Abstract
Background
Disease pathogenesis may result from genetic alterations and/or a more diverse group of epigenetic changes. While events such as DNA methylation are well established, there is significant interest in nucleosome remodeling, RNA interference and histone modifications, as mechanisms that underlie epigenetic effects. While genetic mutations are permanent, epigenetic changes can be transitory. The potential to reverse epigenetic changes has led to the development of therapeutic strategies targeting chromatin-modifying enzymes.
Objective
To review the roles of chromatin-modifying enzymes in gene regulation and to highlight their potentials as therapeutic targets.
Methods
This review is based on recently published literature and online resources.
Results/conclusion
This paper focuses on enzymes responsible for histone acetylation, deacetylation, methylation and demethylation, and their potential as targets for epigenetic therapies. A subsequent paper will do the same for enzymes responsible for histone phosphorylation, ubiquitylation, SUMOylation and poly-ADP-ribosylation as well as ATP-dependent nucleosome remodeling.
Keywords: chromatin, chromatin-modifying enzymes, demethylation, epigenetics, histone acetylation, histone deacetylation, histones, methylation
1. Introduction
Eukaryotic DNA is condensed into chromatin with the core histone proteins, H2A, H2B, H3 and H4. Each histone octamer, consisting of one H3–H4 tetramer and two H2A–H2B dimers, is blanketed with ∼ 146 base pairs of DNA to form a nucleosome [1]. Each core histone can be divided into three segments: a basic N-terminal tail, a histone fold and a C-terminal tail, all of which are subject to various levels of covalent modification such as acetylation, methylation, phosphorylation, ADP ribosylation, ubiquitylation, SUMOylation, biotinylation, carbonylation and glycosylation. However, the positively charged N-terminal tails are the most highly modified as they project through the major and minor grooves formed by the DNA helix [2]. Specific patterns of histone modifications may greatly extend the informative potential of the genetic code into what is often referred to as the ‘histone code’ [2]. An additional histone, H1, is bound to the DNA that links nucleosomes together [3]. The process of DNA condensation and compaction, which is required to efficiently store the DNA in the nucleus, is mediated in part by H1 and the N-terminal tails of the core histones [4,5]. Finally, histone modifications are selectively recognized by specific domains of various chromatin-associated proteins [6]. For example, bromo domains and chromo domains, identified within histone acetyl transferases and lysine methyl transferases, recognize acetylated lysines and methylated lysines, respectively [7,8].
Chromatin regulation is a highly dynamic process and chromatin accessibility is strictly supervised by chromatin-modifying machinery [9]. Depending on the panoply of potential histone modifications a variety of multi-protein complexes will be recruited, thus allowing for tightly regulated transcriptional control. In addition, these enzymes can be triggered to act either locally, recruited by specific transacting factors to individual nucleosomes, or globally, targeting almost all nucleosomes throughout the genome.
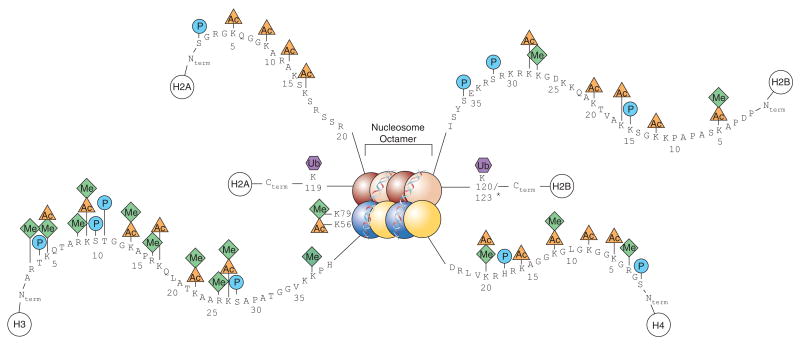
While the chromatin modification pattern that any given nucleosome can adopt may give rise to distinct transcriptional responses, there are a handful of histone- and residue-specific modifications that are generally associated with either transcriptionally active or inactive chromatin. For example, transcriptionally repressed chromatin, or heterochromatin, is densely packed, hypoacetylated and often methylated at lysine 9 on histone H3 and at lysine 20 on histone H4 [10], whereas transcriptionally active chromatin, or euchromatin, is hyperacetylated, namely at lysine 9 and 14, and methylated at lysine 4 and lysine 79 on histone H3 (Figure 1) [11,12]. Typically, modifications of the N-terminal tail on histone H3, which extends further from the nucleosome core than any other histone tail, are associated with regulatory control. Additionally, the N-terminal tail of histone H4 has been shown to be required for chromatin fibre folding [13].
Figure 1. Mammalian core histone modifications.
N- and C-terminal histone tails extend from the globular domains of histones H2A, H2B, H3 and H4. DNA is wrapped around the nucleosome octamer made up of two H2A–H2B dimers (red and pink) and a H3–H4 tetramer (blue and yellow). Post-translational covalent modifications include acetylation (orange triangles), methylation (green diamonds), phosphorylation (blue circles) and ubiquitylation (purple hexagons). Human histone tail amino acid sequences are shown. Corresponding modified residue positions within yeast histones are indicated by asterisks. Lysine positions 56 and 79 on histone H3 are located within the globular domain of the histone.
In many cancers and other diseases, epigenetic mechanisms, working in concert with genetic alterations, may alter histone modifications, sending transcriptional control into disarray. Epigenetic mechanisms of transcriptional control, unlike genetic changes, include changes in gene expression that occur independently of any change in DNA sequence integrity [14]. A number of cellular processes are affected by epigenetic dysregulation including cell growth, proliferation, differentiation, DNA repair, cell cycle control and apoptosis. Therefore, from a therapeutic standpoint, targeting the reversible epigenetic changes may be a more attractive approach than targeting the more stable genetic mutations. In fact, certain cancers, for example, display specific histone modification profiles. Recent studies have shown that specific histone modifications can serve as markers for cancer prognoses [15] and can even be used, with some success, to predict clinical outcome and probability of relapse [16].
Since the discovery of the link between disease pathogenesis and histone modifications, a number of therapies are being developed targeting the various histone modifying enzymes responsible for regulating these modifications. These enzymes include, but are not limited to, histone acetyl transferases (HAT), histone deacetylases (HDAC), histone methyl transferases (HMT), histone demethylases (HDM), kinases, E3 ubiquitin ligases, small ubiquitin-related modifier (SUMO)-cojugating enzymes and ADP-ribosyl transferases (ADPRT). Due to the vast amount of research being done in the field [17-19], we have a rapidly evolving description of how acetylases/deacetylases and methylases/demethylases contribute to precise transcriptional control. This paper will serve as an up-to-date review of these cellular mechanisms, briefly discussing recently identified inhibitors of these enzymes and focusing on new advances in HDAC/HAT and HMT/HDM therapies. The second and final part of this review will focus on ATP-dependent nucleosome remodeling and histone modifications such as phosphorylation, SUMOylation, ubiquitylation and ADP ribosylation and the potential of their respective enzymes as therapeutic targets.
