Abstract
Glycan microarray technology has become a successful tool for studying protein-carbohydrate interactions, but a limitation has been the laborious synthesis of glycan structures by enzymatic and chemical methods. Here we describe a new method to generate quantifiable glycan libraries from natural sources by combining widely used protease digestion of glycoproteins and Fmoc chemistry. Glycoproteins including chicken ovalbumin, bovine fetuin, and horseradish peroxidase (HRP) were digested by pronase, protected by FmocCl, and efficiently separated by 2D-HPLC. We show that glycans from HRP glycopeptides separated by HPLC and fluorescence monitoring retained their natural reducing end structures, mostly core α1,3-fucose and core α1,2-xylose. After simple Fmoc-deprotection, the glycans were printed on NHS-activated glass slides. The glycans were interrogated using plant lectins and antibodies in sera from mice infected with Schistosoma mansoni, which revealed the presence of both IgM and IgG antibody responses to HRP-glycopeptides. This simple approach to glycopeptide purification and conjugation allows for the development of natural glycopeptide microarrays without the need to remove and derivatize glycans and potentially compromise their reducing end determinants.
Keywords: Glycan array, fluorescent labeling, immobilization, functional glycomics
Introductory Statement
The field of functional glycomics [1, 2] has recently seen enormous progress by the development of high throughput methods such as glycan microarrays [3–8]. Glycan structures are immobilized onto activated glass surfaces and interrogated with glycoproteins, antibodies, or whole microorganisms. Many methodologies enabling solid phase immobilization of glycan derivatives onto various activated surfaces have been developed for the purpose of microarray. Non-covalent attachment mechanisms include TLC plate-protein overlay [9, 10], neoglycolipids-nitrocellulose membrane [11, 12] and biotinylated glycoconjugates-streptavidin coated substrates [3]. Covalent attachment generally utilizes inter-reactive pairs, such as activated ester-primary amine [5, 13], epoxy-amine [14–16], sulfhydryl-maleimide [17–19], azide-alkyne [20] etc. While each methodology has its own advantages and disadvantages, their preference depends on the availability of the solid phase substrate and the convenience of the chemistry. The glycan microarray of the Consortium of Functional Glycomics, utilizing commercially available NHS-activated glass slides and including more than 400 glycans, has been one of the most successful glycan microarrays.
Despite the enormous success, the progress of glycan microarray methodology has been limited by the relatively slow expansion of glycan libraries relative to the large size of the glycome. An alternative approach is to develop natural glycan libraries, where glycans from natural sources are released, tagged, and separated. These tagged glycan libraries (TGLs) can be prepared directly from any biological system such as cells, tissues, and microorganisms, and can be interrogated with relevant carbohydrate binding proteins (GBPs), antibodies, and microorganisms. The most crucial component in TGL methodology is the glycan derivatization chemistry, which needs not only to facilitate the separation by adding a sensitive detectable tag, but also to functionalize the glycan for further conjugation such as solid phase immobilization. Widely used fluorescent labels such as 2-aminobenzamide (2-AB) and 2-aminobenzoic acid (2-AA) [21], are good tags for HPLC separation, but are not suitable for further reaction. The bifunctional alkoxyamine linker [22] enables further solid phase immobilization, but it is not fluorescent or UV active, which limits its application to structurally-defined glycans at relatively large scale. To address these questions, we have developed new methods utilizing fluorescent homo- or hetero-bifunctional linkers, i.e., 2,6-diaminopyridine (DAP) [15, 23] and N-aminoethyl 2-aminobenzamide (AEAB) [24] for the generation of TGLs and successfully applied them for the identification of galectin ligands. In this paper, we report a new strategy utilizing well known 9-fluorenylmethyl chloroformate (FmocCl) chemistry to generate TGLs for functional study. Arsequell et. al. reported the derivatization of glycosylamines derivatives of free oligosaccharides with Fmoc-glycine for oligosaccharide fractionation [25]. Here we show that Fmoc serves as a fluorescent tag for glycoamino acids and glycopeptides and facilitates their separation, characterization and quantification. Most importantly, Fmoc is conveniently removed to expose amino groups on the peptide moiety for further reaction such as solid phase immobilization. This new strategy avoids the release of glycans from glycoproteins by hazardous chemicals and expensive enzymes, can be applied to both N- and O-linked glycans as glycopeptides, and serves as a powerful tool for functional glycomics studies.
