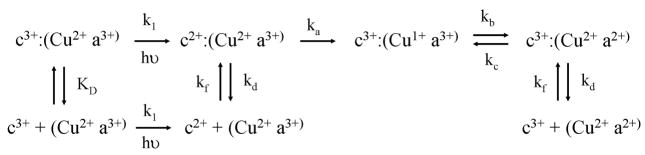Abstract
Ruthenium photoreduction methods are described to study electron transfer from cytochrome c to cytochrome c oxidase, and within cytochrome oxidase. Methods are described to prepare a ruthenium cytochrome c derivative, Ru-39-Cc, by labeling the single sufhydryl group on horse K39C with (4-bromomethyl-4′methylbipyridine) (bis-bipyridine)ruthenium(II). The ruthenium complex attached to Cys-39 on the opposite side of the heme crevice does not interfere with the interaction with cytochrome oxidase. Laser flash photolysis of a 1:1 complex between Ru-39-Cc and bovine cytochrome oxidase results in photoreduction of heme c within 1 μs, followed by electron transfer from heme c to CuA in cytochrome oxidase with a rate constant of 60,000 s−1 and from CuA to heme a with a rate constant of 20,000 s−1. A new ruthenium dimer, Ru2Z, has been developed to reduce CuA within 1 μs with a yield of 60%, followed by electron transfer from CuA to heme a and then to the heme a3/CuB binuclear center. Methods are described to measure the single-electron reduction of each of the intermediates involved in reduction of oxygen to water by cytochrome oxidase, including Pm, F, OH, and E.
1. Introduction
Cytochrome c oxidase (CcO) is the terminal electron transfer complex in the respiratory chains of mitochondria and many prokaryotes (Babcock and Wikstrom, 1992). Electron transfer of four electrons from cytochrome c (Cc) to molecular oxygen is coupled to the uptake of four “chemical” protons to form 2 H2O, and the translocation of four “pumped” protons across the membrane. Cc initially transfers an electron to CuA located in subunit II, followed by electron transfer to heme a and then to the heme a3/CuB binuclear center located in subunit I, where oxygen is reduced to water. The D-channel containing D-132 is thought to be involved in the uptake of both chemical and pumped protons from the matrix side of the membrane, while the K-channel containing K-362 is involved in proton uptake during reduction of the binuclear center (Branden et al., 2006). X-ray crystal structures of bovine (Yoshikawa et al.,1998), Paracoccus denitrificans (Ostermeier et al., 1997) and Rhodocacter sphaeroides (Qin et al. 2006) CcO have revealed the locations of the redox centers and proton channels in great detail. However, determination of the kinetics of the electron and proton transfer steps and the mechanism of coupling between them remains a challenging problem.
One of the most widely used methods to study rapid electron transfer in CcO involves flash photolysis of CO-bound heme a3 in reduced CcO, followed by reaction with oxygen (Hill, 1991; Faxen et al., 2005). In another approach, our laboratory has developed a new ruthenium photoreduction technique to inject single electrons into Cc or CcO and measure the subsequent electron transfer reactions on a rapid time scale (Millett and Durham, 2002). To study electron transfer involving cytochrome c, a polypyridyl ruthenium complex [Ru(II)] is covalently attached to Cys-39 on Cc to form Ru-39-Cc (Geren et al, 1995). Photoexcitation of Ru(II) to the metal-to-ligand charge-transfer state, Ru(II*), a strong reductant, leads to rapid reduction of the ferric heme group in Cc. This new technique has been used to determine that Cc initially transfers an electron into CuA of CcO with a rate constant of 60,000 s−1 (Geren et al., 1995). In another approach, several new ruthenium dimers have been developed which bind with high affinity to cytochrome oxidase and can photoreduce CuA within 1 μs (Zaslavsky et al., 1998). This new technique has been used to measure the kinetics for electron transfer from CuA to heme a and then to the heme a3/CuB binuclear center, as well as coupled proton pumping.