2. HATs
Histone acetylation involves the transfer of the acetyl group from acetyl Coenzyme A to the imino group of lysine. This modification neutralizes the positive charge of lysine, which may disrupt the interaction between DNA and the histone tails, allowing for an open chromatin conformation. The newly acetylated lysine can then be recognized by specific bromo domain-containing proteins, leading to a cascade of additional modifications often culminating in increased transcriptional activity. In opposition to acetylation, deacetylation limits DNA accessibility by revealing the positive charge on lysine, thus allowing for histone tail-DNA interaction and chromatin compaction. Additional condensation is believed to occur as the N-terminal tail of histone H4 interacts with the negatively charged H2A–H2B dimer from an adjacent nucleosome [20].
HATs are categorized into two groups based on their cellular localization. Type A HATs are nuclear and acetylate nucleosomal histones and other chromatin-associated proteins, whereas type B HATs exist in the cytoplasm to acetylate newly synthesized histones. Unlike type A, type B HATs have no direct influence on transcription. Group A HATs may be further grouped into five different families; GNAT (GCN5 (general control of nuclear-5)-related N-acetyl transferases), MYST (MOZ (monocytic leukaemia zinc-finger protein), YBF2 (yeast binding factor 2)/SAS3 (something about silencing 3), SAS2, TIP60 (Tat interactive protein-60)), p300/CBP (CREB (cAMP response element binding protein)-binding protein), nuclear receptor co-activators and general transcription factors (Table 1).
Table 1.
Histone acetyl transferase (HAT) families and specificities.
| HATs | Histone substrate specificity |
|---|---|
| GNAT family | |
| GCN5 | H2B; H3-K9, -K14, -K18, -K23, -K27; H4-K8, -K16 |
| PCAF | H3-K14; H4-K8 |
| MYST family | |
| TIP60 | H2A-K5; H3-K14; H4-K5, -K8, -K12 |
| MOZ | H3; H4 |
| p300/CBP family | |
| p300 | H2A-K5; H2B-K12, -K15, -K20; H3-K14, -K18, -K23; H4-K5, -K8, -K12 |
| CBP | H2A-K5; H2B-K12, -K15, -K20; H3-K14, -K18, -K23; H4-K5, -K8 |
| Nuclear receptor coactivators | |
| AIB1 | H3; H4 |
| General transcription factors | |
| TAF‖250 | H3; H4 |
This table highlights only those HATs discussed in the text and is not fully inclusive of all existing HATs.
AIB1: Amplified in breast cancer-1; CBP: CREB (cAMP response element binding protein)-binding protein; GCN5: General control of nuclear-5; GNAT: GCN5-related N-acetyl transferase; MOZ: Monocytic leukaemia zinc-finger protein; MYST: MOZ, YBF2/SAS3, SAS2, TIP60; PCAF: p300/CBP-associated factor; SAS: Something about silencing; TAF‖250: TATA-box binding protein-associated factor-‖250; TIP60: Tat interactive protein-60; YBF2: Yeast binding factor 2.
Due to their involvement in so many cellular regulation pathways, it has been found that genes encoding a number of HATs are mutated and/or their protein expression levels and activities are altered in many diseases, especially in a variety of cancers. For example, GCN5 dysregulation is found in genetic diseases and cancers and p300 and p300/CBP associated factor (PCAF) are linked to cancer due to their roles in myogenic differentiation (MyoD)-dependent cell cycle arrest [21]. In addition, some HATs, such as p300 and CBP, have been characterized as tumour suppressors and act as transcriptional cofactors for a variety of oncoproteins (p53, pRB, Myb, Jun, Fos) [22]. p300 was first implicated in oncogenic pathways when it was originally identified as an adenovirus E1A oncoprotein binding partner [23]. Not surprisingly, therefore, mutations in the gene encoding p300 are found in breast, colorectal, gastric and epithelial cancers. Glioblastoma and other malignancies can also arise as a result of loss of heterozygosity at the p300 locus [24]. CBP mutations rendering the HAT enzymatically inactive are seen in leukaemogenesis and Rubenstein–Taybi syndrome, a developmental disorder associated with childhood cancers of neural crest origin [25]. Loss of CBP function, caused by mutated polyglutamine proteins, is also found in Huntington's disease, Alzheimer's disease, amytropic lateral schlerosis and spinal and bulbar muscular atrophy [26]. In the MYST family of HATs, Tip60 mutations nullifying acetyl transferase activity disable the cell's DNA double strand break repair system. The resulting cells lose their ability to apoptose, even after gamma-irradiation, thus allowing for rampant proliferation of DNA-damaged cells [27]. Nuclear receptor co-activators possessing HAT activity, such as amplified in breast cancer-1 (AIB1), have also been linked to cancer. AIB1, a member of the steroid receptor coactivator-1 (SRC-1) family of transcriptional coactivators, is amplified and overexpressed in a number of estrogen receptor-positive breast and ovarian cancer cell lines [28]. HAT regulation can also be interrupted as a result of chromosomal translocations. A classic example of such dysregulation is in acute myeloid leukaemia (AML), a hematological malignancy. In AML, a fusion protein is created in which CBP is fused with a MYST domain-containing MOZ [22,29]. This CBP–MOZ chimaera, possessing protein interacting domains from both the p300/CBP and MYST families of HATs, exhibits gain of function characteristics leading to unchecked hyperacetylation and aberrant transcriptional activation.
Despite the involvement of dysregulated HATs in some many disease states, only a modest number of HAT inhibitors have been reported. Initial inhibitors stemmed from the observation that polyamine-CoA conjugates inhibited acetyl transferase activity in cell extracts [30]. Two bisubstrate analogs specific for p300 and PCAF, Lys-CoA and H3-CoA-20, respectively, were synthesized and proved useful in blocking in vitro HAT activity, however, their low metabolic stability and cellular impermeability were significant drawbacks [31]. Naturally-occurring small-molecule HAT inhibitors have also been described. Anacardic acid [32] and garcinol [33] are p300 and PCAF inhibitors and curcumin [34] is a p300-selective inhibitor. Unfortunately anacardic acid, isolated from cashewnut shell liquid, and some synthetic amide derivatives displayed permeability problems in cell culture. On the other hand, garcinol, a polyisoprenylated benzophenone derivative from Garcinia indica fruit rind, was shown to be an equally active HAT inhibitor both in vitro and in vivo. Garcinol inhibited histone acetylation and induced apoptosis in HeLa cells, making it the first ever reported cell permeable HAT inhibitor [33]. Curcumin has also demonstrated therapeutic value as it has been shown to prevent hyperacetylation in cancers and exhibit anti-HIV activity, although its mechanism of action against HIV is not yet fully understood [34]. Additional synthetic HAT inhibitors possessing anticancer activity have been reviewed [17]. Although some of the more recently discovered small molecules have exhibited cell permeability and potencies that rival the natural products, a gold-standard HAT inhibitor has yet to be discovered.
Finally, HATs can also contribute to global levels of histone modifications by acetylating non-histone proteins. p300 and PCAF, for example, have been found to acetylate a number of transcriptional regulators, tumour suppressors, cell cycle regulators and other proteins such as p53, p73, E2F1, c-Jun, MyoD, Rb and HIV Tat, among others [35]. Like the histone code, this pattern of non-histone protein acetylation can be predictive of disease states. When targeting HATs in therapies, therefore, their roles in acetylating both histones and non-histone proteins must be considered.
