.1391. [DOI] [PubMed] [Google Scholar]
- 22.Zimmermann GR, et al. Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug Discov Today. 2007;12:34–42. doi: 10.1016/j.drudis.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 23.Borisy AA, et al. Systematic discovery of multicomponent therapeutics. Proc Natl Acad Sci U S A. 2003;100:7977–7982. doi: 10.1073/pnas.1337088100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qutub AA, et al. Integration of angiogenesis modules at multiple scales: from molecular to tissue. Pac Symp Biocomput. 2009:316–327. [PMC free article] [PubMed] [Google Scholar]
- 25.Klein ML, Shinoda W. Large-scale molecular dynamics simulations of self-assembling systems. Science. 2008;321:798–800. doi: 10.1126/science.1157834. [DOI] [PubMed] [Google Scholar]
- 26.Varshavsky A. Codominant interference, antieffectors, and multitarget drugs. Proc Natl Acad Sci U S A. 1998;95:2094–2099. doi: 10.1073/pnas.95.5.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holly MK, et al. Biomarker and drug-target discovery using proteomics in a new rat model of sepsis-induced acute renal failure. Kidney Int. 2006;70:496–506. doi: 10.1038/sj.ki.5001575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Owen RP, et al. PharmGKB and the International Warfarin Pharmacogenetics Consortium: the changing role for pharmacogenomic databases and single-drug pharmacogenetics. Hum Mutat. 2008;29:456–460. doi: 10.1002/humu.20731. [DOI] [PubMed] [Google Scholar]
- 29.Dudley JT, Butte AJ. Identification of discriminating biomarkers for human disease using integrative network biology. Pac Symp Biocomput. 2009:27–38. [PMC free article] [PubMed] [Google Scholar]
- 30.Chiang AP, Butte AJ. Data-driven methods to discover molecular determinants of serious adverse drug events. Clin Pharmacol Ther. 2009;85:259–268. doi: 10.1038/clpt.2008.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson AE, et al. Failure of serum transforming growth factor-beta (TGF-beta1) as a biomarker of radiographic osteoarthritis at the knee and hip: a cross-sectional analysis in the Johnston County Osteoarthritis Project. Osteoarthritis Cartilage. 2008 doi: 10.1016/j.joca.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaffe AB, et al. Geographic Localization of Knowledge Spillovers as Evidenced by Patent Citations. Quarterly Journal of Economics. 1993;108:577–598. [Google Scholar]
- 33.Bottazzi L, Peri G. Innovation and spillovers in regions: Evidence from European patent data. European Economic Review. 2003;47:687–710. [Google Scholar]
- 34.Wuchty S, et al. The increasing dominance of teams in production of knowledge. Science. 2007;316:1036–1039. doi: 10.1126/science.1136099. [DOI] [PubMed] [Google Scholar]
- 35.Burt RS. Structural holes and good ideas. American Journal of Sociology. 2004;110:349–399. [Google Scholar]
- 36.Fleming L, Sorenson O. Technology as a complex adaptive system: evidence from patent data. Research Policy. 2001;30:1019–1039. [Google Scholar]
- 37.Guimera R, et al. Team assembly mechanisms determine collaboration network structure and team performance. Science. 2005;308:697–702. doi: 10.1126/science.1106340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans JA. Industry Collaboration and Secrecy in Academic Science. Social Studies of Science. 2008 under review. [Google Scholar]
- 39.Evans JA. Industry Collaboration and Theory in Academic Science. American Journal of Sociology. 2008 (forthcoming) [Google Scholar]
- 40.Latour B. Science in action : how to follow scientists and engineers through society. Harvard University Press; 1987. [Google Scholar]
- 41.Dasgupta P, David PA. Toward a New Economics of Science. Research Policy. 1994;23:487–521. [Google Scholar]
- 42.Cohen WM, et al. Protecting their Intelletual Assets: Appropriability conditions and why U.S. manufacturing firms patent (or not) NBER Working Paper 7552 2000 [Google Scholar]
- 43.Anonymous . National, Research Council: Intellectual Property Rights and Research Tools in Molecular Biology. National Academy Press; 1997. [Google Scholar]