2. Preparation of Ruthenium-labeled Cytochrome c Derivatives
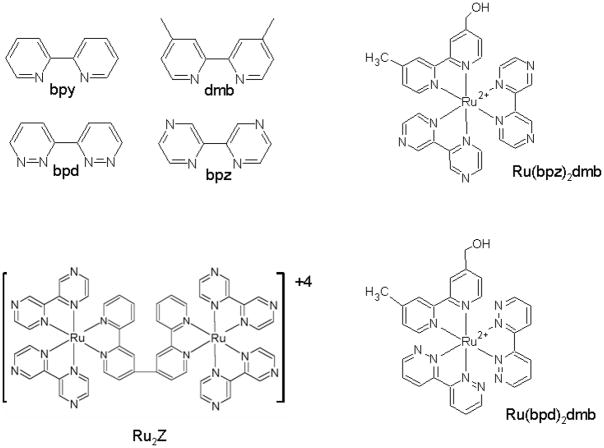
We have developed four different methods for the covalent attachment of a ruthenium complex to a protein (Millett and Durham, 2002). In the method to be described here, a ruthenium bromomethyl reagent is used to label a cysteine sulfhydryl group on the surface of the protein (Geren et al., 1991, 1995). A single free cysteine can be engineered onto the surface of the protein by site-directed mutagenesis to specifically address the goals of the experiment. It is generally desirable to place the ruthenium complex on a surface remote from the binding site with other proteins, so the complex does not interfere with interprotein interactions. It is also important to have a good pathway for electron transfer from the ruthenium complex to the redox center to maximize the rate of electron transfer. Another advantage of this labeling method is that all three chelating ligands can be altered to change the redox potentials of the ruthenium complex, allowing measurement of the reorganization energy, and optimization of the rate and yield of photoinduced electron transfer (Figure 1, Table 1).
Figure 1.
Chemical structures of selected ruthenium complexes
Table I.
Standard Reduction Potentials of Ruthenium Complexes
| Complex | (III)/(II) | (III)/(II*) | (II)/(I) | (II*)/(I) |
|---|---|---|---|---|
| Ru(bpy)3 | 1.27 | −0.87 | −1.31 | 0.83 |
| Ru(bpy)2(dmb) | 1.27 | −0.83 | −1.36 | 0.79 |
| Ru(bpz)2(dmb) | 1.76 | −0.25 | −0.79 | 1.22 |
| Ru(bpd)2(dmb) | 1.49 | −0.49 | −1.00 | 0.98 |
| Ru(bpm)2(dmb) | 1.55 | −0.30 | −0.95 | 0.90 |
| Ru2C | 1.27 | −0.87 | −1.31 | 0.83 |
| Ru2Z | 1.70 | −0.28 | −0.82 | 1.16 |
2.1 Materials
The yeast cytochrome c mutant H39C,C102T is made as described by Geren et al. (1995), and the preparation of horse cytochrome c mutant K39C is described in Patel et al. (2001). The synthesis of (4-bromomethyl-4′methylbipyridine) (bis-bipyridine)ruthenium(II) reagent is described by Geren et al., (1995), with care being taken to maintain dryness and eliminate metal contamination. The bromination method described in that paper has been modified slightly. After reflux in 48 % HBr for 3 hours, the PF6 precipitation was eliminated due to the product’s poor solubility in water. Instead, the excess HBr was allowed to boil off carefully without allowing any overheating of the dried product, with the final drying done under vacuum and low heat. The dry powder was immediately dissolved in very dry DMF and stored frozen under dry N2. Extreme care is required to maintain dryness and prevent hydrolysis of the bromine group.
2.2. Preparation of Ruthenium-labeled Cytochrome c
The protein labeling technique is similar for all the bromo-methyl ruthenium reagents and is similar to that described in Geren et al., (1995), but has been altered somewhat with experience. The protein to be labeled is first treated with a ten fold excess of dithiothreitol for ten minutes to reduce any cross–linked disulfide bonds. The sample is then washed two times as quickly as possible using concentrators or a P-2 column into 50 mM sodium borate, pH 9.0, to remove any remaining dithiothreitol. Three to five mg of cytochrome c in approximately 300 μl of 50 mM sodium borate, pH 9.0, with a concentration of about 1 mM is best. Raising the pH increases the reactivity but decreases the selectivity for cysteines. A pH of 10 to 10.5 will produce labeled lysines as well as cysteines. After degassing under N2, a 0.5 equivalent of dithiothreitol is added and stirred 5 minutes. Using a very dry syringe to protect the ruthenium stock solution and continuing to stir, an approximate 3 fold excess (sometimes more, depending upon the reactivity of the complex) of the ruthenium reagent is slowly added. The ruthenium reagent is very quickly hydrolyzed by water and care must be taken to keep the stock dry and use only syringes which have been rinsed with dry ether or dry DMF. After 5 minutes of stirring the sample is allowed to react overnight (about 18 hours) at room temperature, or 3 hours at 37°C, protected from light. All of the unreacted ruthenium reagent will have hydrolyzed by this time and can then be removed by buffer exchange via dialysis, Amicon concentrators, or Bio-Gel P-2 column. The labeled protein is then purified by ion exchange or HPLC chromatography. The amount of ruthenium reagent used can be adjusted to minimize the amount of unlabeled protein without producing too much multi-labeled material. The reaction may be scaled up or down. If the labeling does not cause any disturbance of the protein structure, the absorbance spectra of the ruthenium complex and the protein should be additive. The spectrum of Ru-39-Cc is the same as that of equimolar Ru(bpy)2(dmb) and horse Cc. The specific residue on the protein labeled with ruthenium should be confirmed with HPLC separation of a trypsin digest, followed by peptide sequencing as described by Geren et al. (1991).