3. HDACs
Unlike HAT inhibitors, however, much more progress has been made in the evolution of HDAC inhibitors. HDACs exist in multi-protein complexes displaying transcriptional repressing capabilities. To date, 18 HDACs have been identified in humans and are categorized into four distinct classes based on their homology to yeast HDACs (Table 2) (reviewed in detail in [17]). Classes I, II and IV have structurally similar, zinc-dependent active sites, whereas Class III, consisting of the sirtuins (SIRTs), is zinc-independent but requires the cofactor nicotinamide adenine dinucleotide (NAD). Class II HDACs, whose functions are regulated by class I HDACs, are also required for developmental stage transcriptional silencing and genomic organization, while class III enzymes have a specific role in gene silencing and serve in acetylation level maintenance [36]. The fourth class of HDACs, consisting solely of HDAC11, has just recently been proposed to have functions similar to those of class I.
Table 2.
Histone deacetylase (HDAC) classes, chelating metal and subcellular localization.
| HDACs | Chelation metal | Localization |
|---|---|---|
| Class I | ||
| HDAC1 | Zn2+ | Nucleus |
| HDAC2 | Zn2+ | Nucleus |
| HDAC3 | Zn2+ | Nucleus/cytoplasm |
| HDAC8 | Zn2+ | Nucleus/cytoplasm |
| Class IIa | ||
| HDAC4 | Zn2+ | Nucleus/cytoplasm |
| HDAC5 | Zn2+ | Nucleus/cytoplasm |
| HDAC7 | Zn2+ | Nucleus/cytoplasm |
| HDAC9 | Zn2+ | Nucleus/cytoplasm |
| Class IIb | ||
| HDAC6 | Zn2+ | Nucleus/cytoplasm |
| HDAC10 | Zn2+ | Nucleus/cytoplasm |
| Class III | ||
| SIRT1 | NAD+ | Nucleus/cytoplasm |
| SIRT2 | NAD+ | Nucleus/cytoplasm |
| SIRT3 | NAD+ | Mitochondria/nucleus |
| SIRT4 | NAD+ | Mitochondria |
| SIRT5 | NAD+ | Mitochondria |
| SIRT6 | NAD+ | Nucleus |
| SIRT7 | NAD+ | Nucleolus |
| Class IV | ||
| HDAC11 | Zn2+ | Nucleus |
NAD: Nicotinamide adenine dinucleotide; SIRT: Sirtuin; Zn: Zinc.
HDACs are known to associate with transcription factors, tumour suppressors and oncogenes to carry out normal cellular functions. HDAC1, for example, exists in complexes with the tumour suppressor retinoblastoma (Rb) and the E2F transcription factor. Rb recruits the deacetylases to help suppress E2F transcriptional activation leading to gene repression and cell cycle arrest [37]. The myocyte enhancer factor-2 (MEF2) transcription factor requires HDAC recruitment and activity in order to carry out its roles in neuronal resistance to excitotoxicity, Epstein-Barr virus (EBV) latency and transcriptional repression, among others [38,39]. SIRT1, a class III HDAC, regulates p53 function [40] and is intimately involved in the inflammatory response, fatty acid metabolism and stress-induced cellular defense and survival. It stands to reason, therefore, that HDAC dysregulation, like HAT dysregulation, is a common theme in a number of disease states. In acute promyelocytic leukaemia (APL), for example, the relationship between HDACs and the retinoic acid receptor (RAR) is altered [41]. In normal cells, in the absence of retinoic acid, RAR–retinoid X receptor (RXR) heterodimers recruit HDAC complexes in order to suppress target gene expression. Upon retinoic acid binding, the heterodimer releases the HDAC complexes in order to recruit the transcriptional activating complexes responsible for initiating haematopoietic differentiation. APL, caused by genetic translocations, is defined by fusions between the retinoic acid receptor (RAR) gene and either the promyelocytic leukaemia (PML) or promyelocytic zinc-finger (PLZF) genes, which produce RAR–PML or RAR–PLZF chimeric proteins. These chimaeras render leukaemic cells insensitive to physiological levels of retinoic acid, thus disallowing differentiation, and subsequently maintain the cells in a constant state of proliferation [42]. Similarly, in acute myelogenous leukaemia (AML), fusion of eight-twenty-one zinc-finger nuclear protein (ETO) to AML, typically a transcriptional activator, converts the protein to a dominant transcriptional repressor. The fusion protein's inability to release HDAC-containing corepressors and/or recruit co-activators blocks myeloid differentiation and aids leukaemic transformation [43]. HDAC2 overexpression is observed in familial-adenomatosis-polyposis-induced tumors [44] and a number of sporadic tumors lack HDAC2 activity due to a truncating mutation [45]. HDAC6 expression may be predictive of clinical outcome in breast cancer [46]. p21, a cyclin-dependent kinase inhibitor and putative tumour suppressor, is epigenetically inactivated in a number of tumours as a result of hypoacetylation at its gene promoter, allowing tumour cells to pass through G1 of the cell cycle unchecked [47].
The present list of HDAC inhibitors include both natural and synthetic compounds and are classified into five categories based on their chemical structures; short-chain fatty acids, benzamides, cyclic peptides, electrophilic ketones, and small-molecule hydroxamic-acid-derived compounds [18,48]. The presence of a metal-binding domain is a common property shared by all categories and serves to block substrate-Zn chelation at the active site [49]. These compounds are well-defined inhibitors of class I, II and IV HDACs. Class III HDACs, however, which require NAD+ as opposed to Zn2+ at their active sites, are unaffected by these types of HDAC inhibitors. Instead, class III HDACs are specifically inhibited by nicotinamide. Detailed descriptions of specific HDAC inhibitors, including their effectiveness in clinical trials, have been reviewed extensively and will therefore only be touched on here [17,18].
HDAC inhibitors have shown promise by inducing cell cycle growth arrest in G1 or G2/M, differentiation and apoptosis. p21 is one of the genes consistently upregulated in a p53-independent manner in response to HDAC inhibitors, a response necessary for cell cycle G1 arrest [47,50]. While this upregulation is partially the result of increased acetylation of histones H3 and H4 near the p21 promoter, some HDAC inhibitors, such as butyrate and trichostatin A, have been shown to stabilize p21 mRNA [51]. Cell cycle arrest in G1 is also induced by HDAC-inhibitor-mediated repression of cyclins A and D and activation of p16 and p27 [52,53]. Although the precise mechanisms of HDAC inhibitor-induced anticancer effects are not fully elucidated, HDAC inhibitors may be ten times more effective in cancerous cells than normal cells and can range in potency from IC50 values in the millimolar range for butyric acid to IC50 values in the nanomolar range for suberoylanilide hydroxamic acid (SAHA), which has been approved by the FDA for the treatment of refractory cutaneous T cell lymphomas (CTCL) [48,54]. This is not to say, however, that HDAC inhibitors cause genome-wide decondensation in tumour cells [47]. In fact, it has been reported that HDAC inhibitors, whose effects can be cell type specific, only affect 2 – 10% of all genes and can induce transcriptional activation or repression [55,56].