Materials and Methods
Materials
HPLC solvents were purchased from Fisher Scientific, Pittsburgh, PA. An Ultraflex-II TOF/TOF system from Bruker Daltonics was used for MALDI-TOF mass spectrometry analysis of glycan conjugates. Pronase was purchased from EMD Chemicals Inc., Darmstadt, Germany. All chemicals were purchased from Sigma-Aldrich, St. Louis, MO. Chicken egg glycopeptide was prepared as described previously [26].
Pronase digestion of glycoproteins
The chicken egg glycopeptide and glycoproteins (200 mg) were dissolved in 0.1 M Tris buffer, pH 8.0 so that the final concentration was 20 mg/mL. Pronase was added to a final concentration of 0.1 mg/mL. At 24 hours and 48 hours of incubation at 37°C, an equivalent amount of Pronase was added, respectively. The final digestion was passed through a 2 g C18 Sep-pak, from which the flow-through was passed through a 1g Carbograph and washed by 6 column volumes (c.v.) water. The glycoamino acids and glycopeptides were eluted from the Carbograph by 3 c.v. 50% acetonitrile with 0.1% trifluoroacetic acid (TFA). The elution was dried by Speed-vac for Fmoc derivatization. Chicken ovalbumin, bovine fetuin, and horseradish peroxidase were digested in a similar fashion.
Fmoc derivatization of glycopeptides
The lyophilized glycopeptides were reconstituted into cold water so that the concentration was 100 mg/mL. An equal volume of 50 mg/mL sodium bicarbonate (NaHCO3) solution, an equal volume of water, and an equal volume of 20 mg/mL FmocCl in tetrahydrofuran (THF) was added. The mixture was then agitated vigorously by vortexing for 30 minutes, after which 3 × 1 volume of ethyl acetate was used to extract the mixture. The aqueous phase was applied on C18 Sep-pak and washed by 6 c.v. water. The Fmoc glycopeptides were eluted by 50% acetonitrile with 0.1% TFA.
High performance liquid chromatography (HPLC)
A Shimadzu HPLC CBM-20A system was used for HPLC analysis and separation of Fmoc-glycopeptides, which was coupled with a UV detector SPD-20A and a fluorescence detector RF-10Axl. UV absorption at 254 nm or fluorescence at 254 nm excitation (Ex) and 340 nm emission (Em) was used to detect and quantify Fmoc-tagged glycopeptides.
For normal phase HPLC separation, a Zorbax NH2 column (250mm × 4.6mm) was used; the mobile phases were acetonitrile, water, and 250 mM ammonium acetate (pH 4.5). The concentration of water increased from 16% to 40% and the concentration of ammonium acetate buffer increased from 4% to 50% over 60 minutes.
For reverse phase HPLC with a C18 column, a Vydec C18 column was used. The mobile phase was acetonitrile and water with 0.1% TFA. The concentration of acetonitrile increased from 15% to 45% in 30 minutes.
Generation of normal and 20-week Schistosoma mansoni infected mice sera
Ten 8 week-old C57BL/6 mice were purchased from Charles River Laboratories (Wilmington, MA) and infected with 50–60 Schistosoma mansoni cercariae (Puerto Rican strain) subcutaneously. Ten age-matched uninfected C57BL/6 mice served as controls. Both groups of mice were housed at the Emory University laboratory animal facility. After 20 weeks, the mice were bled via retro-orbital route and blood collected in microtainer serum separator tubes (BD and Company, Franklin Lakes, NJ). The microtainer tubes with blood were centrifuged at 6,000 rpm in an Eppendorf bench-top microcentrifuge (Westbury, NY) for 10 minutes at room temperature. The sera were aspirated, pooled separately as either infected or uninfected normal mouse sera, and stored at −20°C in small aliquots.