3.0. Measurement of Intraprotein Electron Transfer in Horse Ru-39-Cc
Internal electron transfer between the ruthenium complex and the heme in a ruthenium-labeled protein is measured using laser flash photolysis (Durham et al., 1989). The excitation pulse is the third harmonic of a Nd:YAG laser, with a pulse width of 20 ns and wavelength of 356 nm. The probe source is a pulsed 75 W xenon arc lamp, and the photomultiplier detector has a response time of 10 ns. Photoexcitation of Ru(II) in 10 μM oxidized horse Ru-39-Cc results in electron transfer from Ru(II*) to Fe(III) to form Ru(III) and Fe(II), followed by reverse electron transfer to form Ru(II) and Fe(III) according to Scheme 1 (Figure 2). The reduction of Fe(II) is detected at 550 nm after subtracting a small contribution from Ru(II*) detected at 556 nm, an isobestic for heme c. The photoexcitation and recovery of Ru(II) is detected at 434 nm, a heme c isobestic. Both transients are fit using equations (1) and (2) based on Scheme 1 (Durham et al., 1989).
Scheme 1.
Figure 2.
Photoinduced electron transfer within horse Ru-39-Cc. A solution containing 10 μM Ru-39-horse Cc in 100 mm phosphate buffer pH 7.0 was excited with a 10 ns 356 nm laser flash, and the transients detected at 550 – 556 nm and 434 nm. The smooth lines are fitted to equations 1 and 2 of Scheme 1 with k1 = 0.6 × 106, k2 = 0.7 × 106, kd = 5.5 × 106.
| Eq. (1) |
| Eq. (2) |
where A = k1Δε550C0/(k2-k1-kd), B = (k2-kd)/k2-k1-kd), C = k1/(k2-k1-kd), Δε550=18.6 mM−1, and Δε434=12 mM−1. The best fit for horse Ru-39-Cc is obtained with k1 = (6 ± 1) × 105 sec −1, k2 = (7 ± 1) × 105 sec −1, and kd = (5.5 ± 1.0) × 106 sec −1 (Figure 2). These rate constants are independent of protein concentration and are similar to the rates obtained for the yeast Ru-39-Cc (Geren et al., 1995). Residue 39 was chosen for attachment of the ruthenium complex because it is on the back side of Cc remote from the binding site with CcO. There is an efficient electron transfer pathway between the ruthenium complex and the heme consisting of 13 covalent bonds and one hydrogen bond with a total distance of 12.6 Å. The metal-to-ligand charge-transfer excited state Ru(II*) is a strong reducing agent, and can rapidly transfer an electron to heme c Fe(III) according the scheme 1. Since the driving forces for k1 and k2 of 1.1 and 1.0 eV, respectively, are near the expected reorganizational energy of 0.8 eV, the rate constants k1 and k2 should be near the maximum, activationless rate constant (Durham et al. 1989).
The sacrificial electron donor aniline can be used to reduce Ru(III) and lead to the photoreduction of the heme in Ru-39-Cc through k1 and k6 steps in scheme 1 with a yield of 5%. Aniline does not have a low enough redox potential to directly reduce Ru(II*). Higher photoreduction yields can be obtained using the Ru(bpd)2(dmb) complex, which has a redox potential of 0.98 for the Ru(II*)/Ru(I) transition, allowing the sacrificial donor DMAB to directly reduce Ru(II*) to Ru(I) (Table 1, Scheme 1). The yeast RuD-39-Cc is photoreduced through the k5 and k4 steps in Scheme 1 with a yield of 35%. Reduction potentials for a number of useful ruthenium complexes are listed in Table 1.