One of the more significant therapeutic realizations of late is that HDAC inhibitors can reach a higher potential when used as adjuvants to existing therapies [18,57]. In this way, HDAC inhibitors can increase the efficacy of other drugs by increasing target susceptibility. For example, in breast cancer therapy, the effectiveness of topoisomerase II inhibitors can be increased by pretreating with SAHA [58]. In addition, HDAC inhibitors have been used in combination with DNA demethylating agents in an attempt to reactivate silenced genes involved in tumour suppression. In leukaemia and breast cancer, fusion protein-induced differentiation inhibition caused by RAR–PML, RAR–PLZF or AML–ETO chimaeras can be thwarted with the use of HDAC inhibitors either alone or in conjunction with all-trans retinoic acid (ATRA) [59]. In the fight against HIV, HDAC inhibitors are being used alongside highly active antiretroviral therapy (HAART) in order to force viral expression, thus decreasing the pool of latent HIV reservoirs [60].
Another approach to modulating histone deacetylase activity could be to target specific signal transduction pathways which affect the cellular localization of HDAC proteins. McKinsey et al. unveiled a mechanism of transcriptional control in which myoblast differentiation triggers HDAC5 to relocate from the nucleus to the cytoplasm [61]. Disrupting the cascade of such a pathway cousld prove to be an effective therapeutic approach.
4. HMTs
Histone methyl transferases (HMT) transfer methyl groups to either lysine or arginine residues on histone tails in a highly specific manner. The degree of histone methylation, mono-, di- or tri-methylated lysines and mono- or di-methylated arginines, can vary depending on number of factors including the levels of local histone acetylation and plays a role in promoter accessibility. Most lysine methyl transferases contain a conserved SET (SUV39 (suppressor of variegation 3-9), Enhancer or zeste, Trithorax) domain and can confer either transcriptional silencing or activation. SET domain proteins can be classified into five subfamilies; SET1, SET2, SUV39, RIZ (retinoblastoma protein-interacting zinc-finger) and SMYD3 (SET- and MYND-domain containing protein 3) (Table 3). Enhancer of zeste homologue 2 (EZH2), an H3-K27 specific SET1 HMT, is associated with chromo domain-containing polycomb complexes, which serve to silence homeotic genes and repress transcription and are believed to be involved in the maintenance of cellular memory [62]. Physiologically, EZH2 lies downstream of the Rb-E2F pathway and is required for cellular proliferation. The lysine-specific HMT, SUV39 homologue 1 (SUV39H1), is often recruited to the promoters of cell cycle control genes where it trimethylates lysine 9 on histone H3, thereby recruiting the chromo domain-containing heterochromatin protein 1 (HP1) to repress transcription and delay cell cycle progression [63,64]. RIZ1, an H3-K9 HMT also negatively regulates proliferation and induces cell cycle arrest and apoptosis [65].
Table 3.
Histone methyl transferase (HMT) and histone demethylase (HDM) classifications and specificities.
| Histone lysine methyl transferases | Histone substrate specificity |
|---|---|
| SET domain-containing | |
| SUV39 family | |
| SUV39H1 | H3-K9 |
| SET1 family | |
| SET1 | H3-K4, -K79 |
| EZH2 | H3-K27 |
| MLL | H3-K4 |
| SET2 family | |
| SET2 | H3-K36 |
| NSD1 | H3-K36 |
| RIZ family | |
| RIZ1 | H3-K9 |
| SMYD3 family | |
| SMYD3 | H3-K4 |
| Non-SET domain-containing | |
| DOT1 | H3-K4, -K79 |
| Protein arginine methyl transferases | |
| PRMT1 | H4-R3 |
| PRMT4 | H3-R2, -R17, -R26 |
| PRMT5 | H2A; H4 |
| Histone demethylases | |
| LSD1 | H3-K4, -K9 |
This table highlights only those enzymes discussed in the text and is not fully inclusive of all existing HMTs and HDMs.
DOT1: Disruptor of telomeric silencing-1; EZH2: Enhancer of zeste homologue 2; LSD1: Lysine-specific demathylase-1; MLL: Mixed lineage leukaemia; NSD1: Nuclear receptor-binding SET domain protein-1; PRMT: Protein arginine methyl transferase; RIZ: Retinoblastoma protein-interacting zinc-finger; SET: SuVar39, enhancer of zeste, trithorax; SMYD3: SET- and MYND-domain containing protein-3; SUV39: Suppressor of variegation 3 – 9.
On the other hand, in opposition to EZH2, SUV39H1 and RIZ1, which repress transcription, mixed lineage leukaemia (MLL), SMYD3 and nuclear receptor-binding SET domain protein-1 (NSD1) are HMTs which activate transcription. MLL, an H3K4-specific SET1 HMT, is associated with bromo domain-containing trithorax complexes, which induce chromatin decondensation [66]. SMYD3 exists in a complex with RNA polymerase II and HELZ, an RNA helicase, and is targeted to promoters containing specific DNA binding sequences where it exerts its H3-K4 activity. NSD1, a SET2 subfamily member and H3-K36 HMT, is responsible for developmental regulation and normal Hox gene expression [67].
An increasing number of HMTs are being shown to promote or inhibit tumourigenesis and in many cancers, SET domain proteins are often abnormally regulated. EZH2 gene amplification, for example, has been correlated with a number of cancers [68]. Metastatic prostate cancer, lymphomas and breast cancer all exhibit overexpressed EZH2 levels and concomitant increases in cellular proliferation [69]. In addition, microarray analysis of prostate tumours has revealed a link between elevated EZH2 levels and poor prognosis. Accordingly, RNA interference (RNAi)-induced EZH2 protein repression decreases the rate of proliferation in prostate cancer. SUZ12, a co-repressor in the EZH2-polycomb complex, is also upregulated in colon, breast and liver cancers [70].
Unlike with EZH2, overexpression of SUV39H1 may induce a tumour suppression mechanism [71], whereas SUV39H1 deficiency has been correlated with tumourigenesis [72]. Furthermore, SUV39H1 has been shown to play a role in maintaining senescence and quelling tumour proliferation [71]. Similarly, proliferating cells will arrest and sometimes apoptose upon RIZ1 addition, thereby also implicating RIZ1 in tumour suppression activity [73]. In fact, deletion or inactivation of the RIZ1 gene or silencing of RIZ1 protein expression, all of which affect HMT activity, has been found in a number of cancers including gastric cancer, breast cancer, liver cancer, colon cancer, lung cancer, neuroblastoma, melanoma and osteosarcoma [65].
A well studied example of HMT dysregulation and cancer can be seen in the acute leukaemias that often arise as the result of MLL gene translocations. The resulting fusion oncoproteins lack a functional SET domain, exhibit gain-of-function activities and induce aberrant transcription. In some cases, the chimeric protein will recruit co-activators and basal transcriptional machinery to MLL target genes, resulting in Hox gene overexpression and unabated cellular proliferation [74]. Interestingly, although rare, leukaemogenesis can also occur as the result of loss-of-function MLL mutations [75]. NSD1 translocations also lead to leukaemia. Fusions of NSD1 to nucleoporin 98 (NUP98), found in AML, result in increased myeloid stem cell proliferation and overexpression of Hox A7, Hox A9, Hox A10 and Meis1 proto-oncogenes [67]. NSD1 mutations are also present in overgrowth syndromes, multiple myeloma and lung cancers. The upregulation of SMYD3 in hepatocellular and colorectal carcinomas increases H3-K4 methylation and awakens normally silent oncongenes, thereby classifying SMYD3 as an oncogenic activator [76].
