Abstract
The hTERT core promoter contains a G-rich region of 12 consecutive G-tracts, embracing three Sp1 binding sites, and has the potential to form multiple G-quadruplexes. From the 12 runs of guanine tracts, 9 putative hTERT G-quadruplex-forming sequences were selected to assay for G-quadruplex formation and stability using circular dichroism and a Taq polymerase stop assay. Results from biophysical and chemical assays demonstrate an approximate inverse correlation between total loop size and structure stability. Investigation of the full-length hTERT G-rich sequence using a Taq polymerase arrest assay and dimethyl sulfate footprinting revealed the formation of a unique end-to-end stacked G-quadruplex structure from this sequence. This structure consists of an all parallel G-quadruplex, formed by four consecutive G-tracts, linked to another, atypical G-quadruplex, formed by two pairs of consecutive G-tracts separated by a 26-base loop. This 26-base loop likely forms a stable hairpin structure, which would explain the unexpected stability of this G-quadruplex. Significantly, the formation of this tandem G-quadruplex structure in the full-length sequence masks all three Sp1 binding sites, which is predicted to produce significant inhibition of hTERT promoter activity. Furthermore, our study implies that inhibition of telomerase activity by some G-quadruplex ligands is not only produced by targeting telomeric G-quadruplexes but also by stabilization of the hTERT promoter G-quadruplexes.
INTRODUCTION
The cellular level of the catalytic domain of telomerase (hTERT) is directly related to telomerase activity [1]. Telomerase is expressed in approximately 85–90% of cancer specimens [2] and is a critical element in both tumorigenesis [3–5] and resistance to radiotherapy in cancer cells [6–8]. Due to its role in telomerase activity, hTERT has been targeted to inhibit telomerase activity, and downregulation of hTERT was shown to decrease proliferation cancer cells [9–12] and increase apoptosis [12] and chemosensitivity [13,14].
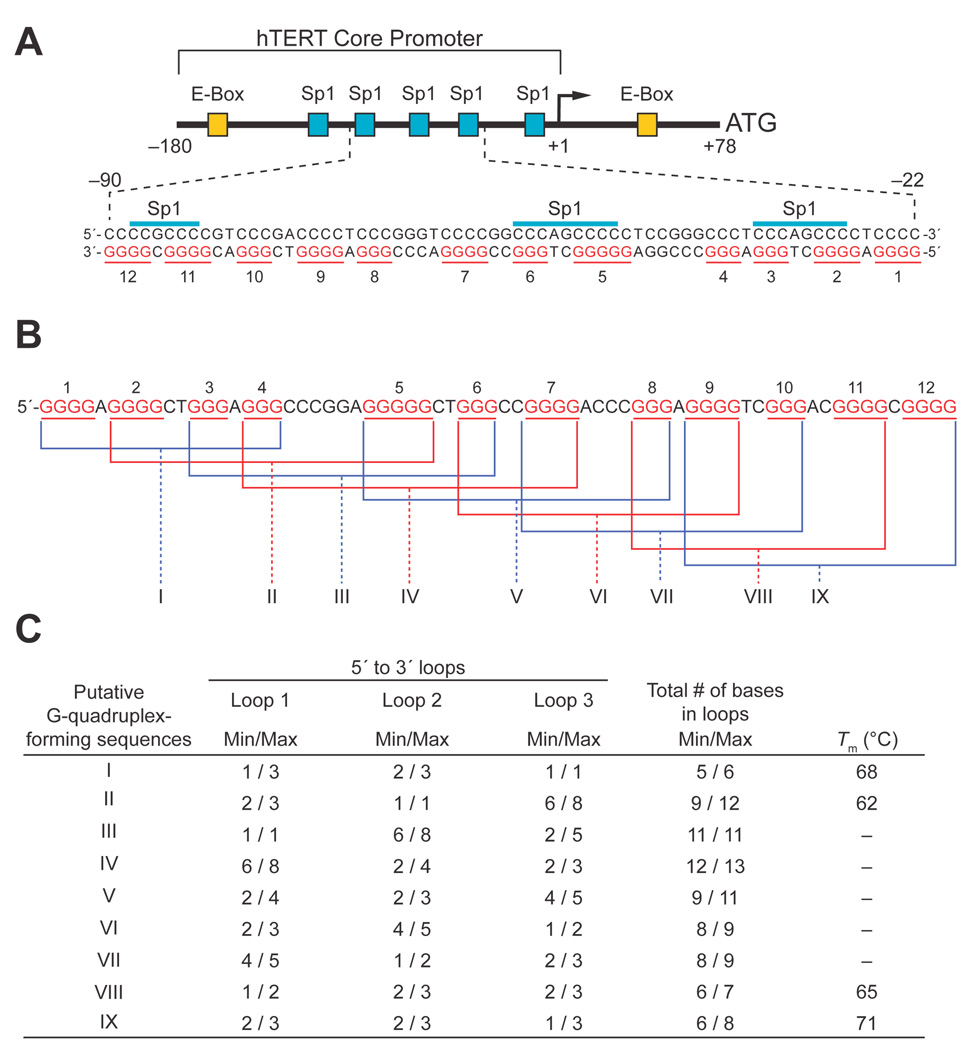
Expression of hTERT is primarily regulated by the core promoter located –180 to +1 relative to the transcription initiation site (+1), and this core promoter contains five Sp1 binding sites and one E-box as a c-myc binding site (Figure 1A) [15]. These binding sites are important for promoter activity, and the middle three Sp1 binding sites have been shown to be essential for hTERT expression in several cancer cell lines [15–18]. In contrast, Mad1 downregulates hTERT promoter activity by competing with c-myc for the E-box of the core promoter activity [19]. Last, the TGF-β/Smad, p53, and BRCA1 signaling pathways can inhibit hTERT expression in an indirect manner via c-myc [20–23].
Figure 1.
The hTERT core promoter and the G-rich region. (A) The hTERT core promoter spanning +1 to −180 contains one E-Box and five Sp1 binding sites that activate the promoter, with the putative G-quadruplex-forming region of the hTERT core promoter shown in inset. Each G-tract is numbered with Arabic numbers. (B) The putative G-quadruplex-forming region of the hTERT core promoter. The region containing the middle three Sp1 binding sites are highly G-rich, consisting of twelve consecutive G-tracts. The putative G-quadruplex-forming sequences are denoted in roman numbers with brackets. (C) Summary of the potential range of loop sizes (minimum [min] and maximum [max] refer to the possible loop isomers that can occur) in sequences I–IX and melting temperatures of selected G-quadruplex-forming regions (see Figure 2B).
The loop sizes in a G-quadruplex are known to have a significant effect on the stability of the structure [24–26]. Studies on libraries of G-quadruplex-forming sequences have demonstrated that loop size can have a major effect on thermal stability of G-quadruplexes [26]. For example, in three-tetrad G-quadruplexes, for loop lengths up to five nucleotides there is a major effect on thermal stability. In naturally occurring intramolecular G-quadruplexes in promoter elements a pattern of two single-nucleotide loops with total loop sizes of four to 11 have been found [27]. In addition, the PDGF-A promoter appears to have a four-tetrad G-quadruplex with two double-chain reversal loops containing two nucleotides and a third loop of 3–5 nucleotides. The region containing the middle three Sp1 binding sites in the hTERT core promoter, located between –22 and –90, contains twelve consecutive G-tracts of three or more guanines (the three Sp1 binding sites are positioned within G-tracts 2/3, 5/6, and 11/12) (Figure 1A). Theoretically, the twelve G-tracts have the potential to form multiple G-quadruplexes, and we assumed that the predominant G-quadruplex structures formed in the full-length hTERT sequence would be those with the smallest loop sizes, possibly having the looping pattern 1:N:1, and that a hierarchy of the stability of the multiple G-quadruplexes would exist according to the size of the combined loops.
Therefore, in this study we set out to determine if a hierarchy of stability exists in the G-quadruplexes formed by the nine putative hTERT G-quadruplex-forming sequences according to the sum of the size of the loops and to partially characterized the G-quadruplex structures formed in the full length G-rich sequence of the hTERT core promoter. Circular dichroic (CD) spectroscopy, Taq polymerase arrest assays, and dimethyl sulfate (DMS) footprinting assays on the nine putative sequences and one full-length G-rich sequence of the hTERT core promoter were carried out. The results from this study show that the overall sum of loop sizes is important for stability of the G-quadruplex structure, but the stability is not in a precise inverse correlation with loop size. Most importantly, the results from the investigation on the full-length hTERT G-rich sequence revealed that the hTERT core promoter adopts a tandem G-quadruplex structure composed of two intramolecular G-quadruplexes that are predicted to interact with each other, one formed by G-tracts 1–4 and the other by G-tracts 5, 6, 11, and 12. The latter structure is striking in that it is stable even in the presence of 26 bases in the middle loop and, surprisingly, more stable than the other more conventional G-quadruplex structure. A mutational study on the middle loop suggests that a hairpin structure is formed in this loop, which would be important for the stability of this G-quadruplex. Significantly, this tandem G-quadruplex structure sequesters all the G-tracts serving as Sp1 binding sites, suggesting that the formation of this structure will completely prevent Sp1 binding to the hTERT core promoter and have a significant effect on promoter activity.
MATERIALS AND METHODS
Oligonucleotides
The oligonucleotides (ODNs) of the hTERT core promoter were synthesized by Operon Biotechnologies, Inc. (Huntsville, AL) and Biosearch Technologies (Novato, CA). All oligonucleotides were gel purified, and the concentrations were determined by absorbance at 260 nm at 95 °C, using a Cary 300 Bio UV-visible spectrophotometer. The sequences of the oligonucleotides are shown in Table 1.
Table 1.
Sequences of the oligonucleotides used in this study.
| ODN Label |
Sequence (5′ to 3′) |
|---|---|
| I | TTTTTGGGGAGGGGCTGGGAGGGTTTTTTTT |
| II | TTTTTGGGGCTGGGAGGGCCCGGAGGGGGTTTTTTTT |
| III | TTTTTGGGAGGGCCCGGAGGGGGCTGGGTTTTTTTT |
| IV | TTTTTGGGCCCGGAGGGGGCTGGGCCGGGGTTTTTTTT |
| V | TTTTTGGGGGCTGGGCCGGGGACCCGGGTTTTTTTT |
| VI | TTTTTGGGCCGGGGACCCGGGAGGGGTTTTTTTT |
| VII | TTTTTGGGGACCCGGGAGGGGTCGGGTTTTTTTT |
| VIII | TTTTTGGGAGGGGTCGGGACGGGGTTTTTTTT |
| IX | TTTTTGGGGTCGGGACGGGGCGGGGTTTTTTTT |
| FL | TTTTTGGGGAGGGGCTGGGAGGGCCCGGAGGGGGCTGGGCCGGGGACCCGGGAGGGGTCGGGACGGGGCGGGGTTTTTTTT |
| XFL | TCCAACTATGTATACTGGGGAGGGGCTGGGAGGGCCCGGAGGGGGCTGGGCCGGGGACCCGGGAGGGGTCGGGACGGGGCGGGGTAGCGGCACGCAATTGCTATAGTGAGTCGTATTA |
| I′ | TCCAACTATGTATACTGGGGAGGGGCTGGGAGGGCGGCACGCAATTGCTATAGTGAGTCGTATTA |
| II′ | TCCAACTATGTATACTGGGGCTGGGAGGGCCCGGAGGGGGTAGCGGCACGCAATTGCTATAGTGAGTCGTATTA |
| VIII′ | TCCAACTATGTATACTGGGAGGGGTCGGGACGGGGTAGCGGCACGCAATTGCTATAGTGAGTCGTATTA |
| IX′ | TCCAACTATGTATACTGGGGTCGGGACGGGGCGGGGCGGCACGCAATTGCTATAGTGAGTCGTATTA |
| 5,12 | TTTTTGGGGGCTGGGCCGGGGACCCGGGAGGGGTCGGGACGGGGCGGGGTTTTTTTT |
| X5,12 | TCCAACTATGTATACTGGGGGCTGGGCCGGGGACCCGGGAGGGGTCGGGACGGGGCGGGGTAGCGGCACGCAATTGCTATAGTGAGTCGTATTA |
NOTE: In ODNs I′, II′, VIII′, IX′, XFL, and X5,12, the G-quadruplex-forming elements are in bold and the primer binding sites are underlined.