4.0. Measurement of Electron Transfer from RuD-39-Cc to Cytochrome c Oxidase
Intracomplex electron transfer from RuD-39-Cc to CcO is studied using laser flash photolysis. A Phase R model DL1400 flash lamp-pumped dye laser emitting a 480 nm light flash of < 0.5 μs duration is used as the excitation source (Geren et al. 1995). The sample contains 5 μM yeast RuD-39-Cc and 5 μM bovine CcO in 5 mM sodium phosphate, pH 7.0, 5 mM DMAB, and 0.1% dodecyl maltoside at 25 °C. Under these low ionic strength conditions, RuD-39-Cc is tightly bound to CcO, allowing measurement of intracomplex electron transfer according to scheme 2. Excitation of Ru(II) to Ru(II*) with a single laser flash results in rapid photoreduction of heme c, followed by electron transfer to CuA and then to heme a. The reaction of heme c is followed at 550 nm, while CuA and heme a are followed at 830 nm and 605 nm, respectively (Figure 3). The decrease in 830 nm absorbance indicates electron transfer from heme c to CuA with a rate constant of 60,000 s−1. The recovery of the 830 nm signal resulting from the reoxidation of CuA is matched by the reduction of heme a seen in the 605 nm transient with a rate of kb = 2 × 104 sec −1. Both ka and kb are independent of protein concentration and ionic strength up to 100 mM, indicating intracomplex electron transfer. Site-directed mutagenesis studies have shown that positively charged lysines surrounding the heme crevice of Cc interact electrostatically with negatively charged carboxylates near CuA on subunit II of CcO (Wang et al., 1999). A highly conserved tryptophan residue on subunit II mediates electron transfer from the heme group of Cc to CuA. Theoretical calculations of the rate of electron transfer based on a computational model of the Cc:CcO complex are in good agreement with the experimental value of 2 × 104 s−1 (Roberts and Pique, 1999; Wang et al., 1999).
Scheme 2.
Figure 3.
Photoinduced electron transfer in RuD-39-Cc:CcO complex. A sample containing 5 μM yeast RuD-39-Cc and 5 μM bovine CcO in 5 mM sodium phosphate, pH 7.0, 5 mM DMAB, and 0.1% dodecyl maltoside at 25 °C was excited with a 480 nm laser flash. CuA was detected at 830 nm, while heme a was detected at 605 nm.
4.1. Measurement of Formation and Dissociation Rate Constants for the Complex between Ru-Cc and CcO
When the ionic strength is increased above 50 mM, the amplitude of the intracomplex phase of electron transfer between Ru-39-Cc and CcO decreases, and a new phase appears due to bimolecular electron transfer between the two proteins according to Scheme 2. At intermediate ionic strengths between 50 and 100 mM, both intracomplex and bimolecular phases are seen in the kinetics, allowing the fraction f of Ru-39-Cc:CcO complex to be determined. Using the total concentrations of CcO (Ot), and Ru-39-cyt c (Cct), KD may be calculated from equation 3.
| Eq. (3) |
The dissociation constant Kd is given by:
| Eq. (4) |
Since the rate constant ka is independent of ionic strength up to 100 mM, the presence of two phases indicates the absence of rapid equilibrium, and kdiss must be much smaller than ka. Using steady state assumptions, the bimolecular reaction is given by equation 5:
| Eq. (5) |
Measurement of k2nd and ka, along with the calculation of KD thus also allows the calculation of kdiss and kform (Geren et al., 1995). For beef oxidase and yeast Ru-39-C c at 100 mM ionic strength these were found to be ka= 6 × 104 s−1, kform = 1.8 × 108 M−1s−1, and kdiss = 8 × 103 s−1 (Scheme 2).