HMTs which transfer methyl groups to the guanidine nitrogen of arginine residues are known as protein arginine methyl transferases (PRMT) (Table 3). PRMT4, for example, positively regulates transcription via H3-R17 methylation as well as interactions with nuclear hormone receptor co-activators. Subsequently, PRMT4 is overexpressed in breast cancer and hormone-dependent prostate cancer [77]. Alternatively, PRMT5 has been found to downregulate cyclin E transcription and arrest proliferating cells in vitro upon H2A and H4 methylation [78]. PRMTs have also been linked to HIV via non-histone protein methylation. The transactivation activity of HIV Tat protein is negatively regulated by PRMT6 methylation, which also restricts HIV replication [79].
Although HMT pathways are just beginning to be elucidated, their relevance in therapy is apparent, as numerous diseases, such as cancer, have been associated with aberrant histone methylation [80]. Few inhibitors have been developed to date but the impetus to find them is growing. A common concern is that anti-HMT drugs will potentially exhibit poor specificity and perturb normal cell homeostasis due to the fact that histone methylation-mediated gene regulation can result in either transcriptional activation or repression. Proof-of-principle studies have revealed that HMTs can be targets for anticancer therapy as small interfering RNAs (siRNAs) that knock down SMYD3 expression, for example, inhibit cell cycle progression in cancer cell lines [76]. SMYD3 in particular is an attractive target due to the fact that it binds to a known DNA binding sequence [81].
Some of the first HMT inhibitors identified were found to compete for the S-adenosyl-L-methionine (AdoMet) methyl donor binding site. The fungal mycotoxin chaetocin, for example, which acts as a competitive inhibitor for AdoMet, was first identified as a lysine-specific HMT inhibitor of SUV3-9 [82]. Such compounds, however, display poor specificity as they target all AdoMet-dependent enzymes. Arginine methyltransferase inhibitor-1 (AMI-1), identified as a specific PRMT inhibitor, is believed to interact with the substrate binding pocket of PRMT1, -3, -4 and -6 [83]. AMI-1 has also been shown to inhibit HIV reverse transcriptase activity and protect cells from further HIV infection [84]. Importantly, AMI-1 is cell-permeable, unlike a number of identified HAT inhibitors, thus making the AMI backbone an important pharmacophore in the search for additional HMT inhibitors. One such derivative, AMI-5, was discovered as an inhibitor of both PRMTs and specific histone lysine methyl transferases [83]. Significantly, a group of AMI-5 analogues were recently found to inhibit the catalytic activity of not only arginine and lysine methyl transferases but also some HATs and SIRTs with similar potency [85]. The chemical structures of these derivatives resemble those of modulators of both methyl transferases and HATs and/or SIRTs, thus spawning the term epigenetic multiple ligands (epi-MLs) [85].
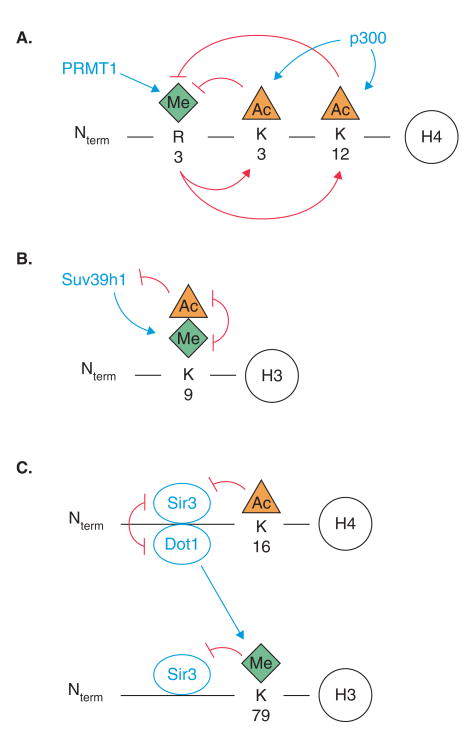
When considering the therapeutic targeting of histone residue-specific modifications, however, the existence of cross-talk between chromatin modifications and chromatin modifying enzymes must be realized. For example, methylation-induced acetylation has been found to occur on histone H4 (Figure 2). PRMT1, an H4-R3 methylase, induces p300-mediated H4 acetylation and transcriptional activation and is involved in nuclear cytoplasmic shuttling (Figure 2A) [86]. This H4 acetylation in turn completes a negative feedback loop by inhibiting H4 methylation.
Figure 2. Cross-regulation between histone acetylation and methylation.
A. Methylation and acetylation are coupled in a negative feedback loop. B. Interplay between acetylation and methylation on the same histone residue determines whether a gene is silenced or activated. C. Acetylation of histone H4 determines methylation of histone H3 as silent information regulator 3 (Sir3) and disruptor of telomeric silencing-1 (Dot1) compete for the same binding sites. Enzymes and their directed action are indicated in blue. Induction and inhibition are indicated in red. PRMT: Protein arginine methyl transferase; Suv39h1: Suppressor of variegation 3 – 9 homologue 1.
In addition, methylating and acetylating enzymes targeting the same histone residue can be indirectly inhibitory towards one another. For example, acetylated lysine 9 on histone H3 will inhibit Suv39h1-mediated methylation of the same residue (Figure 2B). Likewise, methylated H3-K9 will prevent acetylation. These two mutually exclusive modifications have this effect on one another because of their opposing transcriptional outcomes as H3-K9 methylation results in gene silencing and H3-K9 acetylation induces gene activation.
Furthermore, the methyl transferase disruptor of telomeric silencing-1 (Dot1) engages in cross-talk with the heterochromatin protein silent information regulator 3 (Sir3), a member of the SIR complex that also includes Sir4 and the NAD-dependent H4-K16 HDAC Sir2 (Figure 2C) [87]. The SIR complex is associated with transcriptional repression, gene silencing, cell cycle progression and chromosome stability [88]. Dot1, whose misregulation is linked with leukemogenesis [89], is responsible for methylating histone H3 at lysine 79 within the globular domain (Table 3). This methylation, however, is dependent on the ability of Dot1 to concurrently interact with a short basic region on the N-terminal tail of histone H4. Sir3, known to associate with unmodified H3 and H4 tails, competes with Dot1 by interacting with the same basic region on H4 and by binding H3 adjacent to K79. GCN5-mediated H4-K16 acetylation displaces Sir3 on H4, thereby allowing Dot1 to interact with H4 and subsequently methylate H3-K79 (Figure 2C). H3-K79 methylation in turn further blocks Sir3–H3 interactions. This series of events serves to define a heterochromatin boundary and allow for transcriptional elongation.
As is the case with many drugs, combination therapy may be required to evoke the full potential of HMT inhibitors. MLL-fusion protein-induced leukaemias, for example, may not succumb to HMT-inhibitor therapy alone but rather may also require the targeting of both co-activators and chimaera dimerization [90].
5. HDMs
Just as there are HMTs to methylate histones, there exist enzymes responsible for methyl group removal from histones as well. Amine oxidase domain-containing demethylases [91] and Jumonji C (JmjC) domain-containing demethylases [92] antagonize the effects of lysine-specific methyl transferases, whereas peptidylarginine deiminase 4 (PAD4) [93,94] anatagonizes the effects of arginine-specific methyl transferases. However, PAD4, the only identified arginine demethylase, does not convert the methyl-arginine back to arginine but rather to citrulline.