Circular dichroism spectroscopy and Tm determination
For CD spectroscopy, oligonucleotides were prepared at 2.5 µM in 50 mM Tris-HCl (pH 7.5) in the presence or absence of 140 mM KCl and denatured at 95 °C, followed by slow cooling to room temperature to induce G-quadruplex formation. CD spectra were measured by a Jasco-810 spectropolarimeter (Jasco, Easton, MD) using a quartz cell of 1-mm optical path length and an instrument scanning speed of 100 nm/min, with a response of 1 s, over a range of 200–350 nm. A set of three scans was averaged for each sample at 25 °C. For the determination of Tm, G-quadruplex formation was induced and molar ellipticity at 264 nm was measured over a temperature range of 20–100 °C. The Tm values were calculated by the computer program Prism4 (GraphPad Software, La Jolla, CA).
Electrophoretic mobility shift assay (EMSA) and DMS protection assay
Oligonucleotides were 5′-end labeled, diluted to 50 nM in 40 µL of 10 mM Tris-HCl (pH 8.0), denatured at 95 °C, and slowly cooled down to room temperature in the presence or absence of 140 mM KCl. Two µg of dIdC was added to each sample, and the samples were treated with 0.5% DMS as a final concentration for 5 or 15 min at room temperature. The reaction was quenched by adding 13.5 µL of a stop solution (3.5 µL of 14.3 M β-mercaptoethanol and 10 µL of 10 % glycerol) to each sample, and the samples were immediately loaded on a 10% native gel. The unimolecular oligonucleotide species were separated by EMSA, excised and extracted in dH2O, and precipitated at –20 °C in the presence of 70% EtOH, 97 mM NaOAc (pH 5.2), and 6.5 mg/mL yeast tRNA as final concentrations. The DMS-treated oligonucleotides were cleaved by 10% (v/v in 10% Tris-HCl [pH 8.0]) piperidine and separated on a sequencing gel.
Taq polymerase arrest assays
Taq polymerase arrest assays were performed as previously described [28]. Briefly, a primer was annealed to the template bearing the sequence of interest, and the asymmetric primer–template duplex was gel-purified. The asymmetric duplex was incubated in KCl at 0–140 mM overnight at room temperature, and primer extension was conducted with 2.5 U of Taq polymerase (Fermentas, Hanover, MD) at 57 °C for 25 min. In the case of compound treatment, the samples were incubated with G-quadruplex-interactive compounds for 20 min at room temperature in the presence of 40 mM KCl, and primer extension was performed as described above. The reaction was stopped by adding denaturing loading dye (93% formamide, 10 mM EDTA [pH 8.0], 0.25 mg/mL xylene cyanol, 0.25 mg/mL bromophenol blue), and the primer extension products were separated on a 12% denaturing gel.
RESULTS
The truncated G-rich sequences at the 5′- and 3′-ends of the hTERT G-rich promoter region form stable G-quadruplexes
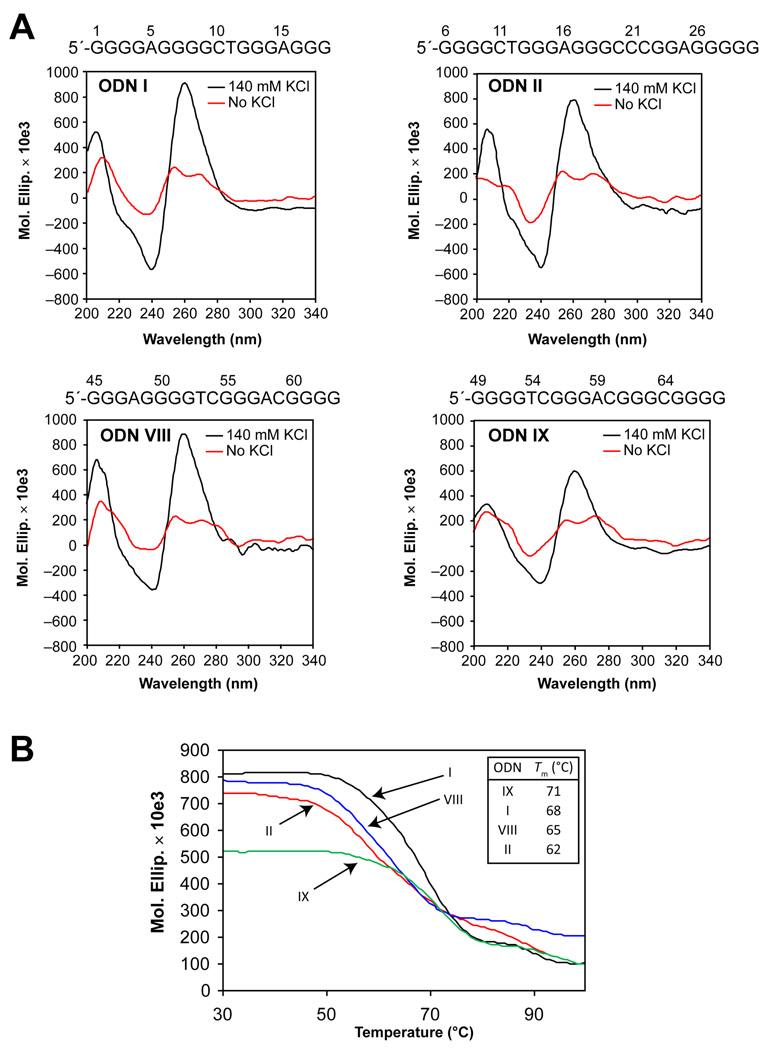
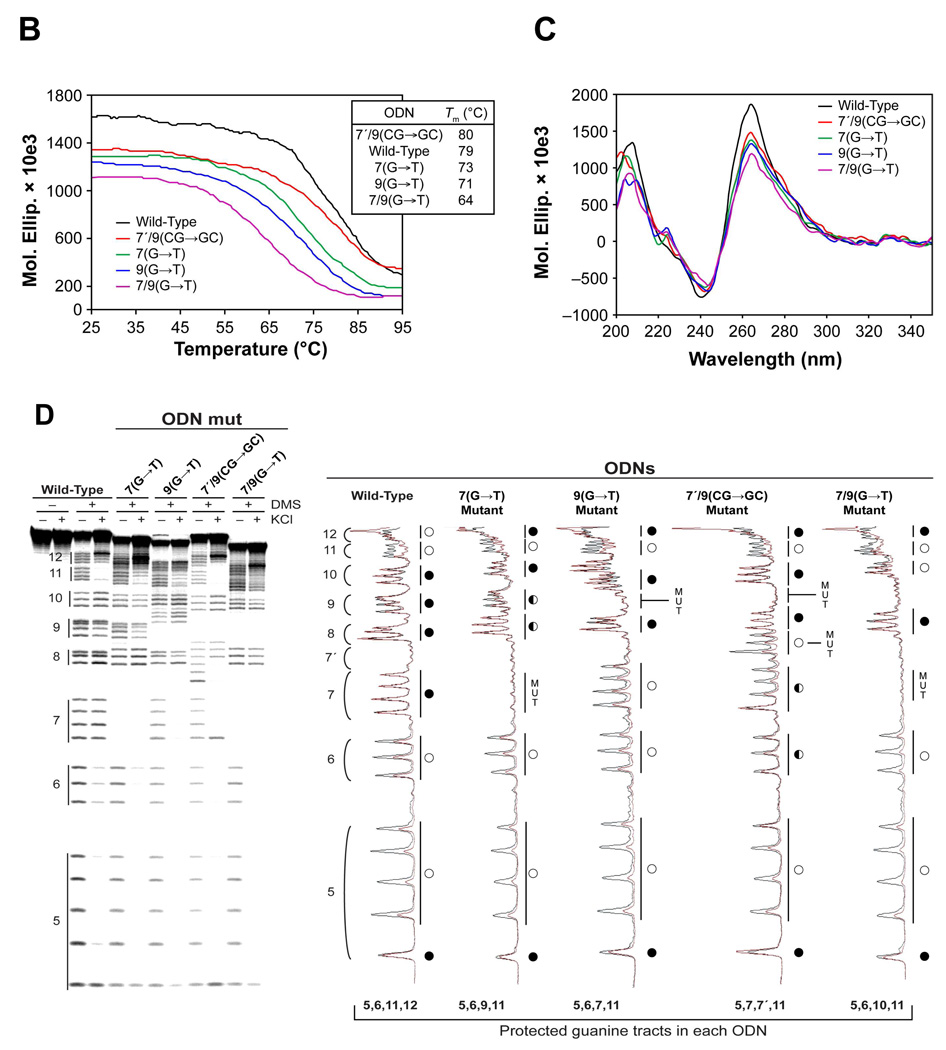
The hTERT core promoter contains twelve G-tracts between –22 and –90 relative to the transcription initiation site (Figure 1A). Due to the presence of a redundant number of G-tracts, the G-rich region has the potential to form multiple G-quadruplexes, with the assumption that each G-tract has an equal opportunity to be part of a G-quadruplex structure. We selected nine putative G-quadruplex-forming sequences with a total number of 5 (minimum) to 13 (maximum) bases in the loops (I to IX) to investigate G-quadruplex formation (Figure 1, B and C). T-tails were added at each end of sequences I–IX so we could isolate the effect of loop size from the variable effect of different flanking bases. CD spectroscopy was initially carried out on ODNs I through IX (Table 1) containing the putative G-quadruplex-forming sequences shown in Figure 1B in the presence or absence of 140 mM KCl to determine if a G-quadruplex is formed by a given DNA sequence and, if so, the likely topology of guanine strands in the structure. The results show that only four of the nine sequences (ODNs I, II, VIII, and IX in Figure 1, B and C) produced CD spectra characteristic of a G-quadruplex structure in a potassium-dependent manner (Figure 2A). ODNs I, II, and IX produced a positive maximum at 264 nm and a negative minimum at 240 nm, similar to the signature CD spectrum of a parallel G-quadruplex, while ODN VIII exhibited a positive maximum at 264 nm and a small positive shoulder adsorption at 295 nm, indicative of a potential mixed parallel/antiparallel G-quadruplex, such as in the bcl-2 G-quadruplex promoter sequence [29]. With the exception of ODN II, which has a total of between 9 and 12 bases in the loops (Figure 1C), the ODNs determined to be G-quadruplex-positive contain the minimum total number of 5 or 6 bases in the loops, which is clearly shorter than those of ODNs III to VII (minimum total number of 8–12 bases). This shows that loop size is an important factor for determining the stability of the G-quadruplexes formed by the hTERT G-rich sequences. With this preliminary evidence of G-quadruplex formation, we further examined the relationship between stability and loop size by measuring the thermal stability of ODNs I, II, VIII, and IX. Figure 2B shows the melting curves of the ODNs in the presence of 140 mM KCl, and the Tm values calculated on the basis of the curves are in order of ODNs IX (71 °C) > I (68 °C) > VIII (65 °C) > II (62 °C), which shows a good but not complete inverse correlation with the total loop size of the ODNs (Figure 1C).
Figure 2.
CD spectra and melting curves of ODNs I, II, VIII, and IX. (A) CD analyses were performed on ODNs I, II, VIII, and IX in the absence (red line) and presence (black line) of 140 mM KCl (CD spectra of ODNs III, IV, V, VI, and VII are shown in Supporting Information). (B) Thermal stability of the G-quadruplexes formed by ODNs I, II, VIII, and IX. The melting temperatures (Tm) of the G-quadruplexes formed are in order of ODNs IX (71 °C) > I (68 °C) > VIII (65 °C) > II (62 °C).