5.0 Measurement of Electron Transfer and Oxygen Reduction in Cytochrome c Oxidase using Electrostatically Bound Ruthenium Complexes
A number of positively charged ruthenium complexes have been developed that bind to CcO and directly photoreduce CuA (Figure 1). The ruthenium dimer Ru2C has a charge of +4 that allows it to bind to the acidic patch on subunit II of CcO (Zaslavsky et al, 1998). The same laser flash photolysis instrumentation described in section 4.0 above is used to photoreduce CcO by Ru2C. Laser flash photolysis of 5 μM oxidized bovine CcO and 25 μM Ru2C in 5 mM TrisHCl, pH 8.0 with 10 mM aniline and 1 mM 3CP results in electron transfer from the excited state Ru(II*) of Ru2C to CuA in CcO within 1 μs, followed by electron transfer from CuA to heme a with a rate constant of 20,000 s−1. The mechanism of photoreduction of CuA follows the k1 and k6 steps of Scheme 1, and the total yield of photoreduced heme a is 6.2%. Another ruthenium dimer, Ru2Z, has recently been developed which can photoreduce CcO with a yield of 60% (Brand et al., 2007). Replacement of the bipyridine ligands in Ru2C with the bipyrazine ligands in Ru2Z changed all four redox potentials (Table 1). The redox potential of the Ru(II*)/Ru(I) transition was increased from 0.83 V to 1.16 V, allowing the sacrificial donor aniline to reduce the excited state Ru(II*) to Ru(I), followed by electron transfer from Ru(I) to CuA(II) along the k5 and k4 steps in scheme 1. CuA is 16% reduced and heme a is 52% reduced after electron transfer equilibrium is established between them. The equilibrium constant K for electron transfer between CuA and heme a is thus 3.2, and the redox potential of heme a is 30 mV more positive than that of CuA (Brand et al., 2007).
5.1. Measurement of Single Electron Reduction of CcO state Pm to F
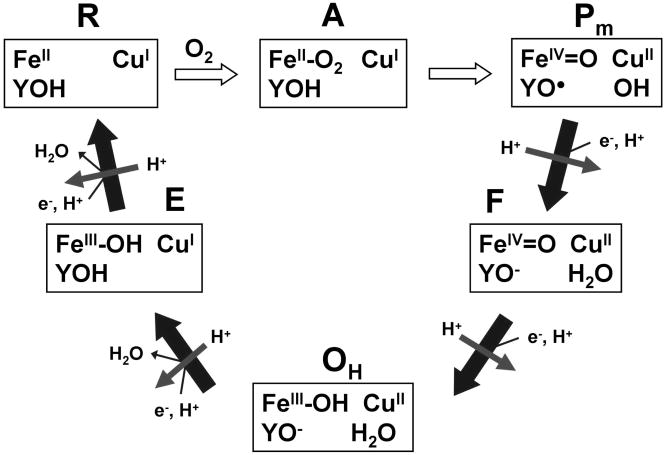
A mechanism for the reduction of O2 by CcO is shown in Figure 4 (Branden et al, 2006). State O containing a fully oxidized heme a3/CuB binuclear center is reduced by one electron to state E and then by a second electron to state R2, which rapid binds oxygen to from state A. State A is reduced to state Pm in a rapid 4-electron reaction in which electrons come from heme a3, CuB, and a neighboring tyrosine. The tyrosine radical in state Pm is reduced to form state F, and then oxyferryl heme a3 is reduced to form state O. There is growing agreement that each of the one-electron transfers to the heme a3/CuB binuclear center is coupled to pumping one proton across the membrane, as indicated in Figure 4 (Verkhovsky et al., 2006; Bloch et al., 2004). Methods have been developed to study each of the one-electron reduction steps in the mechanism using the ruthenium photoreduction technique.
Figure 4.
Cytochrome c oxidase catalytic cycle showing states of the heme a3/CuB binuclear center and conserved tyrosine. Reactions steps indicated by solid arrows are coupled to proton pumping.
The Pm state is formed by treating resting state CcO with CO under aerobic conditions (Siletsky et al, 2006). A solution containing 10 μM oxidized bovine CcO and 30 μM Ru2C in 5 mM Tris-HCl, pH 8.0 is gently bubbled with CO under aerobic conditions for 60 seconds and incubated until the difference spectrum indicates formation of Pm. The yield of the Pm state is calculated from Δε607–630 = 11 mM−1 cm−1, and is typically about 75%. After the Pm state is completely formed, 10 mM aniline and 1 mM 3CP are added as sacrificial donors, followed by laser flash photolysis. Photoreduced CuA transfers an electron to heme a, followed by electron transfer from heme a to the tyrosine radical near the binuclear center with a rate constant of 4,000 s−1 (Siletsky et al., 2006). The reduction and reoxidation of heme a is followed at 650 nm, while the reduction of the binuclear center is followed at 580 nm. Measurement of transmembrane voltage generation under the same conditions indicated two phases of electrogenic proton transfer with rate constants of 3300 s−1 and 770 s−1 (Siletsky et al., 2006). The fast phase of proton transfer is coincident with electron transfer to the tyrosine radical, and is associated with proton delivery to CuB bound hydroxide to form water. The slow phase of proton transfer could be due to proton translocation across the membrane.