The first lysine demethylase to be discovered was lysine-specific demathylase-1 (LSD1) [19,95]. LSD1, an amine oxidase flavoenzyme [91], belongs to a variety of transcriptional corepressor complexes. JmjC domain-containing demethylases 1 (JHDM1), an iron-dependent oxygenase, was discovered shortly thereafter [92]. Unlike LSD1, which shares sequence homology to only one other human protein (LSD2), there exists about 30 JmjC domain-containing proteins, each displaying unique histone- and residue-specific demethylating capabilities. LSD1, typically existing as an H3-K4 demethylase, is believed by some scientists to be capable of altering its substrate specificity toward H3-K9 demethylation in the presence of androgen receptor (Table 3) [96]. This ability is not well-defined but is most probably made possible via a protein interaction-induced conformational change. LSD1 also works synergistically with and enhances HDAC1 activity. For example, LSD1 overexpression in HEK 293 cells leads to a decrease in H3-K4 methylation and a concurrent decrease in H3 acetylation, thereby silencing transcription [97]. Alternatively, inhibiting HDAC1 increased both H3 acetylation and H3-K4 methylation [97].
Interesting new relationships are beginning to surface between demethylases and disease. For example, LSD1 overexpression has just recently been purported to be a predictive biomarker for prostate cancer [98]. The first attempts to inhibit LSD1 involved the use of known inhibitors of monoamine oxidases (MAO), which share homology with LSD1. Two non-selective MAO inhibitors, phenelzine and tranylcypromine, were found to effectively inhibit LSD1-mediated demethylation of nucleosomal substrates [99]. Currently, additional LSD1 inhibitors are being synthesized and studied for their therapeutic potential [100]. As is the case in the search for any therapeutic drug, there exists an issue with LSD1 inhibitor selectivity due to the fact that the LSD1-mediated oxidation reaction mechanism is potentially similar to those of other physiologically important cellular flavin-dependent amine oxidases, such as MAOs. Furthermore, the fact that LSD1 regulates both transcriptional repression, via H3-K4 demethylation, and activation, via H3-K9 demethylation, poses an additional specificity problem. The active site of LSD1, unlike those of other amine oxidases, however, has been shown to be very large and highly accessible and should make the challenge of finding a highly selective inhibitor easier. Identifying highly selective inhibitors for the plethora of JmjC domain-containing demethylases, on the other hand, is a daunting task and has yet to be undertaken.
6. Expert opinion and conclusions
The finely tuned mechanisms of transcriptional regulation are vital for most physiological processes and any interruption in their pathways typically results in a number of disease states amenable to therapeutic intervention. To this end, chromatin-modifying enzymes are already being targeted therapeutically. HDAC inhibitors, for example, have led the charge in the search for novel therapies with promising results, including a number of FDA-approved drugs. The majority of HDAC inhibitors display pleiotropic effects, inducing cell cycle arrest, differentiation and apoptosis. However, due to the abundance of cellular acetylation and deacetylation machinery and the fact that most of these enzymes target a diverse set of other nuclear proteins in addition to histones, issues such as drug specificity, toxicity and bioavailability have been cumbersome.
Since the initial search for chromatin-modifying-enzyme-specific drugs, the intricate web of not only individual histone modifications but also combinatorial modifications has been further clarified. Multiple tiers of cross-talk mechanisms highlight the importance of histone modifications in the control and regulation of transcription. As we have seen, cross-talk can exist not only between different modifications but also between different histone tails (Figure 2C). Additionally, the mechanisms and involvement of less well described histone modifications, such as phosphorylation, ubiquitylation, SUMOylation and ADP-ribosylation, are proving to be equally important to gene regulation and are also found to engage in cross-talk with histone acetylation and methylation. A complete understanding of the cooperation between all of these modifications will be required to develop the best therapeutic approaches. These additional modifications will be discussed in the second and final part of this review.
Footnotes
Declaration of interest: The author declares no conflict of interest and has received no payment for the preparation of this manuscript.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Kornberg RD, Thomas JO. Chromatin structure; oligomers of the histones. Science. 1974;184(139):865–8. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]; •• A comprehensive review of the ‘histone code’ hypothesis.
- 3.Kornberg RD, Lorch Y. Chromatin structure and transcription. Ann Rev Cell Biol. 1992;8:563–87. doi: 10.1146/annurev.cb.08.110192.003023. [DOI] [PubMed] [Google Scholar]
- 4.Luger K, Mader AW, Richmond RK, et al. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389(6648):251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]; •• The X-ray crystal structure of the nucleosome core is revealed.
- 5.Van Holde K, Zlatanova J. What determines the folding of the chromatin fiber? Proc Natl Acad Sci USA. 1996;93(20):10548–55. doi: 10.1073/pnas.93.20.10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bottomley MJ. Structures of protein domains that create or recognize histone modifications. EMBO Rep. 2004;5(5):464–9. doi: 10.1038/sj.embor.7400146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang XJ. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays. 2004;26(10):1076–87. doi: 10.1002/bies.20104. [DOI] [PubMed] [Google Scholar]
- 8.Brehm A, Tufteland KR, Aasland R, Becker PB. The many colours of chromodomains. Bioessays. 2004;26(2):133–40. doi: 10.1002/bies.10392. [DOI] [PubMed] [Google Scholar]
- 9.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Ann Rev Biochem. 1998;67:545–79. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 10.Schotta G, Lachner M, Sarma K, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18(11):1251–62. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci USA. 1999;96(26):14967–72. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang G, Lin JC, Wei V, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci USA. 2004;101(19):7357–62. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol. 2003;327(1):85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 14.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3(6):415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]; •• An excellent review on cancer epigenetics.
- 15.Kurdistani SK. Histone modifications as markers of cancer prognosis: a cellular view. Br J Cancer. 2007;97(1):1–5. doi: 10.1038/sj.bjc.6603844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seligson DB, Horvath S, Shi T, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435(7046):1262–6. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 17.Mai A. The therapeutic uses of chromatin-modifying agents. Expert Opin Ther Targets. 2007;11(6):835–51. doi: 10.1517/14728222.11.6.835. [DOI] [PubMed] [Google Scholar]; • A good review focusing on histone acetylating and methylating enzymes and their inhibitors.
- 18.Rasheed WK, Johnstone RW, Prince HM. Histone deacetylase inhibitors in cancer therapy. Expert Opin Investig Drugs. 2007;16(5):659–78. doi: 10.1517/13543784.16.5.659. [DOI] [PubMed] [Google Scholar]; •• A detailed review of common HDAC inhibitors, including their therapeutic outcomes.
- 19.Stavropoulos P, Hoelz A. Lysine-specific demethylase 1 as a potential therapeutic target. Expert Opin Ther Targets. 2007;11(6):809–20. doi: 10.1517/14728222.11.6.809. [DOI] [PubMed] [Google Scholar]; • A comprehensive review of LSD1.