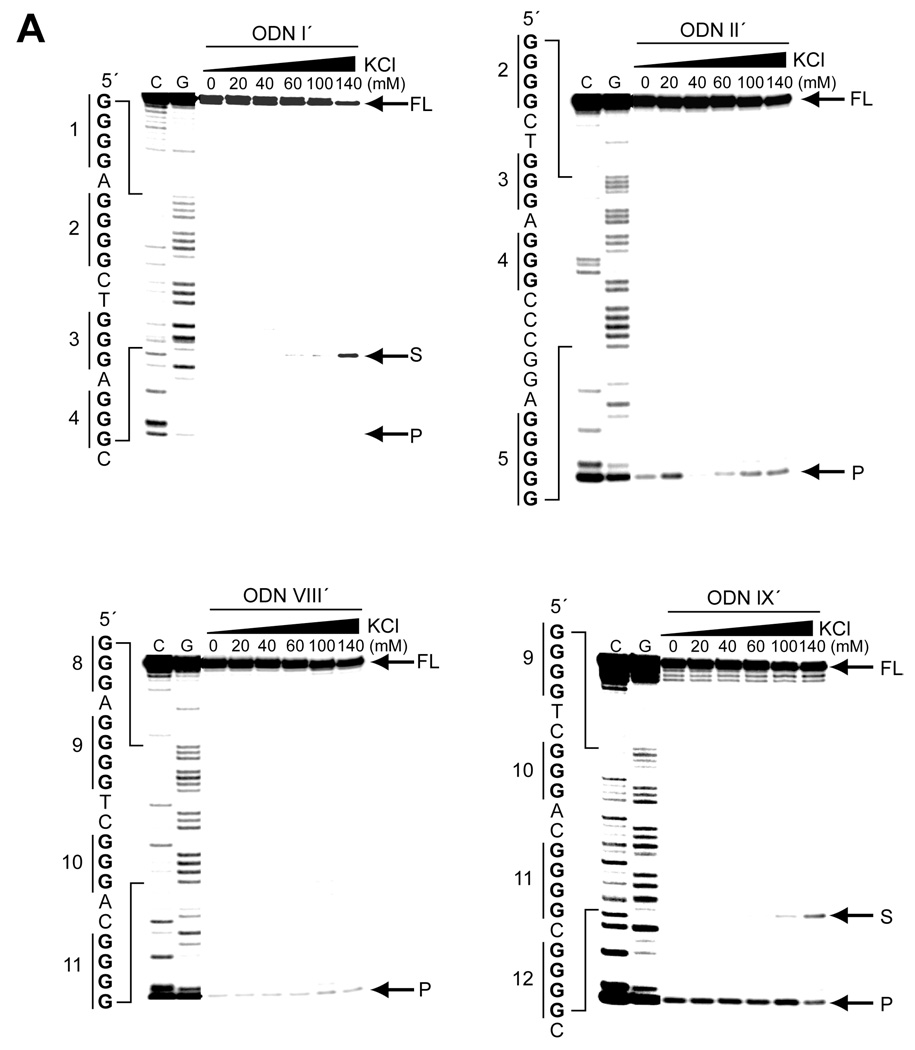
Further evidence for the stability of G-quadruplexes formed by the ODNs was provided by a Taq polymerase arrest assay. Figure 3A shows the results of the assay performed with increasing concentrations of KCl (0–140 mM) on ODNs I′, II′, VIII′, and IX′ (Table 1) containing the G-quadruplex-forming sequences I, II, VIII, and IX, respectively. ODNs I′ and IX′ produced a premature primer extension product at the cytosines upstream of G18 and G68, respectively, in a potassium-dependent manner (Figure 3A). The arrest products first appeared around 60 and 100 mM KCl in ODNs I′ and IX′ respectively, which is in agreement with the thermal stability of the ODNs evaluated by the Tm values (Figure 2C). Unlike ODNs I′ and IX′, ODNs II′ and VIII′ did not produce such premature products, implying that the hTERT G-quadruplex-forming sequences II and VIII form less stable G-quadruplexes. These results support the findings from the thermal stability study, which showed that the hTERT G-quadruplex-forming sequences I and IX form more stable structures than sequences II and VIII.
Figure 3.
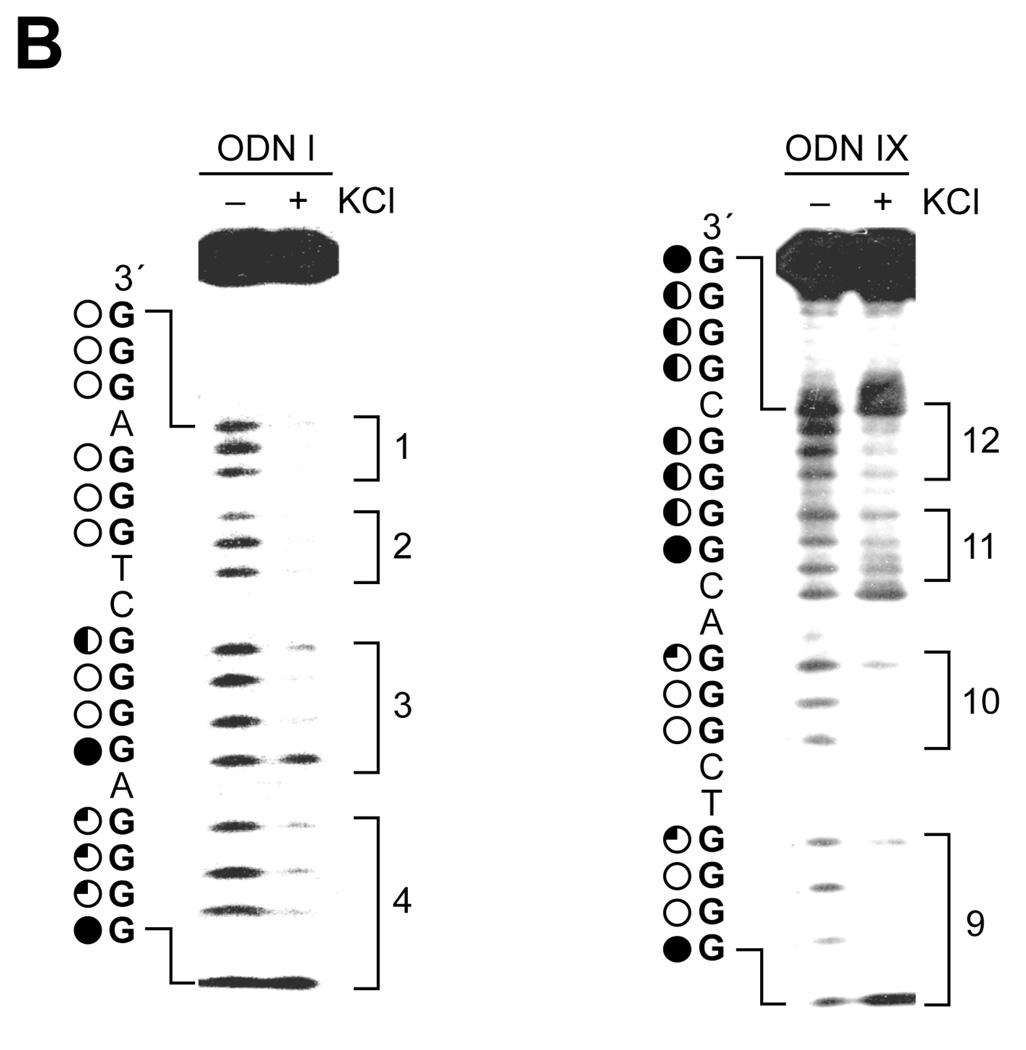
Taq polymerase arrest assays on ODNs I′, II′, VIII′, and IX′ and DMS footprinting assays on ODNs I and IX. (A) Taq polymerase arrest assays (see Materials and Methods) were performed on ODNs I′, II′, VIII′, and IX′ with increasing concentrations of KCl. The C and G sequencing lanes are shown to the left of the gels together with the sequence and the G-tracts. The locations of the primer start site (P), quadruplex pausing site (S), and full-length product (FL) are shown to the right of the gels. (B) DMS footprinting assays of ODNs I and IX in the absence and presence of 140 mM KCl. DMS footprinting was as described in Materials and Methods. The sequences of ODNs I and IX are shown to the left of the gels together with the degree of DMS protection of guanines (complete protection = open circles, no protection = filled circles, and partial protection = half or quarter filled circles). The brackets to the right of the gels indicate the guanine tracts.
The G-quadruplexes formed by sequences I and IX were further characterized by performing a DMS footprinting assay. Figure 3B shows clear protection in the four G-tracts of ODNs I, while in sequence IX G-tracts 9 and 10 are clearly protected (G50–G52 and G55–G57), but there is only partial protection of G-tracts 11 and 12 (G61–G63 and G65–G67), demonstrating G-quadruplex formation by these ODNs. Almost complete protection occurred in the first, second, third, and fourth G-tracts of ODN I, with complete protection on G7/G8, G12–G14, and G16–G18. These partial protection patterns indicate loop isomer formation by ODNs due to the alternative use of guanines in the G-tracts. Moreover, our data demonstrate that the G-quadruplex-forming sequences I and IX predominantly form a 2:2:1 loop isomer [5′-(A5G6:C10T11:A15)-3′] and a 2:3:1 loop isomer [5′-(T53C54:A58C59G60:C64)-3′], respectively, which correspond to the minimum number of bases required in the loops of the structures. Collectively, these results show that total loop size in the G-quadruplexes formed by the nine putative sequences of the hTERT core promoter is important for evaluating stability of the structures.
The secondary structure formed in the full-length hTERT G-rich region consists of tandem end-to-end stacked intramolecular G-quadruplexes
The data reported so far on the truncated hTERT G-rich sequences demonstrate that only the 5′- and 3′-end sequences (ODNs I and IX, respectively) appear to form stable G-quadruplexes. Since the twelve G-tracts in the full-length G-rich sequence of the hTERT core promoter will all compete for G-quadruplex formation, which could result in the formation of other more stable intramolecular G-quadruplexes that had not been tested, it was necessary to confirm if the same G-quadruplexes are formed in the full-length G-rich sequence. The same techniques were used to characterize the G-quadruplex structures formed in the full-length sequence.
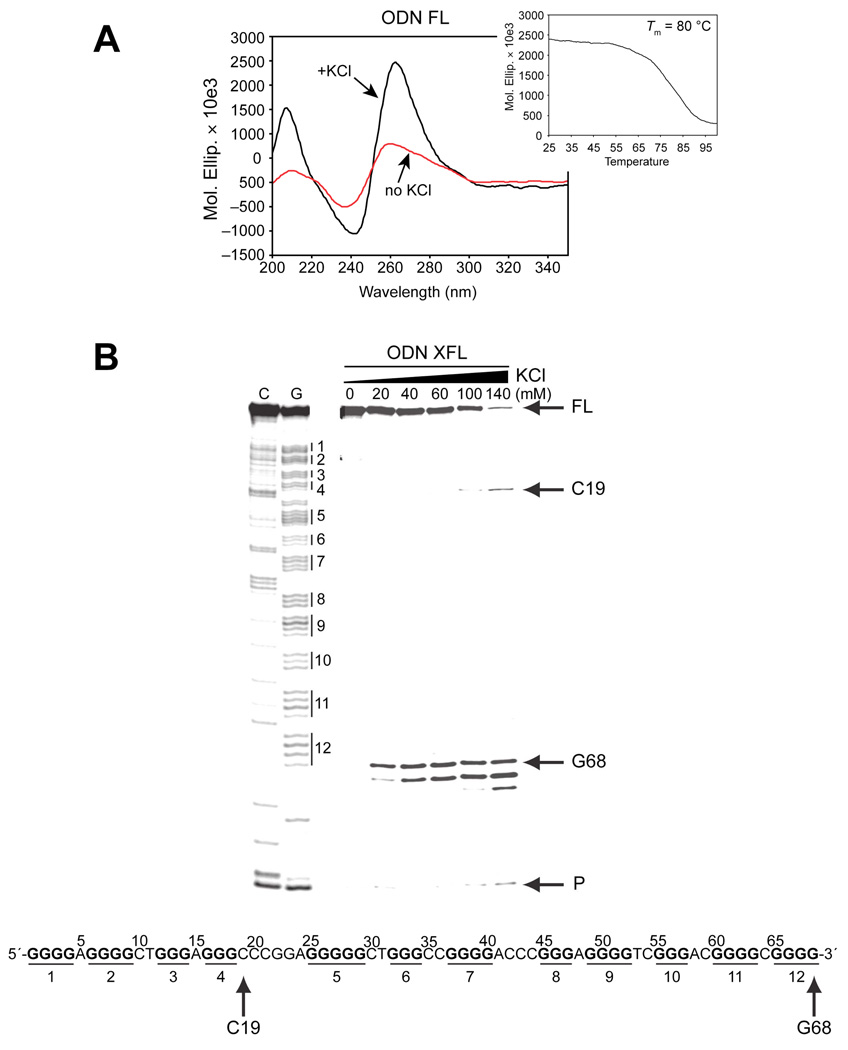
The CD spectra of ODN FL (sequence shown in Table 1), containing the full-length G-rich sequence of the hTERT core promoter, shows a large positive maximum at 264 nm and a small positive shoulder at 290 nm (Figure 4A), which is suggestive of a mixed parallel/antiparallel G-quadruplex structure. To determine the stability of the G-quadruplex structure(s) formed by ODN FL, a melting curve of ODN FL was generated, and the Tm was found to be 80 °C (Figure 4A, inset). Significantly, when compared to those of ODNs I and IX, the molar ellipticity at 264 nm of ODN FL (approximately 2,500,000) is 2.5 and 5 times higher, and the Tm of ODN FL is 12 °C and 9 °C higher, respectively (compare Figure 2A and Figure 4A). This dramatic increase in molar ellipticity and Tm of ODN FL was unexpected and prompted us to examine this full sequence in more detail.