5.2 Measurement of Single Electron Reduction of F state to O state
The F state is generated by treating resting CcO with 4 mM H2O2 (Fabian and Palmer, 1995). A solution containing 8 μM bovine CcO and 22 μM Ru2C in 5 mM Tris-HCl, pH 8.0 with 10 mM aniline and 1 mM 3CP is treated with 4 mM H2O2 and incubated for 5 minutes to form state F. The yield of F is determined from the peak in the difference spectrum (F – O) at 583 nm using extinction coefficients of Δε583–630 = 5.3 mM−1 cm−1 and Δε436–414 = 67 mM−1 cm−1 (Fabian and Palmer, 1995). Laser flash photolysis results in photoreduction of CuA, followed by electron transfer to heme a with a rate constant of 20,000 s−1, and electron transfer from heme a to oxyferryl heme a3 to form state O with a rate constant of 470 s−1. The reduction and reoxidation of the heme a is followed at 605 nm, while the reduction of oxyferryl heme a3 is followed at 580 nm (Zaslavsky et al., 1998). A deuterium isotope effect of 4.3 indicats a proton-transfer rate-limiting step. Measurement of transmembrane voltage generation indicated two phases of electrogenic proton transfer with rate constants of 830 s−1 and 220 s−1 in a 1:3 ratio (Konstantinov, 1998). The fast phase is coincident with reduction of oxyferryl heme, and is associated with proton transfer to the oxygen on the oxyferryl heme to form water. The slow phase of proton transfer is associated with proton translocation (Konstantinov, 1998). A fast phase of proton release is also observed coincident with reduction of oxyferryl heme (Zaslavsky et al., 2004).
5.3. Measururement of Single Electron Reduction of State OH to state E
There are several forms of the fully oxidized state of CcO, including the resting state O (Moody, 1996), and the state OH which is formed immediately after fully reduced state R4 reacts with oxygen (Bloch et al., 2004). If an additional electron is not injected into the OH state within approximately 30 seconds, then it relaxes back to the resting state O. It has been reported that reduction of state OH is coupled to proton pumping, while reduction of state O is not (Bloch et al., 2004; Belevich et al., 2007; Verkhovsky et al., 2006). A stopped-flow-flash technique is used to study 1-electron reduction of state OH (Brand et al., 2007). 5 μM bovine CcO is anaerobically reduced with 2 mM ascorbate and 1 μM PMS in 5 mM HEPES buffer, pH 7.9 in one syringe of the stopped-flow, and then rapidly mixed with oxygenated buffer containing 20 μM Ru2Z, 10 mM aniline, and 1 mM 3CP. State R4 reacts with oxygen within 5 msec to form state OH, and then the laser is fired to excite Ru2Z to the Ru(I) state, which injects a single electron into CuA. An electron is transferred from CuA to heme a with a rate constant of 20,000 s−1, followed by biphasic electron transfer from heme a to CuB with rate constants of 750 s−1 and 110 s−1 and relative amplitudes of 25% and 75% (Brand et al., 2007). There is no change in absorbance at 436 nm, indicating that the electron acceptor in the binuclear site is CuB rather than heme a3. One electron reduction of P. denitrificans CcO state OH has been studied by Belevich et al. (2007) using Ru(bpy)3 as photoreductant. Correlation of the kinetics of electron transfer and transmembrane voltage generation led to a mechanism for coupling between electron transfer and proton translocation.
5.4. Measurement of Single Electron Reduction of State E to States R2 and Pm
The one-electron reduction of state E is studied by a stopped-flow-flash technique. 5 μM bovine CcO and 5 μM horse heart cytochrome c mutant K13E is anaerobically reduced with 2 mM ascorbate and 1 μM PMS in 5 mM HEPES, pH 8.0, 0.1% LM in one syringe of the stopped-flow. This syringe is then mixed rapidly with the other syringe of the stopped-flow containing 20 μM Ru2Z, 10 mM aniline, and 1 mM 3CP in buffer. After mixing, the reduced CcO which is fully oxidized to the OH state within 5 ms, is then reduced by the ferrous K13E Cc within 10 ms. The laser is flashed 20 ms after mixing to excite Ru2Z and inject a single electron into CuA, which transfers an electron the heme a. Biphasic electron transfer from heme a to heme a3 to form state R2 then occurs with rate constants of 1100 s−1 and 90 s−1, and relative amplitudes of 11% and 89%. The 436 nm transient indicates that O2 rapidly binds to state R2 to form state P. Additional laser flashes result in transients with a rate constant of 3000 s−1, consistent with the sequential PM to F transition (Siletsky et al., 2006), demonstrating the advantage of using higher yield photoreductants.