- 20.Luger K, Richmond TJ. The histone tails of the nucleosome. Curr Opin Genet Dev. 1998;8(2):140–6. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 21.Puri PL, Sartorelli V, Yang XJ, et al. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997;1(1):35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 22.Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32(3):959–76. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stein RW, Corrigan M, Yaciuk P, et al. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNA synthesis-inducing activity. J Virol. 1990;64(9):4421–7. doi: 10.1128/jvi.64.9.4421-4427.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gayther SA, Batley SJ, Linger L, et al. Mutations truncating the EP300 acetylase in human cancers. Nat Genet. 2000;24(3):300–3. doi: 10.1038/73536. [DOI] [PubMed] [Google Scholar]
- 25.Petrij F, Giles RH, Dauwerse HG, et al. Rubinstein–Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376(6538):348–51. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 26.Rouaux C, Loeffler JP, Boutillier AL. Targeting CREB-binding protein (CBP) loss of function as a therapeutic strategy in neurological disorders. Biochem Pharmacol. 2004;68(6):1157–64. doi: 10.1016/j.bcp.2004.05.035. [DOI] [PubMed] [Google Scholar]
- 27.Ikura T, Ogryzko VV, Grigoriev M, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102(4):463–73. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 28.Anzick SL, Kononen J, Walker RL, et al. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277(5328):965–8. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 29.Borrow J, Stanton VP, Jr, Andresen JM, et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14(1):33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 30.Cullis PM, Wolfenden R, Cousens LS, Alberts BM. Inhibition of histone acetylation by N-[2-(S-coenzyme A)acetyl] spermidine amide, a multisubstrate analog. J Biol Chem. 1982;257(20):12165–9. [PubMed] [Google Scholar]
- 31.Lau OD, Kundu TK, Soccio RE, et al. HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol Cell. 2000;5(3):589–95. doi: 10.1016/s1097-2765(00)80452-9. [DOI] [PubMed] [Google Scholar]; • The first report, to the author's knowledge, of a synthetic HAT inhibitor.
- 32.Balasubramanyam K, Swaminathan V, Ranganathan A, Kundu TK. Small molecule modulators of histone acetyltransferase p300. J Biol Chem. 2003;278(21):19134–40. doi: 10.1074/jbc.M301580200. [DOI] [PubMed] [Google Scholar]
- 33.Balasubramanyam K, Altaf M, Varier RA, et al. Polyisoprenylated benzophenone, garcinol, a natural histone acetyltransferase inhibitor, represses chromatin transcription and alters global gene expression. J Biol Chem. 2004;279(32):33716–26. doi: 10.1074/jbc.M402839200. [DOI] [PubMed] [Google Scholar]
- 34.Balasubramanyam K, Varier RA, Altaf M, et al. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J Biol Chem. 2004;279(49):51163–71. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- 35.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Ann Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 36.Denu JM. Linking chromatin function with metabolic networks: Sir2 family of NAD+-dependent deacetylases. Trends Biochem Sci. 2003;28(1):41–8. doi: 10.1016/s0968-0004(02)00005-1. [DOI] [PubMed] [Google Scholar]
- 37.Brehm A, Miska EA, McCance DJ, et al. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391(6667):597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 38.McKinsey TA, Zhang CL, Olson EN. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci. 2002;27(1):40–7. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- 39.Gruffat H, Manet E, Sergeant A. MEF2-mediated recruitment of class II HDAC at the EBV immediate early gene BZLF1 links latency and chromatin remodeling. EMBO Rep. 2002;3(2):141–6. doi: 10.1093/embo-reports/kvf031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaziri H, Dessain SK, Ng Eaton E, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107(2):149–59. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 41.Weston AD, Blumberg B, Underhill TM. Active repression by unliganded retinoid receptors in development: less is sometimes more. J Cell Biol. 2003;161(2):223–8. doi: 10.1083/jcb.200211117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong SH, David G, Wong CW, et al. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor alpha (RARα) and PLZF-RARα oncoproteins associated with acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94(17):9028–33. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scandura JM, Boccuni P, Cammenga J, Nimer SD. Transcription factor fusions in acute leukemia: variations on a theme. Oncogene. 2002;21(21):3422–44. doi: 10.1038/sj.onc.1205315. [DOI] [PubMed] [Google Scholar]
- 44.Zhu P, Martin E, Mengwasser J, et al. Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis. Cancer Cell. 2004;5(5):455–63. doi: 10.1016/s1535-6108(04)00114-x. [DOI] [PubMed] [Google Scholar]
- 45.Ropero S, Fraga MF, Ballestar E, et al. A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition. Nat Genet. 2006;38(5):566–9. doi: 10.1038/ng1773. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z, Yamashita H, Toyama T, et al. HDAC6 expression is correlated with better survival in breast cancer. Clin Cancer Res. 2004;10(20):6962–8. doi: 10.1158/1078-0432.CCR-04-0455. [DOI] [PubMed] [Google Scholar]
- 47.Gui CY, Ngo L, Xu WS, et al. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci USA. 2004;101(5):1241–6. doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Espino PS, Drobic B, Dunn KL, Davie JR. Histone modifications as a platform for cancer therapy. J Cell Biochem. 2005;94(6):1088–102. doi: 10.1002/jcb.20387. [DOI] [PubMed] [Google Scholar]
- 49.Miller TA, Witter DJ, Belvedere S. Histone deacetylase inhibitors. J Med Chem. 2003;46(24):5097–116. doi: 10.1021/jm0303094. [DOI] [PubMed] [Google Scholar]
- 50.Vrana JA, Decker RH, Johnson CR, et al. Induction of apoptosis in U937 human leukemia cells by suberoylanilide hydroxamic acid (SAHA) proceeds through pathways that are regulated by Bcl-2/Bcl-XL, c-Jun, and p21CIP1, but independent of p53. Oncogene. 1999;18(50):7016–25. doi: 10.1038/sj.onc.1203176. [DOI] [PubMed] [Google Scholar]
- 51.Hirsch CL, Bonham K. Histone deacetylase inhibitors regulate p21WAF1 gene expression at the post-transcriptional level in HepG2 cells. FEBS Lett. 2004;570(13):37–40. doi: 10.1016/j.febslet.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 52.Sandor V, Senderowicz A, Mertins S, et al. P21-dependent G1 arrest with downregulation of cyclin D1 and upregulation of cyclin E by the histone deacetylase inhibitor FR901228. Br J Cancer. 2000;83(6):817–25. doi: 10.1054/bjoc.2000.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wharton W, Savell J, Cress WD, et al. Inhibition of mitogenesis in Balb/c-3T3 cells by Trichostatin A. Multiple alterations in the induction and activation of cyclin-cyclin-dependent kinase complexes. J Biol Chem. 2000;275(43):33981–7. doi: 10.1074/jbc.M005600200. [DOI] [PubMed] [Google Scholar]
- 54.Kelly WK, Richon VM, O'Connor O, et al. Phase I clinical trial of histone deacetylase inhibitor: suberoylanilide hydroxamic acid administered intravenously. Clin Cancer Res. 2003;9(10 Pt 1):3578–88. [PubMed] [Google Scholar]
- 55.Mitsiades CS, Mitsiades NS, McMullan CJ, et al. Transcriptional signature of histone deacetylase inhibition in multiple myeloma: biological and clinical implications. Proc Natl Acad Sci USA. 2004;101(2):540–5. doi: 10.1073/pnas.2536759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Lint C, Emiliani S, Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 1996;5(45):245–53. [PMC free article] [PubMed] [Google Scholar]
- 57.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5(9):769–84. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]; • An excellent review describing the cellular mechanisms responsible for the anticancer effects of HDAC inhibitors.