Figure 4.
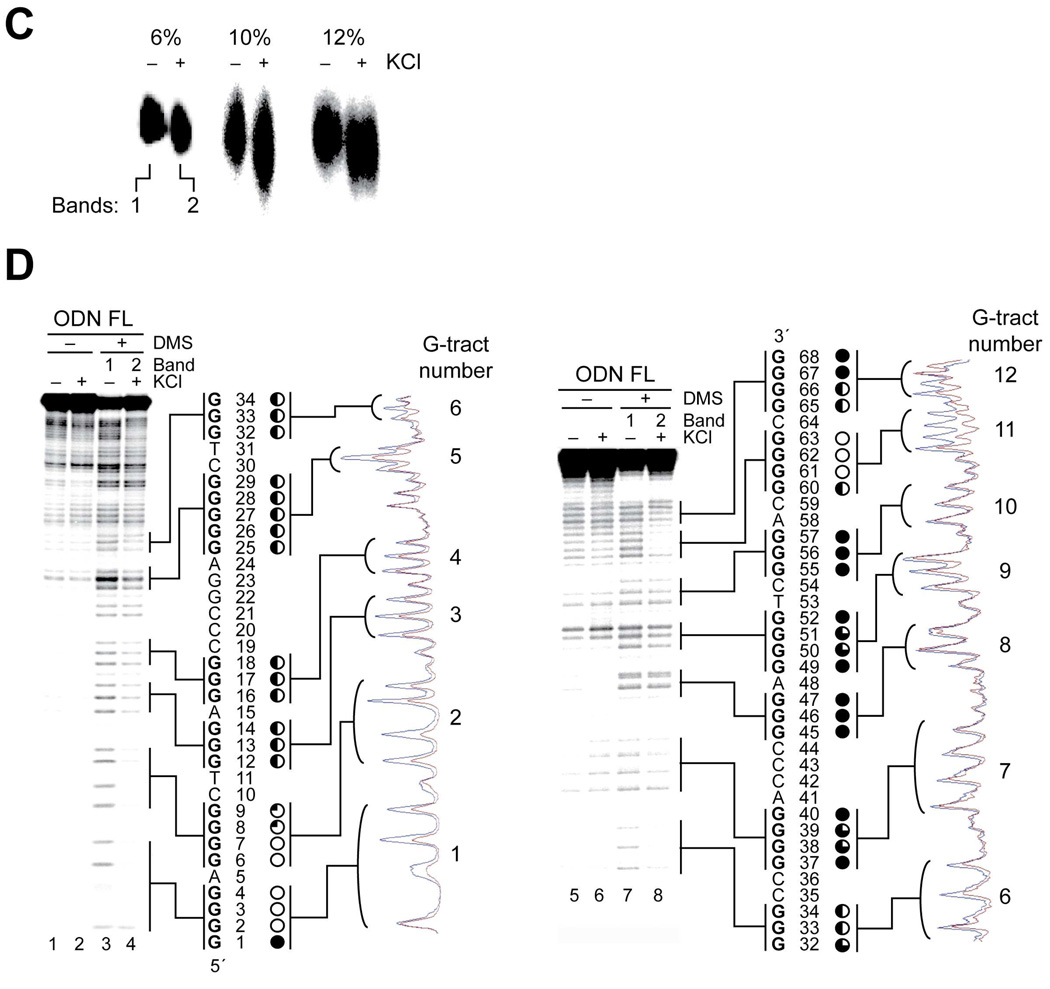
Characterization of G-quadruplex formation in the full-length hTERT G-rich sequence by CD, Taq polymerase stop assay, and DMS footprinting. (A) CD spectroscopy was performed on ODN FL containing the full-length hTERT G-rich sequence in the presence (black line) and absence (red line) of 140 mM KCl. The melting curve is shown as an inset. (B) A Taq polymerase arrest assay was carried out on ODN XFL with increasing concentrations of KCl. CG sequencing lanes are shown to the left of the gel. The guanine tracts are shown to the right of the sequencing lanes. The stop site at the primer (P), two additional stop sites (G68 and C19), and the full-length product (FL) are shown to the right of the gel. The FL sequence and corresponding stop sites are shown at the bottom of the figure. (C) DMS-treated samples in the presence or absence of 140 mM KCl were run on 6%, 10%, and 12% native gels (full gels are shown in Supporting Information). (D) DMS footprinting assays, as described in Materials and Methods, were carried out on bands 1 and 2 from (C). The two gels were run separately to resolve the 5′ (left) and 3′ (right) ends of the sequence. Lanes 1 and 2 and 5 and 6 are treated only with piperidine, whereas lanes 3 and 4 and 7 and 8 were also treated with DMS. To the far right of each gel are scans of each gel showing a comparison of lanes 3 and 4 and 7 and 8 (blue and red lines, respectively). Between each of the gels and scans are the DNA sequence, base numbers, and degree of DMS protection of guanines (complete protection = open circles, no protection = filled circles, and partial protection = one- to three-quarters filled circles).
A Taq polymerase arrest assay was carried out on ODN XFL (the hTERT full-length G-rich sequence) (Table 1) with increasing concentrations of KCl (0–140 mM). ODN XFL produced two premature primer extension products, the first at C19 immediately upstream of G-tract 4 and the second at G68 (Figure 4B), which is the 3′-end guanine of G-tract 12. This result suggests that the full-length sequence forms at least two stable G-quadruplex structures. Significantly, the premature product at G68 first appeared at 20 mM KCl, which is a much lower concentration than those at which the premature products of ODNs I′ and IX′ first occurred (60 and 100 mM, respectively, in Figure 3A), confirming that the G-quadruplex structure formed by the full-length sequence is much more stable than those formed by the truncated hTERT G-rich sequences I and IX. The premature product at C19 is presumably due to the intramolecular G-quadruplex formed by G-tracts 1–4 (ODN I in Figure 1B), and the premature product at G68 could correspond to G-tract IX formed by G-tracts 9–12, although on the basis of the comparative CD, Tm, and polymerase stop assay results on the sequences of ODN IX and ODN FL, this seemed unlikely. Instead, the latter premature product is more likely caused by a different G-quadruplex involving four different G-tracts between G-tracts 5 and 12 in the full-length sequence.
To further probe the structures responsible for the premature primer extension products at C19 and G68 in ODN FL, a DMS footprinting assay was carried out on ODN FL. In the assay, EMSA was carried out on the DMS-treated ODN FL samples on a native gel to isolate the random-coil and intramolecular G-quadruplex-structured species of ODN FL formed in the absence and presence of KCl, respectively. As expected, a slightly higher mobility was observed in the G-quadruplex-structured ODN FL samples than in the unfolded form of ODN FL (compare (+) and (–) KCl lanes in Figure 4C), suggesting the formation of an intramolecular G-quadruplex structure in the full-length hTERT G-rich sequence in the presence of KCl. These DMS-treated unfolded and folded DNA species (bands 1 and 2 in Figure 4C) were extracted from the gel and subjected to piperidine cleavage. Since it is difficult to completely resolve the full-length sequence in a single lane due to its long length, we ran the same samples twice on the same gel and focused on resolving either the 5′-end (Figure 4D lanes 1–4) or the 3′-end sequence (Figure 4D lanes 5–8). Also, the ODN FL samples treated by piperidine but not by DMS are included as controls for background cleavage by piperidine in the presence or absence of KCl to ensure that an equal background cleavage occurs regardless of KCl in the reaction (Figure 4D, lanes 1 and 2 and lanes 5 and 6). The results from this assay show clear protection in G-tracts 1–4 at the 5′-end of the full-length sequence, with complete protection of G2–G4 and G6/G7 in G-tracts 1 and 2 and partial protection in G-tracts 2–4 (G8/G9, G12–G14 and G16–G18) (Figure 4D, lanes 3 and 4), demonstrating intramolecular G-quadruplex formation by these four G-tracts. Furthermore, complete cleavage on G1 and slight cleavage on G8 and G9 in G-tracts 1 and 2 indicate the formation of loop isomers by G-tracts 1–4 due to the redundant number of guanines in these tracts (Figure 4D, lanes 3 and 4). However, the predominant isomer form is in a 1:3:1 loop conformation [5′-(A5:G9C10T11:A15)-3′] in contrast to that in ODN I, which is 2:2:1. These results demonstrate that G-tracts 1–4 form a stable G-quadruplex in the full-length hTERT G-rich sequence and caused the premature primer extension product at C19 in the Taq polymerase assay.
In accordance with our suspicion that the G-quadruplex responsible for the premature primer extension product at G68 (Figure 4B) was not formed by G-tracts 9–12, the DMS footprinting pattern in the full-length sequence showed methylation protection in G-tracts 5, 6, 11, and 12, with almost complete protection in G-tract 11 (G61–G63 demonstrate complete protection) and partial protection in G-tracts 5, 6, and 12 (G25–G29, G32–G34, and G65/G66, respectively) (Figure 4D). This protection pattern strongly suggests that G-tracts 5, 6, 11, and 12 form the stable G-quadruplex in the full-length sequence that caused the premature primer extension product at G68 in the Taq polymerase assay (Figure 4B). Moreover, the partial protection in G-tracts 5 and 11 (G25–G29 and G60), which contain a redundant number of guanines, implies loop isomer formation in the G-quadruplex. This G-quadruplex is unexpectedly stable, even with the 26-base loop comprising bases 35–60. One notable observation is that a small but reproducible amount of DMS protection also occurred in G-tracts 7 and 9, suggesting the formation of a secondary DNA structure in the middle loop. This small amount of DMS protection suggests a duplex environment for these guanines that would be predicted to be less susceptible to cleavage than guanines in a single-stranded region. Since this G-quadruplex is so unique, we decided to confirm G-quadruplex formation by G-tracts 5, 6, 11, and 12 by focusing on G-tracts 5–12 of the hTERT G-rich sequence.
hTERT G-tracts 5–12 form an unusual G-quadruplex structure with a 26-base middle loop
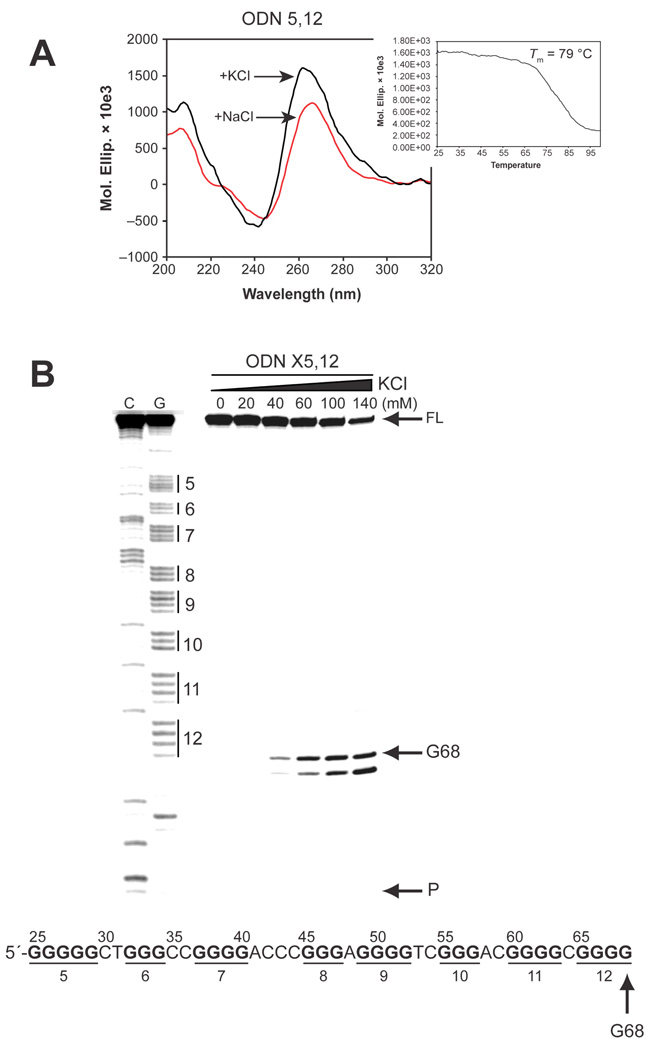
To more precisely characterize the G-quadruplex formed by G-tracts 5–12, we performed CD spectroscopy, a Taq polymerase arrest assay, and a DMS footprinting assay on the wild-type sequence (ODN 5,12) (Table 1). The CD spectra of ODN 5,12 in KCl showed a positive maximum at 264 nm and a small positive shoulder at 290 nm, a CD spectrum again suggestive of a mixed parallel/antiparallel G-quadruplex, and the structure had a Tm of 79 °C, based on the melting curve (Figure 5A). Significantly, the molar ellipticity at 264 nm (1, 600, 000) and the Tm (79 °C) of ODN 5,12 are much higher than those of ODN I (90,000 and 68 °C, respectively) but slightly lower than those of ODN FL (2,500,000 and 80 °C, respectively), suggesting that the G-quadruplex structure formed by G-tracts 5–12 is not only stable but is the structure that is responsible for the overall stability of the secondary structures formed in the full-length hTERT sequence. Further evidence for this finding was provided by a Taq polymerase arrest assay on ODN X5,12 (Table 1 and Figure 5B). The assay was performed with increasing concentrations of KCl (0–140 mM) and resulted in a single premature primer extension product at G68, indicating that a single intramolecular G-quadruplex is formed by G-tracts 5–12. The premature product in ODN X5,12 first appeared at 40 mM KCl, which is a lower concentration than where the premature product first occurred in ODN I′ (60 mM in Figure 3A) but higher than that in ODN XFL (20 mM in Figure 4B).