Acknowledgments
Supported by NIH grants GM20488 and NCRR COBRE RR15569
Abbreviations
- Cc
cytochrome c
- CcO
cytochrome c oxidase
- DMAB
dimethylaminobenzoate
- 3CP
3-carboxy-2,2,5,5-tetramethyl-1-pyrrolidinyloxy free radical
- PMS
phenazine methosulfate
- bpy
2,2′-bipyridine
- dmb
4,4′-dimethyl-2,2′-bipyridine
- bpz
2,2′-bipyrazine
- bphb
1,4-bis(2,2′-bipyridyl-4′-yl)-benzene
- bpd
3,3′-bipyridazine
- Ru2C
[Ru(bpy)2]2(bphb)
- Ru2Z
[Ru(bpz)2]2(qpy)
- Ru-39-Cc
cytochrome c labeled at Cys-39 with Ru(bpy)2(dmb)
- RuD-39-Cc
cytochrome c labeled at Cys-39 with Ru(bpd)2(dmb)
References
- Babcock GT, Wikstrom M. Oxygen Activation and the Conservation of Energy in Cell Respiration. Nature. 1992;356:301–309. doi: 10.1038/356301a0. [DOI] [PubMed] [Google Scholar]
- Belevich Ilya, Bloch Dmitry A, Belevich Nikolai, Wikstrom Marten, Verkhovsky Michael I. Exploring the proton pump mechanism of cytochrome c oxidase in real time. Proc Natl Acad Sci U S A. 2007;104:2685–2690. doi: 10.1073/pnas.0608794104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch D, Belevich I, Jasaitis A, Ribacka C, Puustinen A, Verkhovsky MI, Wikstrom M. The catalytic cycle of cytochrome c oxidase is not the sum of its two halves. Proc Natl Acad Sci U S A. 2004;101:529–533. doi: 10.1073/pnas.0306036101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand SE, Rajagukguk S, Ganesas K, Geren L, Fabian M, Han D, Gennis R, Durham B, Millett F. A New Ruthenium Complex to Study Single-Electron Reduction of the Pulsed OH State of Detergent-Solubilized Cytochrome Oxidase. Biochemistry. 2007;46:14610–14618. doi: 10.1021/bi701424d. [DOI] [PubMed] [Google Scholar]
- Branden G, Gennis RB, Brzezinski P. Transmembrane proton translocation by cytochrome c oxidase. Biochim Biophysc Acta. 2006;175(7):1052–1063. doi: 10.1016/j.bbabio.2006.05.020. [DOI] [PubMed] [Google Scholar]
- Durham B, Pan LP, Long J, Millett F. Photoinduced Electron-Transfer Kinetics of Singly Labeled Ruthenium Bis-(bipyridine) Dicarboxybipyridine Cytochrome c Derivatives. Biochemistry. 1989;28:8659–8665. doi: 10.1021/bi00447a057. [DOI] [PubMed] [Google Scholar]
- Fabian M, Palmer G. The Interaction of Cytochrome Oxidase with Hydrogen Peroxide: The Relationship of Compounds P and F. Biochemistry. 1995;34:13802–13810. doi: 10.1021/bi00042a011. [DOI] [PubMed] [Google Scholar]
- Faxen K, Gilderson G, Adelroth P, Brzezinski P. A mechanistic principle for proton pumping by cytochrome c oxidase. Nature. 2005;437:286–289. doi: 10.1038/nature03921. [DOI] [PubMed] [Google Scholar]
- Geren LM, Beasley JR, Fines BR, Saunders AJ, Hibdon S, Pielak GJ, Durham B, Millett F. Design of a Ruthenium-Cytochrome c Derivative to Measure the Electron Transfer to the Initial Acceptor in Cytochrome c Oxidase. J Biol Chem. 1995;270:2466. doi: 10.1074/jbc.270.6.2466. [DOI] [PubMed] [Google Scholar]
- Geren L, Hahm S, Durham B, Millett F. Photoinduced Electron Transfer Between Cytochrome c Peroxidase and Yeast Cytochrome c Labeled at Cys 102 with (4-Bromomethyl-4′-methylbipyridine)[bis(bipyridine)]ruthenium2+ Biochemistry. 1991;30:9450. doi: 10.1021/bi00103a009. [DOI] [PubMed] [Google Scholar]
- Hill B. The Reaction of the Electrostatic Cytochrome c-Cytochrome Oxidase Complex with. 1991 [PubMed] [Google Scholar]
- Konstantinov AA. Cytochrome c oxidase as a proton-pumping peroxidase: reaction cycle and electrogenic mechanism. J Bioenerg Biomembranes. 1998;30:121–130. doi: 10.1023/a:1020571930850. [DOI] [PubMed] [Google Scholar]