- 58.Marchion DC, Bicaku E, Daud AI, et al. Sequence-specific potentiation of topoisomerase II inhibitors by the histone deacetylase inhibitor suberoylanilide hydroxamic acid. J Cell Biochem. 2004;92(2):223–37. doi: 10.1002/jcb.20045. [DOI] [PubMed] [Google Scholar]
- 59.Johnstone RW, Licht JD. Histone deacetylase inhibitors in cancer therapy: is transcription the primary target? Cancer Cell. 2003;4(1):13–8. doi: 10.1016/s1535-6108(03)00165-x. [DOI] [PubMed] [Google Scholar]
- 60.Demonte D, Quivy V, Colette Y, Van Lint C. Administration of HDAC inhibitors to reactivate HIV-1 expression in latent cellular reservoirs: implications for the development of therapeutic strategies. Biochem Pharmacol. 2004;68(6):1231–8. doi: 10.1016/j.bcp.2004.05.040. [DOI] [PubMed] [Google Scholar]
- 61.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408(6808):106–11. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valk-Lingbeek ME, Bruggeman SW, Van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118(4):409–18. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 63.Bannister AJ, Zegerman P, Partridge JF, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410(6824):120–4. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 64.Lachner M, O'Carroll D, Rea S, et al. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410(6824):116–20. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 65.He L, Yu JX, Liu L, et al. RIZ1, but not the alternative RIZ2 product of the same gene, is underexpressed in breast cancer, and forced RIZ1 expression causes G2-M cell cycle arrest and/or apoptosis. Cancer Res. 1998;58(19):4238–44. [PubMed] [Google Scholar]
- 66.Milne TA, Briggs SD, Brock HW, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10(5):1107–17. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 67.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9(7):804–12. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 68.Bracken AP, Pasini D, Capra M, et al. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22(20):5323–35. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kleer CG, Cao Q, Varambally S, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100(20):11606–11. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kirmizis A, Bartley SM, Farnham PJ. Identification of the polycomb group protein SU(Z)12 as a potential molecular target for human cancer therapy. Mol Cancer Ther. 2003;2(1):113–21. [PubMed] [Google Scholar]
- 71.Braig M, Lee S, Loddenkemper C, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436(7051):660–5. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 72.Peters AH, O'Carroll D, Scherthan H, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107(3):323–37. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 73.Derunes C, Briknarova K, Geng L, et al. Characterization of the PR domain of RIZ1 histone methyltransferase. Biochem Biophys Res Commun. 2005;333(3):925–34. doi: 10.1016/j.bbrc.2005.05.190. [DOI] [PubMed] [Google Scholar]
- 74.So CW, Lin M, Ayton PM, et al. Dimerization contributes to oncogenic activation of MLL chimeras in acute leukemias. Cancer Cell. 2003;4(2):99–110. doi: 10.1016/s1535-6108(03)00188-0. [DOI] [PubMed] [Google Scholar]
- 75.Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20(40):5695–707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- 76.Hamamoto R, Furukawa Y, Morita M, et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6(8):731–40. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 77.Hong H, Kao C, Jeng MH, et al. Aberrant expression of CARM1, a transcriptional coactivator of androgen receptor, in the development of prostate carcinoma and androgen-independent status. Cancer. 2004;101(1):83–9. doi: 10.1002/cncr.20327. [DOI] [PubMed] [Google Scholar]
- 78.Fabbrizio E, El Messaoudi S, Polanowska J, et al. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 2002;3(7):641–5. doi: 10.1093/embo-reports/kvf136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boulanger MC, Liang C, Russell RS, et al. Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J Virol. 2005;79(1):124–31. doi: 10.1128/JVI.79.1.124-131.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schneider R, Bannister AJ, Kouzarides T. Unsafe SETs: histone lysine methyltransferases and cancer. Trends Biochem Sci. 2002;27(8):396–402. doi: 10.1016/s0968-0004(02)02141-2. [DOI] [PubMed] [Google Scholar]
- 81.Sims RJ, 3rd, Reinberg D. From chromatin to cancer: a new histone lysine methyltransferase enters the mix. Nat Cell Biol. 2004;6(8):685–7. doi: 10.1038/ncb0804-685. [DOI] [PubMed] [Google Scholar]; • The first report, to the author's knowledge, of an HKMT inhibitor.
- 82.Greiner D, Bonaldi T, Eskeland R, et al. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat Chem Biol. 2005;1(13):143–5. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]; • The first report, to the author's knowledge, of PRMT inhibitors.
- 83.Cheng D, Yadav N, King RW, et al. Small molecule regulators of protein arginine methyltransferases. J Biol Chem. 2004;279(23):23892–9. doi: 10.1074/jbc.M401853200. [DOI] [PubMed] [Google Scholar]
- 84.Skillman AG, Maurer KW, Roe DC, et al. A novel mechanism for inhibition of HIV-1 reverse transcriptase. Bioorg Chem. 2002;30(6):443–58. doi: 10.1016/s0045-2068(02)00502-3. [DOI] [PubMed] [Google Scholar]
- 85.Mai A, Cheng D, Bedford MT, et al. Epigenetic multiple ligands: mixed histone/protein methyltransferase, acetyltransferase, and class III deacetylase (sirtuin) inhibitors. J Med Chem. 2008;51(7):2279–90. doi: 10.1021/jm701595q. [DOI] [PubMed] [Google Scholar]
- 86.An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117(6):735–48. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]; •• An important report on histone acetylation-methylation cross-talk.
- 87.Altaf M, Utley RT, Lacoste N, et al. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol Cell. 2007;28(6):1002–14. doi: 10.1016/j.molcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• An important report on inter-histone acetylation-methylation cross-talk.
- 88.Brachmann CB, Sherman JM, Devine SE, et al. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9(23):2888–902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 89.Okada Y, Feng Q, Lin Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121(2):167–78. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 90.So CW, Cleary ML. Dimerization: a versatile switch for oncogenesis. Blood. 2004;104(4):919–22. doi: 10.1182/blood-2004-03-0992. [DOI] [PubMed] [Google Scholar]
- 91.Forneris F, Binda C, Vanoni MA, et al. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett. 2005;579(10):2203–7. doi: 10.1016/j.febslet.2005.03.015. [DOI] [PubMed] [Google Scholar]; • Discovery of the JmjC class of HDMs.
- 92.Tsukada Y, Fang J, Erdjument-Bromage H, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439(7078):811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 93.Cuthbert GL, Daujat S, Snowden AW, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118(5):545–53. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 94.Wang Y, Wysocka J, Sayegh J, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306(5694):279–83. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]; •• Discovery of the first HDM.
- 95.Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 96.Metzger E, Wissmann M, Yin N, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437(7057):436–9. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]; •• An important paper identifying a link between demethylation and deacetylation.
- 97.Lee MG, Wynder C, Bochar DA, et al. Functional interplay between histone demethylase and deacetylase enzymes. Mol Cell Biol. 2006;26(17):6395–402. doi: 10.1128/MCB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kahl P, Gullotti L, Heukamp LC, et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 2006;66(23):11341–7. doi: 10.1158/0008-5472.CAN-06-1570. [DOI] [PubMed] [Google Scholar]
- 99.Lee MG, Wynder C, Schmidt DM, et al. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem Biol. 2006;13(6):563–7. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 100.Culhane JC, Szewczuk LM, Liu X, et al. A mechanism-based inactivator for histone demethylase LSD1. J Am Chem Soc. 2006;128(14):4536–7. doi: 10.1021/ja0602748. [DOI] [PubMed] [Google Scholar]