Figure 5.
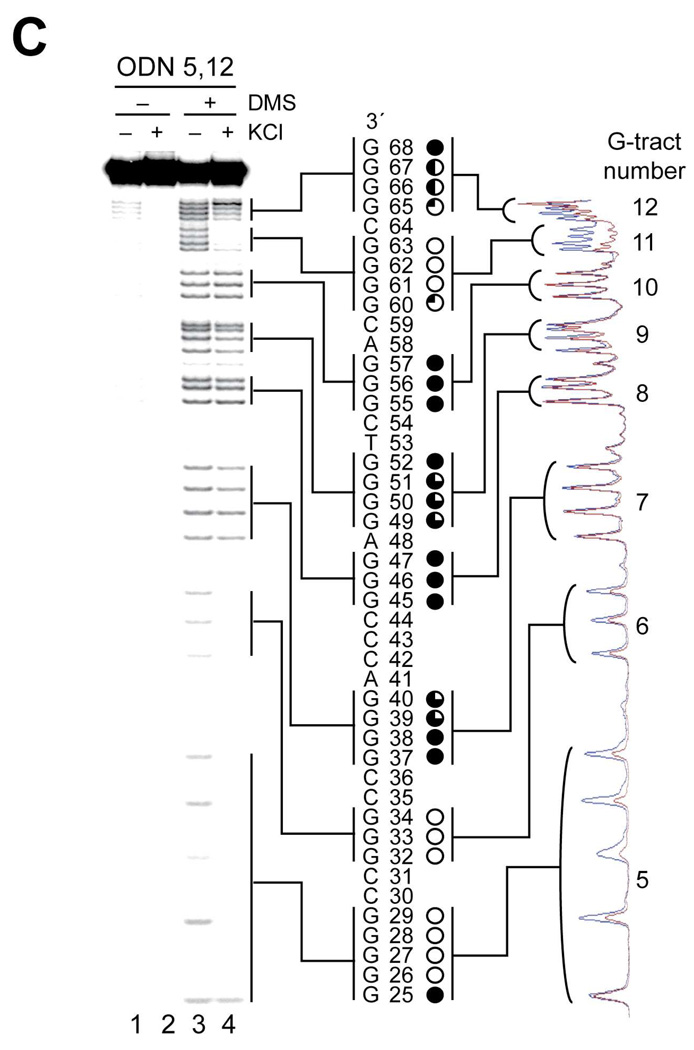
Characterization of G-quadruplex formation in hTERT G-tracts 5–12 by CD, Taq polymerase stop assay, and DMS footprinting. (A) Comparative CD spectroscopy of ODN 5,12 with KCl (black line) and with NaCl (red line). The inset shows the melting curve of ODN 5,12. (B) A Taq polymerase stop assay was carried out on ODN X5,12 with increasing concentrations of KCl, as described in Materials and Methods See legend of Figure 4B for description of figure. (C) DMS footprinting of isolated bands as described for Figure 4, C and 4D). For description of the footprinting gel and scans, please refer to Figure 4D.
The DMS protection pattern of ODN FL in Figure 4D strongly suggests the formation of an unusual G-quadruplex by G-tracts 5, 6, 11, and 12 in the full-length hTERT G-rich sequence. To confirm the formation of this unusual structure in the full-length sequence, a DMS footprinting assay was carried out on ODN 5,12. Figure 5C shows clear protection in G-tracts 5, 6, 11, and 12, with complete protection of G26–G29, G32–G34, and G61–G63 of G-tracts 5, 6, and 11, respectively, and partial protection in G-tract 12 (G65–G67). This result confirms that the G-quadruplex formed in G-tracts 5–12 is composed of G-tracts 5, 6, 11, and 12 (Figure 4D). Partial protection in G-tracts 11 and 12 (G60 and G65–G67, respectively), which contain a redundant number of guanines, suggests the formation of loop isomers in the G-quadruplex. However, the predominant loop isomer of the G-quadruplex formed in ODN 5,12 is in a 2:26:1 conformation [5′-(C30T31:C35–G60:C64)-3′] or a 3:26:1 conformation [5′-(G29C30T31:C35–G60:C64)-3′], in which 26 bases are located in the middle loop.
A hairpin structure is predicted to form in the central loop of the G-quadruplex formed by G-tracts 5–12
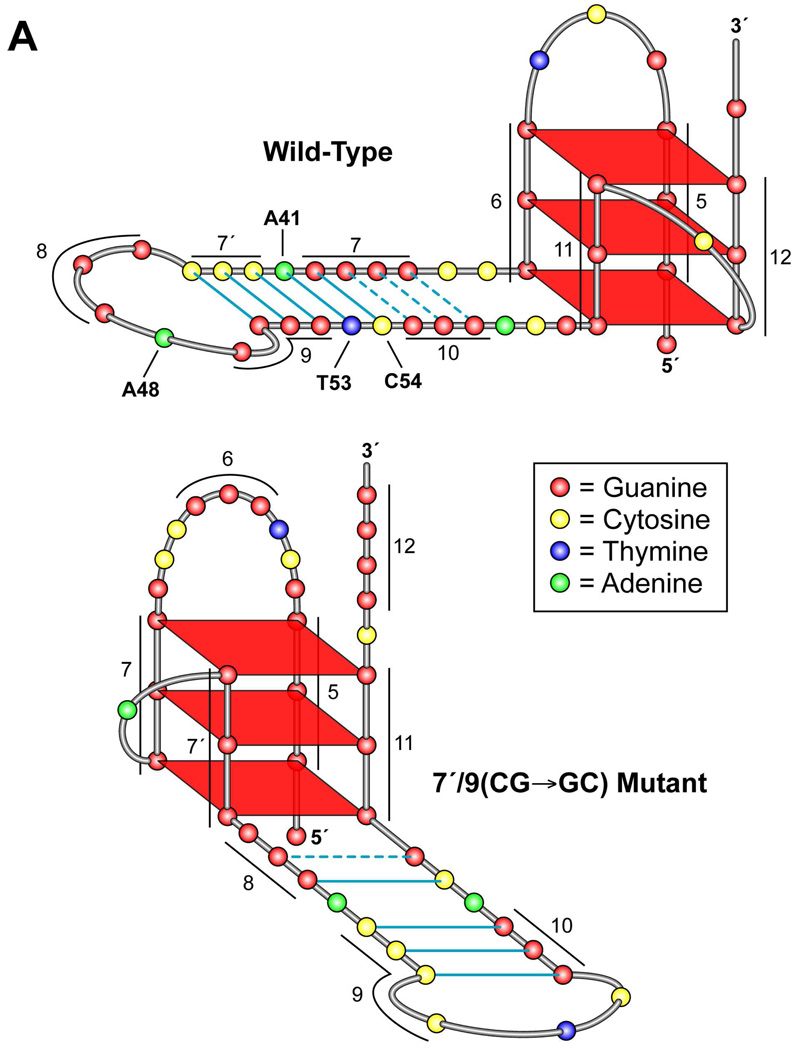
Significantly, the slight protection in G-tracts 7 and 9 previously shown in DMS footprinting of ODN FL (Figure 4D) was also observed in ODN 5,12 (see G39/G40 and G49–G51 in Figure 5C). This result suggests that a secondary DNA structure is formed in the middle loop (C35–G60) of the G-quadruplex, most likely a hairpin structure. Indeed, a hairpin structure could be formed by the middle loop sequence (C35–G60) based on the DMS protection pattern shown in Figure 5C and predicted base-parings in the structure, in which G-tract 9 base-pairs with C-tract 7′ (three G–C base pairs) and G-tract 7 base-pairs with G-tract 10 and cytosine 54 to form three G–G base pairs and one G–C base pair (Figure 6A, wild-type structure). An additional A–T base pair (bases 41 and 53) is also predicted to occur. Finally, G-tract 8, A48, and G49 are proposed to form the five-base loop in the hairpin structure.
Figure 6.
Characterization of the effects of mutants formed by substitution of guanine tracts 7, 9, and 10 with thymines and interchanging of C-tract 7′ and G-tract 9, using CD, Taq polymerase stop assay, and DMS footprinting (the wild-type G- and C-tracts are shown in Figure 6A, top). (A) Proposed folding patterns of the wild-type and the 7′/9(CG→GC) mutant sequences, based upon CD and DMS footprinting results. The G- and C-tracts are indicated with numbers. Solid lines between GC and AT base pairs indicate Watson-Crick base pairing. Dashed lines indicate G–G base pairing. (B) Comparative thermal stability of wild-type, 7(G→T), 9(G→T), 7/9(G→T), and 7′/9(CG→GC) mutants. (C) Comparative CD spectroscopy of the same mutants in (B). (D) DMS footprinting of ODN 5,12 (wild-type) and the mutants in the stem of the hairpin. DMS-treated samples in the presence and absence of 140 mM were treated as described in Materials and Methods. The guanine runs are indicated by bars to the left of the gel. The scans to the right show the pattern of DMS protection and cleavage of the wild-type and each mutant oligomer. The degree of DMS footprinting is indicated by open circles (protected) and closed circles (cleaved). Where ambiguity exists in the extent of cleavage [7(G→T) and 7′/9(CG→GC)], both similarly cleaved G-tracts are denoted with half-filled circles. The mutated guanine or cytosine tract is indicated by MUT. The predominant G-tracts that are protected from DMS footprinting are shown for each ODN at the bottom of the figure.
To further examine the predicted hairpin structure and its role in stability of the G-quadruplex formed by G-tracts 5, 6, 11, and 12, we introduced mutations in G-tracts 7 and/or 9 of ODN 5,12, where the slight DMS protection was observed, and evaluated the stability of the G-quadruplexes formed in the mutants. Significantly, a G-to-T mutation of G-tract 7 or 9 [7(G→T) and 9(G→T) mutants in Figure 6A], each of which disrupting the hairpin structure in the stem, resulted in a significant decrease in Tm of the G-quadruplex (6 or 8 °C, respectively) (Figure 6B). Furthermore, G-to-T mutations in both G-tracts 7 and 9 [7/9(G→T) mutant in Figure 6A] caused a greater decrease in Tm (15 °C) (Figure 6B) compared to either 7(G→T) or 9(G→T) mutants. If the predicted hairpin structure is correct, switching the bases of G-tract 9 (G49–G51) and C-tract 7′ (C42–C44), resulting in C49–C51 and G42–G44, would be anticipated not to have a significant effect on the formation of the hairpin structure in the middle loop, and therefore result in minimal change in Tm when compared to that of the G-quadruplex formed in the wild-type sequence (ODN 5,12). Indeed, the thermal stability of the switch mutant 7′/9(CG→GC) was found to be 80 °C, which is very similar to that of the G-quadruplex formed by ODN 5,12 (79 °C) (Figure 6B). This result is in accordance with our prediction of the hairpin structure. These data suggest that the formation of the hairpin structure in the middle loop of the G-quadruplex formed by G-tracts 5, 6, 11, and 12 plays a critical role in the stability of the structure. We also performed CD spectroscopy on the same mutants, and they had very similar CD spectra to that of the wild-type (Figure 6C).