Printing, binding assay, and scanning
NHS-activated slides were purchased from Schott, Louisville, KY. Epoxy slides were purchased from Corning, Lowell, MA. Non-contact printing was performed using a Piezoarray printer from Perkin Elmer. The average spot volume was within 10% variation of 1/3 nL. All the samples were printed in phosphate buffer (300 mM sodium phosphates, pH 8.5). After printing, the slides were boxed loosely and put in a high moisture chamber at 50°C and incubated for 1 h. The slides were then washed and blocked with 50 mM ethanolamine in 0.1 M Tris buffer (pH 9.0) for 1 h. The slides were dried by centrifugation and stored desiccated at −20°C for future use. Before assay, the slides were rehydrated for 5 minutes in TSM buffer (20 mM Tris- HCL, 150 mM sodium chloride (NaCl), 0.2 mM calcium chloride (CaCl2) and 0.2 mM magnesium chloride (MgCl2). Biotinylated lectins were used in the binding assay and the bound lectins were detected by a secondary incubation with cyanine 5-streptavidin. For multi-panel experiment on a single slide, the array layout was designed using Piezoarray software according to the dimension of a standard 16-chamber adaptor. The adaptor was applied on the slide to separate a single slide into 16 chambers sealed from each other during the assay.
The slides were scanned with a Perkin Elmer ProScanarray microarray scanner equipped with 4 lasers covering an excitation range from 488 nm to 637 nm. The scanned images were analyzed with the ScanArray Express software. Detection of bound biotinylated lectins was carried out by incubation with cyanine5-streptavidin. Detection of bound mouse sera antibodies was carried out by incubation with Alexa568 labeled goat anti-mouse IgG and Alexa488 labeled goat anti-mouse IgM. For cyanine5 fluorescence, 649 nm (Ex) and 670 nm (Em) were used. For Alexa488, 495 nm (Ex) and 519 nm (Em) were used. For Alexa568, 579 nm (Ex) and 604 nm (Em) were used. All images obtained from the scanner were in grayscale and colored for easy discrimination.
Results
Fmoc derivatization of glycoamino acids and glycopeptides
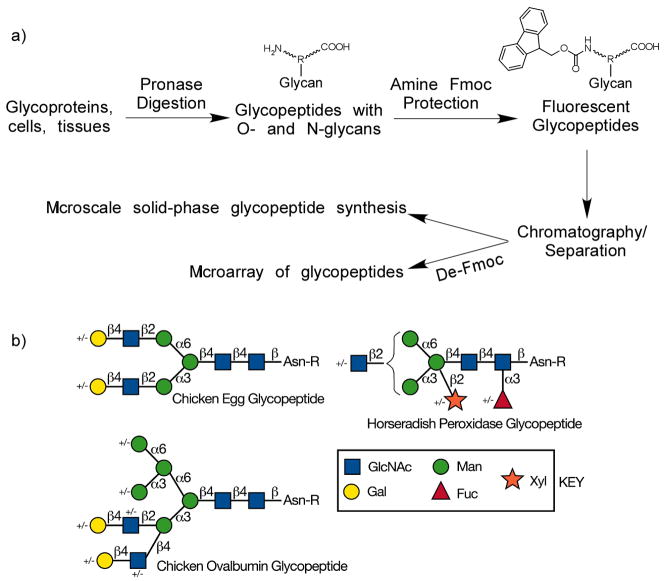
The general strategy is schematically represented in Fig. 1a. Glycoproteins can be digested with Pronase to glycopeptides bearing O- or N-linked glycan, and in most cases either a single Asn residue, or one or two additional residues [27–29]. The amino groups of the glycopeptides are reactive with FmocCl under mild conditions. The resulting fluorescent glycopeptide mixture can easily be separated chromatographically and monitored by fluorescence to generate a tagged-glycan library or TGL. This Fmoc-protected TGL can be used as building blocks in microscale glycopeptide synthesis by prevalent solid phase peptide synthesis (SPPS). After Fmoc deprotection, this TGL can be immobilized onto various solid surfaces such as microarray slides for further functional studies.
Figure 1.
a) The general strategy of utilization of Fmoc-tag for fluorescent glycopeptide library construction and its possible applications. b) The types of glycopeptides with N-glycans used in this study, labeled chicken egg glycopeptide, ovalbumin glycopeptide, and horseradish peroxidase glycopeptide. The symbols used for monosaccharides are shown in the key.