- Millett F, Durham B. Design of Photoactive Ruthenium Complexes to Study Interprotein Electron Transfer. Biochemistry. 2002;41:11315–11324. doi: 10.1021/bi0262956. [DOI] [PubMed] [Google Scholar]
- Moody AJ. As-prepared forms of fully oxidised haem/Cu terminal oxidases. Biochimica Biophysica Acta. 1996 Aug 7;1276(1):6–20. doi: 10.1016/0005-2728(96)00035-7. [DOI] [PubMed] [Google Scholar]
- Ostermeier C, Harrenga A, Ermler U, Michel H. Structure at 2.7 A Resolution of the Paracoccus denitrificans two-subunit cytochrome c oxidase complexed with an antibody FV fragment. Proc Nat Acad Sci USA. 1997;94:10547–10553. doi: 10.1073/pnas.94.20.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel CN, Lind MC, Pielak GJ. Characterization of horse cytochrome c expressed in Escherichia coli. Protein Expr Purif. 2001;22:220–224. doi: 10.1006/prep.2001.1438. [DOI] [PubMed] [Google Scholar]
- Qin L, Hiser C, Mulichak A, Garavito R, Ferguson-Miller S. Identification of conserved lipid/detergent-binding sites in a high-resolution sstructure of the membrane protein cytochrome c oxidase. Proc Natl Acad Sci USA. 2006;103:16117–1622. doi: 10.1073/pnas.0606149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts VA, Pique ME. Definition of the Interaction Domain for Cytochrome c on Cytochrome c Oxidase. III Prediction of the docked complex by a complete systematic search. J Biol Chem. 1999;274:38051–38060. doi: 10.1074/jbc.274.53.38051. [DOI] [PubMed] [Google Scholar]
- Siletsky SA, Han D, Brand S, Morgan JE, Fabian M, Geren L, Millett F, Durham B, Konstantinov AA, Gennis RB. Single-electron photoreduction of the PM intermediate of cytochrome c oxidase. Biochimica et Biophysica Acta. 2006;1757:1122–1132. doi: 10.1016/j.bbabio.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Verkhovsky MI, Belevich I, Bloch DA, Wikstrom M. Elementary steps of proton translocation in the catalytic cycle of cytochrome oxidase. Biochimica Biophysica Acta. 2006 May-June;1757(5–6):401–407. doi: 10.1016/j.bbabio.2006.05.026. [DOI] [PubMed] [Google Scholar]
- Wang K, Zhen Y, Sadoski R, Grinnell S, Geren L, Ferguson-Miller S, Durham B, Millett F. Definition of the interaction domain for cytochrome c on cytochrome c oxidase: II. Rapid kinetic analysis of electron transfer from cytochrome c to Rhodobacter sphaeroides cytochrome oxidase surface mutants. J Biol Chem. 1999;274:38042–38050. doi: 10.1074/jbc.274.53.38042. [DOI] [PubMed] [Google Scholar]
- Yoshikawa S, Shinzawa-Iroh K, Nakashima R, Yaono R, Yamashita E, Inoue N, Yao M, Fei MJ, Libeu CP, Mizushima T, Yamagichi H, Tomizaki T, Tsukihara T. Redox-Coupled Structural Changes in Bovine Heart Cytochrome c Oxidase. Science. 1998;280:1723–1729. doi: 10.1126/science.280.5370.1723. [DOI] [PubMed] [Google Scholar]
- Zaslavsky D, Sadoski RC, Wang K, Durham B, Gennis RB, Millett F. Single Electron Reduction of Cytochrome c Oxidase Compound F: Resolution of Partial Steps by Transient Spectroscopy. Biochemistry. 1998;37:14910–14916. doi: 10.1021/bi981490z. [DOI] [PubMed] [Google Scholar]
- Zaslavsky D, Sadoski RS, Rajagukguk S, Geren L, Millett F, Durham B, Gennis RB. Direct measurement of proton release by cytochrome c oxidase in solution during the F to O transition. Proc Natl Acad Sci USA. 2004;101:10544–10547. doi: 10.1073/pnas.0401521101. [DOI] [PMC free article] [PubMed] [Google Scholar]