Although the Tm and CD data support our hypothesis, DMS footprinting experiments were conducted on the mutants to determine if the folding patterns were affected by these changes. The results shown in Figure 6D surprisingly demonstrate that for all four mutants the DMS footprinting pattern and G-quadruplex folded form were different to that of the wild-type sequence. For example, in the case of the 7′/9(CG→GC) mutant in which we had predicted that a transversion of the proposed base-pairing guanines and cytosines in the stem would have minimal effect on the structure and stability of the hairpin, a new folding pattern occurs such that the new run of guanines (7′) is now incorporated into the G-quadruplex structure and G-tract 12 is moved outside of the G-quadruplex (see lower folded mutant in Figure 6A). Concurrently, although some ambiguity exists (see G-tracts 6 and 7 in Figure 6D), G-tract 7 is the second 5′-end G-tract to be used in this G-quadruplex, which then sets up a favored one-base double-chain reversal loop for the 7/7′ face of the G-quadruplex (Figure 6A). This still allows for considerable base pairing in the stem, which could account for the equivalent stability of this mutant sequence to the wild-type (see Figure 6B). A similar rationale can be used to propose different folding patterns and base pairings for the other mutants examined in this study, which results in larger lateral loops and less base pairing in the stems (unpublished results), and this could account for the lower stability of these structures (Figure 6B).
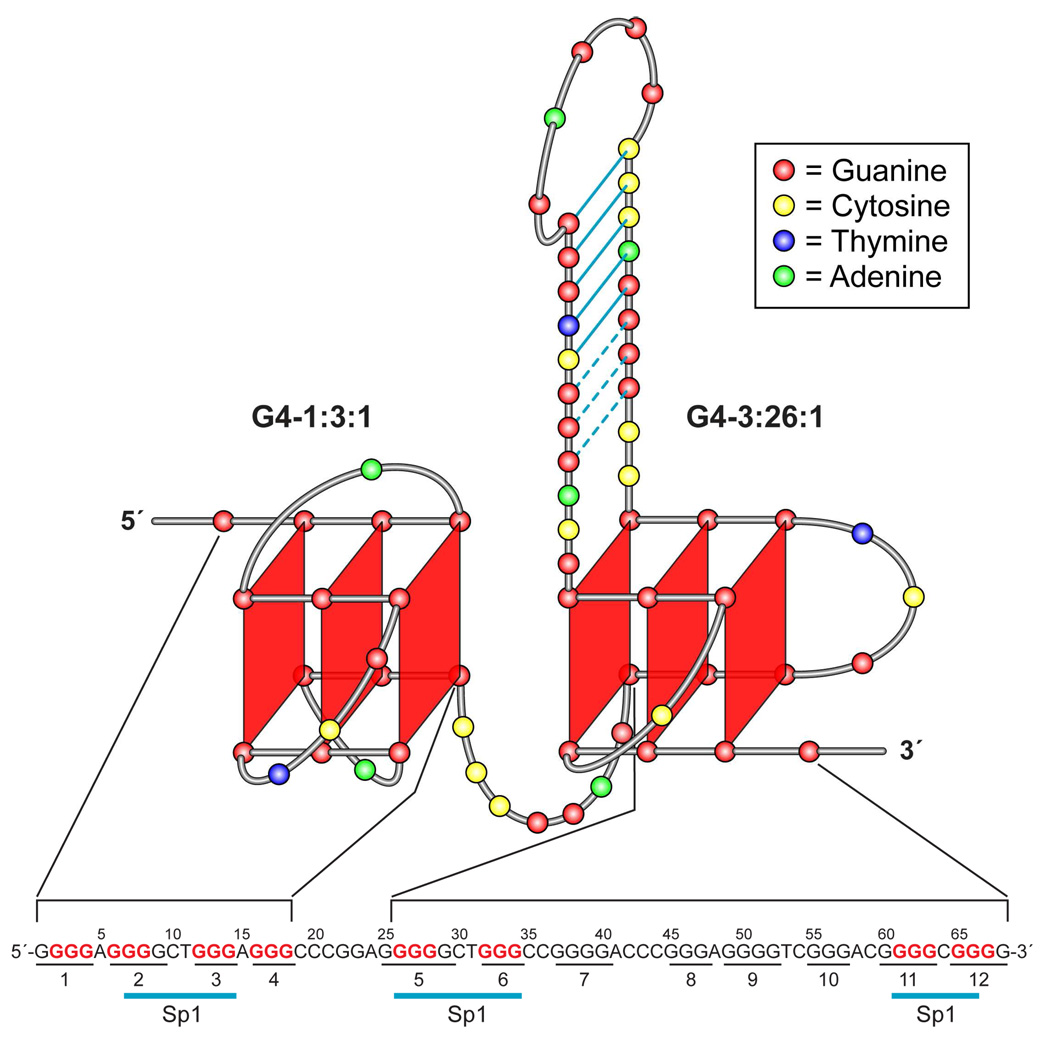
Taken together, the data in Figure 4–Figure 6 demonstrate that the G-rich region in the hTERT core promoter forms a stable tandem G-quadruplex structure (Figure 7). One G-quadruplex (G4-1:3:1) is formed by G-tracts 1–4 at the 5′-end of the G-rich region, and the other (G4-3:26:1) is formed by G-tracts 5, 6, 11, and 12 at the 3′-end of the G-rich region and contains a 26-base middle loop. Surprisingly, the latter G-quadruplex is more stable than the former and provides the overall stability of the tandem G-quadruplex structure formed in the full-length G-rich sequence of the hTERT core promoter.
Figure 7.
The proposed model of the tandem G-quadruplex structure formed in the hTERT core promoter. The two G-quadruplexes formed by G-tracts 1–4 (G4-1:3:1) and G-tracts 5, 6, 11, and 12 (G4-3:26:1) are linked by the sequence C19–G25. The hTERT promoter sequence, with the Sp1 binding sites indicated, is shown below the proposed model. G-tracts involved in the G-quadruplex structures are shown in red and numbered.
The hTERT G-quadruplex structures in the full-length G-rich sequence are stabilized by the G-quadruplex-interactive compounds TMPyP4 and telomestatin
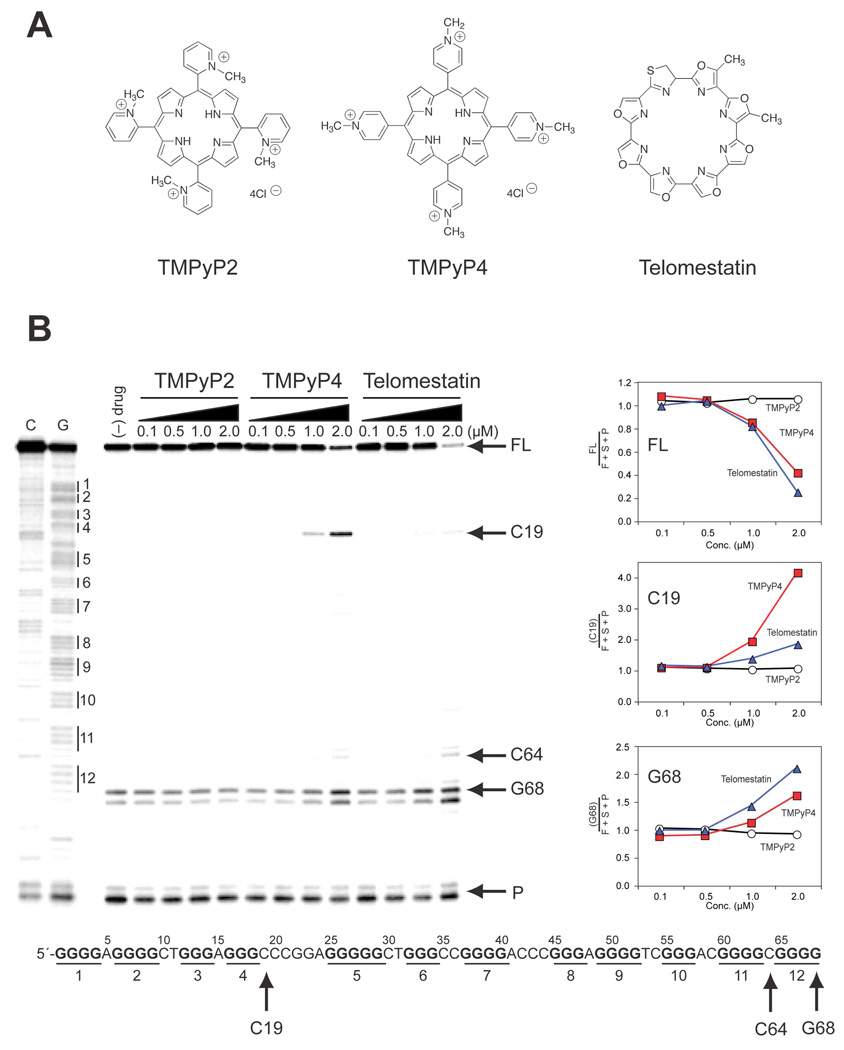
The unique structure of the tandem G-quadruplexes found in the hTERT promoter could provide sites that are selectively recognized by G-quadruplex-interactive compounds. For this reason, we examined if the hTERT G-quadruplex structure is stabilized by G-quadruplex-interactive ligands in a ligand-specific manner. A Taq polymerase arrest assay was performed with two well-known G-quadruplex ligands, the cationic porphyrin TMPyP4 and the antitumor antibiotic telomestatin. TMPyP2, a positional isomer of TMPyP4 that lacks the ability to interact with a G-quadruplex structure, was also included in the assay as a negative control compound (Figure 7A). A similar decrease in the full-length product was observed with increasing concentrations of TMPyP4 and telomestatin (Figure 7B). However, as expected, TMPyP2 did not change the amount of full-length or premature primer extension products (Figure 7B). One notable observation is that the amount of premature product at C19 was selectively increased by TMPyP4. These data suggest that TMPyP4 binds between the two tandem hTERT G-quadruplexes, while telomestatin preferentially recognizes the external tetrads of the tandem structure (compare middle and lower line graphs in Figure 7B). A minor premature primer extension product appeared at C64 at high concentrations of the ligands, indicating that TMPyP4 and telomestatin may also interact with minor G-quadruplexes formed in the full-length hTERT G-rich sequence.
DISCUSSION
Activation of telomerase is a critical step for immortalization of cancerous cells during tumorigenesis [3–5]. The catalytic subunit of telomerase (hTERT) is the rate-limiting factor of telomerase activity [1] and is expressed in about 90% of cancers, but not in normal somatic cells [2]. Due to its cancer-specific expression and significant role in cancer development, hTERT has been an attractive target for the development of therapeutics to treat a broad spectrum of cancers. The two major therapeutic approaches are immunotherapy that uses hTERT as a cancer-specific antigen to discriminately kill hTERT-positive cancer cells [30–33] and the use of antisense oligonucleotides to directly target expression of hTERT [7,9–13].
In this study we investigated the putative G-quadruplex-forming sequence in the hTERT core promoter containing twelve consecutive G-tracts (Figure 1). We demonstrated that the hTERT G-rich region forms a tandem G-quadruplex structure. One of the G-quadruplexes is formed by G-tracts 1–4 and has a more conventional 1:3:1 loop pattern, which is known to form a stable folding pattern (Figure 7A). In contrast, the other G-quadruplex, formed from G-tracts 5, 6, 11, and 12, has an unusual 2:26:1 or 3:26:1 pattern, with a total number of 29 or 30 bases in the loops (Figure 7A). Studies on the two individual G-quadruplexes revealed that the G-quadruplex formed by G-tracts 5, 6, 11, and 12 is responsible for the overall stability of the G-quadruplex structure formed in the full-length hTERT G-rich sequence (insets in Figure 2B and Figure 5A). The importance of loop size to G-quadruplex stability has been reported in several studies, demonstrating a major effect on thermal stability for total loop lengths of up to five nucleotides in three-tetrad G-quadruplexes [24–26]. Also, where there are more than four consecutive G-tracts (e.g., in the promoters of c-myc, bcl-2, HIF-1α, VEGF, and mouse KRAS), the naturally occurring G-quadruplexes have the propensity to contain fewer bases in the loops by employing four contiguous G-tracts for G-quadruplex formation [28,34–37,38]. In contrast, the G4-3:26:1 places G-tracts 7–10 in the middle loop, resulting in the formation of an unconventional G-quadruplex structure with a 26-base middle loop (Figure 4D and Figure 5C). This hTERT G-quadruplex is the first example that has been shown to be so stable in the presence of such a long loop. Furthermore, the slight DMS protection of G-tracts 7 and 9 of this middle loop implies the formation of base-pairing (Figure 4D and Figure 5C), which supports the formation of a loosely organized secondary DNA structure in the loop, most likely a hairpin structure, and therefore provides a possible explanation for the formation of a stable G-quadruplex even in the presence of 26 bases in the loop.