- 44.Merton RK. Priorities in Scientific Discovery: A Chapter in the sociology of science. American Sociological Review. 1957;22:635–659. [Google Scholar]
- 45.Anonymous Disease and demography. Health Aff (Millwood) 2008;27:1051. doi: 10.1377/hlthaff.27.4.1051. [DOI] [PubMed] [Google Scholar]
- 46.Berndt C. Manufacturing culture: the institutional geography of industrial practice. Environment and Planning A. 2006;38:990–992. [Google Scholar]
- 47.Powell WW. Interorganizational Collaboration in the Biotechnology Industry. Journal of Institutional and Theoretical Economics. 1996;120:197–215. [Google Scholar]
- 48.Heller MA, Eisenberg RS. Can Patents Deter Innovation? The anticommons in biomedical research. Science. 1998;280:698. doi: 10.1126/science.280.5364.698. [DOI] [PubMed] [Google Scholar]
- 49.Goldwasser E. Erythropoietin: a somewhat personal history. Perspect Biol Med. 1996;40:18–32. doi: 10.1353/pbm.1996.0005. [DOI] [PubMed] [Google Scholar]
- 50.Jelkmann W. Erythropoietin after a century of research: younger than ever. Eur J Haematol. 2007;78:183–205. doi: 10.1111/j.1600-0609.2007.00818.x. [DOI] [PubMed] [Google Scholar]
- 51.Jones BF. The Burden of Knowledge and the “Death of the Renaissance Man”: Is Innovation Getting Harder? Review of Economic Studies. 2009;76:283–317. [Google Scholar]
- 52.Korbel JO, et al. Systematic association of genes to phenotypes by genome and literature mining. PLoS Biol. 2005;3:e134. doi: 10.1371/journal.pbio.0030134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heller MA. The tragedy of the anticommons: Property in the transition from Marx to markets. Harvard Law Review. 1998;111:621–688. [Google Scholar]
- 54.Turner JS. The nonmanufacturing patent owner: Toward a theory of efficient infringement. California Law Review. 1998;86:179–210. [Google Scholar]
- 55.Murray F, Stern S. Do formal intellectual property rights hinder the free flow of scientific knowledge? An empirical test of the anticommons hypothesis. Journal of Economic Behavior & Organization. 2007;63:648–687. [Google Scholar]
- 56.Lipkus AH, et al. Structural diversity of organic chemistry. A scaffold analysis of the CAS Registry. J Org Chem. 2008;73:4443–4451. doi: 10.1021/jo8001276. [DOI] [PubMed] [Google Scholar]
- 57.Klein TE, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eichelbaum M, et al. New feature: pathways and important genes from PharmGKB. Pharmacogenet Genomics. 2009;19:403. doi: 10.1097/FPC.0b013e32832b16ba. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yildirim MA, et al. Drug-target network. Nat Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 60.Garber KB, et al. Fragile X syndrome. Eur J Hum Genet. 2008;16:666–672. doi: 10.1038/ejhg.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mulligan LM, et al. Genetic mapping of DNA segments relative to the locus for the fragile-X syndrome at Xq27.3. Am J Hum Genet. 1985;37:463–472. [PMC free article] [PubMed] [Google Scholar]
- 62.Gedeon AK, et al. Fragile X syndrome without CCG amplification has an FMR1 deletion. Nat Genet. 1992;1:341–344. doi: 10.1038/ng0892-341. [DOI] [PubMed] [Google Scholar]
- 63.Anonymous Fmr1 knockout mice: a model to study fragile X mental retardation. The Dutch-Belgian Fragile X Consortium. Cell. 1994;78:23–33. [PubMed] [Google Scholar]
- 64.de Vrij FM, et al. Rescue of behavioral phenotype and neuronal protrusion morphology in Fmr1 KO mice. Neurobiol Dis. 2008;31:127–132. doi: 10.1016/j.nbd.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pecknold JC, et al. Treatment of anxiety using fenobam (a nonbenzodiazepine) in a double-blind standard (diazepam) placebo-controlled study. J Clin Psychopharmacol. 1982;2:129–133. [PubMed] [Google Scholar]
- 66.Jacob W, et al. The anxiolytic and analgesic properties of fenobam, a potent mGlu5 receptor antagonist, in relation to the impairment of learning. Neuropharmacology. 2009 doi: 10.1016/j.neuropharm.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 67.Berry-Kravis E, et al. A pilot open label, single dose trial of fenobam in adults with fragile X syndrome. J Med Genet. 2009;46:266–271. doi: 10.1136/jmg.2008.063701. [DOI] [PMC free article] [PubMed] [Google Scholar]