The mutational studies demonstrated that even apparently minor changes (e.g., a G-to-C transversion) in the stem of the hairpin produce significant changes in the location of guanine tracts in the stem versus those found in the G-quadruplex. While this was not expected, upon examination of the resulting folding patterns, these changes can be rationalized. For example, in the 7′/9(CG→GC) mutant the proposed folding pattern retains the two lateral loops and the one-base double-chain reversal loop found in the wild-type sequence, but the G-quadruplex faces upon which these loops are found are different [compare wild-type and 7′/9(CG→GC) in Figure 6A]. In both the wild-type and G-to-C transversion mutant, base pairing in the hairpin loop is possible. Overall, these changes are consistent with the minor differences found in Tm and CD results (see Figure 6, B and C). While it is clear that the guanine tracts in the G-quadruplex are important for the folding pattern, the precise relative positions of the G-tracts in the hairpin also determine the overall structure and folding pattern of this unusual G-quadruplex. This may be important in recognition of proteins involved in stabilizing or resolving these structures.
Comparison of the hTERT G-quadruplex-forming sequences (G-tracts 1–4 and 5– 12) with previously characterized G-quadruplex-forming sequences reveals a similarity between the hTERT G4-1:3:1 and the parallel G-quadruplexes (including the NMR-determined parallel G-quadruplex of c-myc [39]) and between the hTERT G4-3:26:1 (G-tracts 5–12) and the mixed parallel/antiparallel G-quadruplex of bcl-2 defined by NMR (Figure 7A) [40]. CD analysis and DMS footprinting on the full-length hTERT G-rich sequence and two individual G-quadruplex-forming sequences support that the G4-1:3:1 is a parallel G-quadruplex with three double-chain-reversal loops, and that G4-3:26:1 is a mixed parallel/antiparallel G-quadruplex with a 3:26:1 loop conformation (Figure 7). The identical number of bases in the first and third loops between G4-3:26:1 and the bcl-2 G-quadruplex (3:7:1 loop conformation) (Figure 8A) suggests that the hTERT G-quadruplex folding pattern may be similar to the bcl-2 G-quadruplex, in which the first and middle loops are lateral and the third loop is a double-chain reversal [29,40]. However, we recognize that relying solely on comparative CD and DMS footprinting patterns to conclude topologies of G-quadruplexes can be misleading, and NMR studies will be needed to confirm these predictions.
Figure 8.
Stabilization of the hTERT G-quadruplex structure by G-quadruplex ligands. (A) Structures of TMPyP2, TMPyP4, and telomestatin. (B) A Taq polymerase stop assay was performed on ODN XFL with increasing concentrations of TMPyP4 and telomestatin, as well as the control compound TMPyP2. The polymerase stop assay was carried out as described in Materials and Methods. The stop sites at the primer (P), the G-quadruplex drug-stabilized sites (G68, C64, and C19), and the full-length DNA (FL) are shown to the right of the gel. All the samples, including the (–) drug sample, contain 40 mM KCl. Quantification of the relative stop sites at FL, C19, and G68 for the three compounds from the autoradiogram is shown to the right of the gel. The FL sequence with the determined drug-stabilized sites is shown at the bottom of the gel.
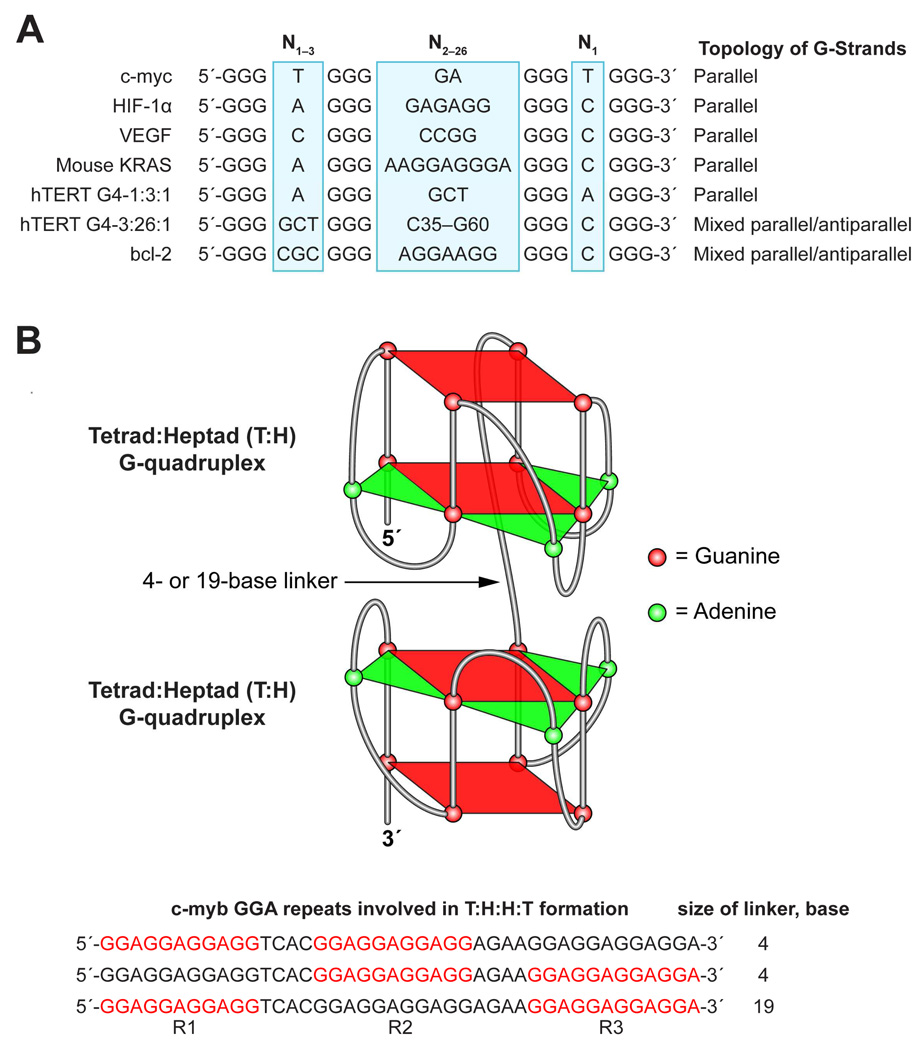
There are several lines of evidence that suggest an interaction between the two hTERT G-quadruplexes, which are located 7 or 8 bases apart, including Tm, CD spectra, and Taq polymerase arrest assay on the full-length hTERT G-rich sequence. The presence of a single phase in the melting curve of the full-length sequence, rather than two phases, which would represent two non-interacting G-quadruplexes (Figure 4A), together with an increase in the molar ellipticity of the two G-quadruplexes when in a single oligomer, suggests an interaction between the two G-quadruplexes (compare Figure 4A, insert, and Figure 5A, insert). Previously, interaction of tandem G-quadruplexes was found in the G-quadruplex structures formed in the GGA repeat region of the c-myb promoter, in which two unstable tetrad:heptad (T:H) G-quadruplexes, positioned 4 or 19 bases apart, intramolecularly dimerize to form a very stable T:H:H:T G-quadruplex structure (Figure 9B) [28]. The dimerization of the two T:H G-quadruplexes that are 19 bases apart suggests the potential of the two hTERT G-quadruplexes to interact with each other, and the interface between the two hTERT G-quadruplexes can serve as a unique drug recognition site (Figure 7). Although not a promoter G-quadruplex, human telomeric quadruplexes have been proposed to form dimeric or higher number end-to-end stacked compact stacking structures in the form of both parallel [41] or hybrid-type telomeric structures [42,43]. In both cases the interface between the end-to-end stacking of G-quadruplexes could provide small molecule binding sites.
Figure 9.
(A) Comparison of G-quadruplex-forming sequences found in selected gene promoters [38] and the hTERT G4-1:3:1 and G4-3:26:1 determined in this study. The proposed topology is shown on the right. (B) The topology of the dimer (T:H:H:T) G-quadruplex formed in the c-myb promoter [28]. The sequences shown at the bottom of the figure show the c-myb GGA repeat region with the three combinations of T:H:H:Ts, each using two copies of (GGA)4. The (GGA)4 copies involved in the formation are shown in red.
G-quadruplex-interactive ligands have been reported to inhibit telomerase activity, which causes telomere shortening [44–55]. The main mechanism for this inhibition is presumed to be due to the stabilization of telomeric G-quadruplexes by the ligands, which inhibits the access of telomerase to telomeres [44–55]. Although one might expect growth inhibition by this mechanism to have a long lag phase, due to the requirements for progressive telomere shortening over many cell replications, growth inhibition is observed much earlier than when telomere shortening can be detected in the cells [51,52]. One plausible mechanism for this early response is the disruption of the telomere-protective protein complex by the interaction of G-quadruplex ligands with telomeric G-quadruplexes [56–59], and indeed anaphase bridges have been shown to occur within 24–48 hours [60]. The data presented in this study allow us to propose an additional mechanism that may explain some of the anticancer effects caused by G-quadruplex ligands. We have demonstrated that the G-quadruplex ligands (Figure 7) TMPyP4 and telomestatin can target and stabilize the hTERT G-quadruplex structure, which suggests an alternative mechanism of action for these compounds. The stabilization of the hTERT G-quadruplex structure by these ligands would cause downregulation of hTERT expression due to the disruption of Sp1 binding sites in the core promoter and subsequently result in telomere shortening in the cell. Significantly, the results from previous hTERT antisense oligonucleotide studies, in which hTERT expression is specifically downregulated, are very similar to those from previous studies on the role of telomeric G-quadruplexes and G-quadruplex ligands [9–12]. In both cases, early response growth inhibition and delayed telomere shortening were observed, which suggests that growth inhibition and telomere shortening by G-quadruplex ligands may be due to downregulation of hTERT expression. In at least three distinct cases, TMPyP4, ligand 12459, and BRACO-19 downregulation of hTERT has been observed [53,60,61]. For TMPYP4 a 60% downregulation of hTERT mRNA was detected after 12 h and persisted up to 48 h [53]. However, this was rationalized, at least in the case of TMPyP4, to be due to the stabilization of a G-quadruplex in the c-myc promoter by G-quadruplex ligands, which is known to silence c-myc expression [53,62,63]. For BRACO-19 an almost complete lack of protein expression of the catalytic subunit of telomerase, hTERT was found in vitro and in vivo. In this case the absence was rationalized by ubiquitin-mediated response following BRACO-19 stabilizing the G-quadruplex formation at the 3′ telomeric DNA overhang and displacement of hTERT from the capping structure [61]. Last, in the case of ligand 12459 a novel mechanism involving an hTERT RNA alternative splicing caused by 12459 stabilization of G-quadruplex located in the hTERT intron 6. Significantly because of the different G-quadruplex selectivities BRACO-19 and telomestatin did not produce the same effects on hTERT splicing. Therefore, in view of the results reported here, direct targeting of the hTERT G-quadruplex structure in the core promoter may be the major contributor in downregulation of hTERT expression. G-quadruplex-interactive compounds that selectively target the G-quadruplexes in the promoters of hTERT and c-myc and the telomeric G-quadruplex will be required to separate the cellular consequences of targeting the different structure. In view of the observations reported here and those reported by Gomez et al. [60] it may be important to compare the binding of telomere-interactive compounds with those G-quadruplex-forming sequences found in the hTERT promoter and the hTERT intron 6 as well as measure effect on hTERT mRNA expression and its splicing variants. Meanwhile, it may be advantageous to have multi-targeted G-quadruplex-interactive compounds.
In conclusion, we provide evidence for the formation of a unique G-quadruplex structure in the hTERT core promoter, in which the mixed parallel/antiparallel G-quadruplex formed by G-tracts 5, 6, 11, and 12 (G4-3:26:1), with a hairpin structure of G-tracts 7–10, stacks on the parallel G-quadruplex formed by G-tracts 1–4 (G4-1:3:1), and furthermore we propose the hTERT G-quadruplex structure as a possible target of G-quadruplex ligands in telomere shortening caused by these ligands.
Supplementary Material
CD spectra of ODNs not shown in Figure 2 and original gels from Figure 4C. This material is available free of charge via the Internet at http://pubs.acs.org.
ACKNOWLEDGEMENT
This research was supported by grants from the National Institutes of Health (CA94166) and the Arizona Biomedical Research Commission (0021). We would like to thank Daekyu Sun for his scientific insights on the project, and we are grateful to David Bishop for preparing, proofreading, and editing the final version of the manuscript and figures.
REFERENCES
- 1.Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. Dev. Genet. 1996;18:173. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 2.Dhaene K, Van Marck E, Parwaresch R. Virchows Arch. 2000;437:1. doi: 10.1007/s004280000189. [DOI] [PubMed] [Google Scholar]
- 3.Miyazu YM, Miyazawa T, Hiyama K, Kurimoto N, Iwamoto Y, Matsuura H, Kanoh K, Kohno N, Nishiyama M, Hiyama E. Cancer Res. 2005;65:9623. doi: 10.1158/0008-5472.CAN-05-0976. [DOI] [PubMed] [Google Scholar]
- 4.Sampedro Camarena F, Cano Serral G, Sampedro Santaló F. Clin. Transl. Oncol. 2007;9:145. doi: 10.1007/s12094-007-0028-1. [DOI] [PubMed] [Google Scholar]
- 5.Harley CB, Kim NW. Important Adv. Oncol. 1996:57. [PubMed] [Google Scholar]
- 6.Nakamura M, Masutomi K, Kyo S, Hashimoto M, Maida Y, Kanaya T, Tanaka M, Hahn WC, Inoue M. Hum. Gene Ther. 2005;16:859. doi: 10.1089/hum.2005.16.859. [DOI] [PubMed] [Google Scholar]
- 7.Ji XM, Xie CH, Fang MH, Zhou FX, Zhang WJ, Zhang MS, Zhou YF. Acta Pharmacol. Sin. 2006;27:1185. doi: 10.1111/j.1745-7254.2006.00417.x. [DOI] [PubMed] [Google Scholar]
- 8.Parsch D, Brassat U, Brummendorf TH, Fellenberg J. Cancer Invest. 2008;26:590. doi: 10.1080/07357900802072905. [DOI] [PubMed] [Google Scholar]
- 9.Kraemer K, Fuessel S, Schmidt U, Kotzsch M, Schwenzer B, Wirth MP, Meye A. Clin. Cancer Res. 2003;9:3794. [PubMed] [Google Scholar]
- 10.Teng L, Specht MC, Barden CB, Fahey TJ., 3rd J. Clin. Endocrinol. Metab. 2003;88:1362. doi: 10.1210/jc.2002-021222. [DOI] [PubMed] [Google Scholar]
- 11.Fu XH, Zhang JS, Zhang N, Zhang YD. World J. Gastroenterol. 2005;11:785. doi: 10.3748/wjg.v11.i6.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Folini M, Brambilla C, Villa R, Gandellini P, Vignati S, Paduano F, Daidone MG, Zaffaroni N. Eur. J. Cancer. 2005;41:624. doi: 10.1016/j.ejca.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Kraemer K, Fuessel S, Kotzsch M, Ning S, Schmidt U, Wirth MP, Meye A. J. Urol. 2004;172:2023. doi: 10.1097/01.ju.0000138157.46464.6e. [DOI] [PubMed] [Google Scholar]
- 14.Kraemer K, Fuessel S, Meye A. Methods Mol. Biol. 2007;405:9. doi: 10.1007/978-1-60327-070-0_2. [DOI] [PubMed] [Google Scholar]
- 15.Takakura M, Kyo S, Kanaya T, Hirano H, Takeda J, Yutsudo M, Inoue M. Cancer Res. 1999;59:551. [PubMed] [Google Scholar]
- 16.Horikawa I, Cable PL, Afshari C, Barrett JC. Cancer Res. 1999;59:826. [PubMed] [Google Scholar]
- 17.Cong YS, Wen J, Bacchetti S. Hum. Mol. Genet. 1999;8:137. doi: 10.1093/hmg/8.1.137. [DOI] [PubMed] [Google Scholar]
- 18.Kyo S, Takakura M, Taira T, Kanaya T, Itoh H, Yutsudo M, Ariga H, Inoue M. Nucleic Acids Res. 2000;28:669. doi: 10.1093/nar/28.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grandori C, Cowley SM, James LP, Eisenman RN. Annu. Rev. Cell Dev. Biol. 2000;16:653. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 20.Yagi K, Furuhashi M, Aoki H, Goto D, Kuwano H, Sugamura K, Miyazono K, Kato M. J. Biol. Chem. 2002;277:854. doi: 10.1074/jbc.M104170200. [DOI] [PubMed] [Google Scholar]
- 21.Chen CR, Kang Y, Massague J. Proc. Natl. Acad. Sci. U.S.A. 2001;98:992. doi: 10.1073/pnas.98.3.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu D, Wang Q, Gruber A, Bjorkholm M, Chen Z, Zaid A, Selivanova G, Peterson C, Wiman KG, Pisa P. Oncogene. 2000;19:5123. doi: 10.1038/sj.onc.1203890. [DOI] [PubMed] [Google Scholar]
- 23.Zhou C, Liu J. Biochem. Biophys. Res. Commun. 2003;303:130. doi: 10.1016/s0006-291x(03)00318-8. [DOI] [PubMed] [Google Scholar]
- 24.Hazel P, Huppert J, Balasubramanian S, Neidle S. J. Am. Chem. Soc. 2004;126:16405. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 25.Risitano A, Fox KR. Nucleic Acids Res. 2004;32:2598. doi: 10.1093/nar/gkh598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bugaut A, Balasubramanian S. Biochemistry. 2008;47:689. doi: 10.1021/bi701873c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walsh K, Gualberto A. J. Biol. Chem. 1992;267:13714. [PubMed] [Google Scholar]
- 28.Palumbo SL, Memmott RM, Uribe DJ, Krotova-Khan Y, Hurley LH, Ebbinghaus SW. Nucleic Acids Res. 2008;36:1755. doi: 10.1093/nar/gkm1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dexheimer TS, Sun D, Hurley LH. J. Am. Chem. Soc. 2006;128:5404. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel KP, Vonderheide RH. Cytotechnology. 2004;45:91. doi: 10.1007/s10616-004-5132-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beatty GL, Vonderheide RH. Expert Rev. Vaccines. 2008;7:881. doi: 10.1586/14760584.7.7.881. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Katik I, Liu JP. Methods Mol. Biol. 2007;405:61. doi: 10.1007/978-1-60327-070-0_7. [DOI] [PubMed] [Google Scholar]
- 33.Wenandy L, Sorensen RB, Sengelov L, Svane IM, thor Straten P, Andersen MH. Clin. Cancer Res. 2008;14:4. doi: 10.1158/1078-0432.CCR-07-4590. [DOI] [PubMed] [Google Scholar]
- 34.De Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW. Biochemistry. 2005;44:16341. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 35.Guo K, Gokhale V, Hurley LH, Sun D. Nucleic Acids Res. 2008;36:4598. doi: 10.1093/nar/gkn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phan AT, Modi YS, Patel DJ. J. Am. Chem. Soc. 2004;126:8710. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cogoi S, Xodo LE. Nucleic Acids Res. 2006;34:2536. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin Y, Hurley LH. Biochimie. 2008;90:1149. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Biochemistry. 2005;44:2048. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 40.Dai J, Chen D, Jones RA, Hurley LH, Yang D. Nucleic Acids Res. 2006;34:5133. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haider SM, Neidle S. Biochem. Soc. Trans. 2009;37:583. doi: 10.1042/BST0370583. [DOI] [PubMed] [Google Scholar]
- 42.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Nucleic Acids Res. 2006;34:2723. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petraccone L, Trent JO, Chaires JB. J. Am. Chem. Soc. 2008;120:16530. doi: 10.1021/ja8075567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun D, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, Jenkins TC, Neidle S, Hurley LH. J. Med. Chem. 1997;40:2113. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 45.Koeppel F, Riou JF, Laoui A, Mailliet P, Arimondo PB, Labit D, Petitgenet O, Hélène C, Mergny JL. Nucleic Acids Res. 2001;29:1087. doi: 10.1093/nar/29.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shin-ya K, Wierzba K, Matsuo K, Ohtani T, Yamada Y, Furihata K, Hayakawa Y, Seto HJ. J. Am. Chem. Soc. 2001;123:1262. doi: 10.1021/ja005780q. [DOI] [PubMed] [Google Scholar]
- 47.Shammas MA, Shmookler Reis, R. J., Li C, Koley H, Hurley LH, Anderson KC, Munshi NC. Clin. Cancer Res. 2004;10:770. doi: 10.1158/1078-0432.ccr-0793-03. [DOI] [PubMed] [Google Scholar]
- 48.Kelland LR. Eur. J. Cancer. 2005;41:971. doi: 10.1016/j.ejca.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 49.Burger AM, Dai F, Schultes CM, Reszka AP, Moore MJ, Double JA, Neidle S. Cancer Res. 2005;65:1489. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
- 50.Reed JE, Arnal AA, Neidle S, Vilar R. J. Am. Chem. Soc. 2006;128:5992. doi: 10.1021/ja058509n. [DOI] [PubMed] [Google Scholar]
- 51.Gunaratnam M, Greciano O, Martins C, Reszka AP, Schultes CM, Morjani H, Riou JF, Neidle S. Biochem. Pharmacol. 2007;74:679. doi: 10.1016/j.bcp.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 52.Gowan SM, Harrison JR, Patterson L, Valenti M, Read MA, Neidle S, Kelland LR. Mol. Pharmacol. 2002;61:1154. doi: 10.1124/mol.61.5.1154. [DOI] [PubMed] [Google Scholar]
- 53.Grand CL, Han H, Muñoz RM, Weitman S, Von Hoff DD, Hurley LH, Bearss D. J. Mol. Cancer Ther. 2002;1:565. [PubMed] [Google Scholar]
- 54.Tauchi T, Shin-Ya K, Sashida G, Sumi M, Nakajima A, Shimamoto T, Ohyashiki JH, Ohyashiki K. Oncogene. 2003;22:5338. doi: 10.1038/sj.onc.1206833. [DOI] [PubMed] [Google Scholar]
- 55.Tauchi T, Shin-ya K, Sashida G, Sumi M, Okabe S, Ohyashiki JH, Ohyashiki K. Oncogene. 2006;25:5719. doi: 10.1038/sj.onc.1209577. [DOI] [PubMed] [Google Scholar]
- 56.Veldman T, Etheridge KT, Counter CM. Curr. Biol. 2004;14:2264. doi: 10.1016/j.cub.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 57.Zaug AJ, Podell ER, Cech TR. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10864. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gomez D, O’Donohue MF, Wenner T, Douarre C, Macadre J, Koebel P, Giraud-Panis MJ, Kaplan H, Kolkes A, Shin-ya K, Riou JF. Cancer Res. 2006;66:6908. doi: 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- 59.Tahara H, Shin-Ya K, Seimiya H, Yamada H, Tsuruo T, Ide T. Oncogene. 2006;25:1955. doi: 10.1038/sj.onc.1209217. [DOI] [PubMed] [Google Scholar]
- 60.Gomez D, Lemarteleur T, Lacroix L, Mailliet P, Mergny J-L, Riou J-F. Nucleic Acids Res. 2004;32:371. doi: 10.1093/nar/gkh181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burger AM, Dai F, Schultes CM, Reszka AP, Moore MJ, Double JA, Neidle S. Cancer Res. 2005;65:1489. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
- 62.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11593. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mikami-Terao Y, Akiyama M, Yuza Y, Yanagisawa T, Yamada O, Yamada H. Cancer Lett. 2008;261:226. doi: 10.1016/j.canlet.2007.11.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CD spectra of ODNs not shown in Figure 2 and original gels from Figure 4C. This material is available free of charge via the Internet at http://pubs.acs.org.