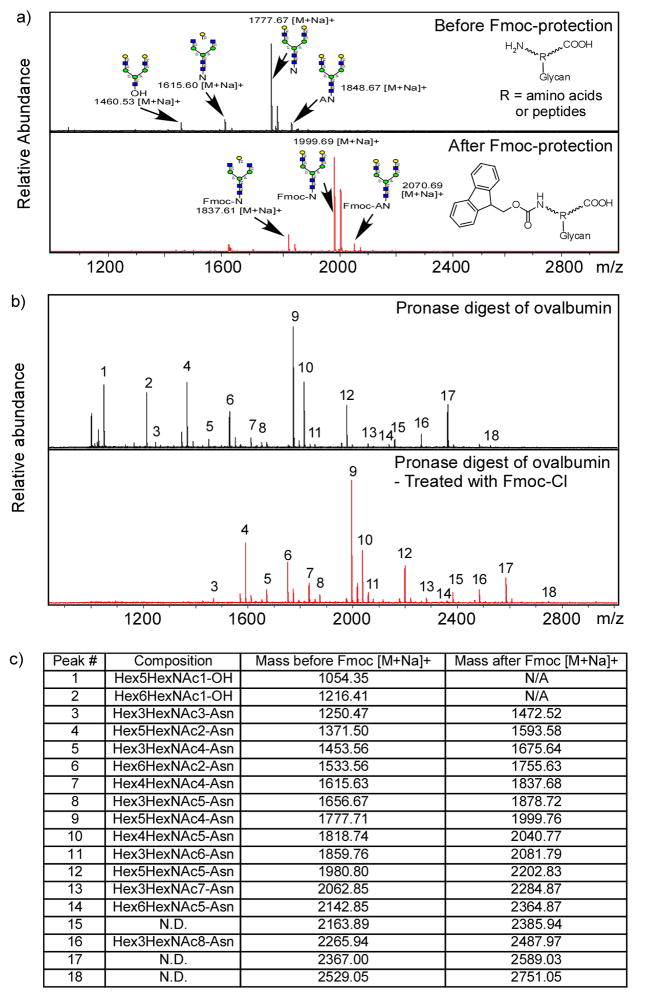
A glycopeptide prepared from chicken egg yolk termed chicken egg glycopeptide (Fig. 1b) [26], a disialo-biantennary N-glycan attached to peptide KVANKT, was utilized in the preliminary study because of its availability and relative homogeneity. After Pronase digestion and mild acid hydrolysis to remove all sialic acids, it was treated with FmocCl. MALDI-TOF profiles of the mixture before and after FmocCl treatment are shown in Fig. 2a. The major component (Hex)5(HexNAc)4-Asn (NA2-N) and two minor components (Hex)4(HexNAc)4-Asn, Ala-((Hex)5(HexNAc)4-)Asn were successfully derivatized with Fmoc-group, as evidenced by a mass shift of 222 Daltons. In contrast, one minor free oligosaccharide component, (Hex)5(HexNAc)3-OH, was not derivatized with Fmoc-group and shows no corresponding mass shift after FmocCl treatment. Thus, the Fmoc-derivatization is specific for amino groups and suitable for labeling glycoamino acids and glycopeptides.
Figure 2.
Fmoc-protected of glycoamino acids and glycopeptides: a) MALDI-TOF spectra of Pronase treated egg yolk glycopeptide before (top) and after (bottom) FmocCl treatment; b) MALDI-TOF spectra of Pronase treated chicken ovalbumin before (top) and after (bottom) FmocCl treatment; c) The MALDI-TOF spectra mass lists of Pronase treated chicken ovalbumin before and after FmocCl treatment. Quantitative Fmoc-protection of glycoamino acids and glycopeptides are shown by a mass increase of 222.
This strategy was further tested on a more complex system. Chicken ovalbumin is an abundant egg white glycoprotein with a single N-glycosylation site [30]. After extensive Pronase digestion, solid phase extraction columns, including C18 Sep-pak and carbograph, were used to purify the glycoamino acids and glycopeptides. The mixture was treated with FmocCl and profiled by MALDI-TOF. The MALDI-TOF profiles of ovalbumin glycopeptides before and after Fmoc-derivatization are shown in Fig. 2b. Except for several free reducing glycans present as contaminants of the commercial glycoprotein, all of the glycopeptides generated from ovalbumin were successfully derivatized and showed a Δ222 Dalton mass shift (Fig. 2c). The very similar MALDI-TOF profiles suggested a non-selective and quantitative derivatization of Fmoc. Furthermore, most of the peaks can be assigned to a glycan attached to a single asparagine, suggesting high efficiency of Pronase digestion of ovalbumin.
Solid phase immobilization of Fmoc derivatized glycoconjugates
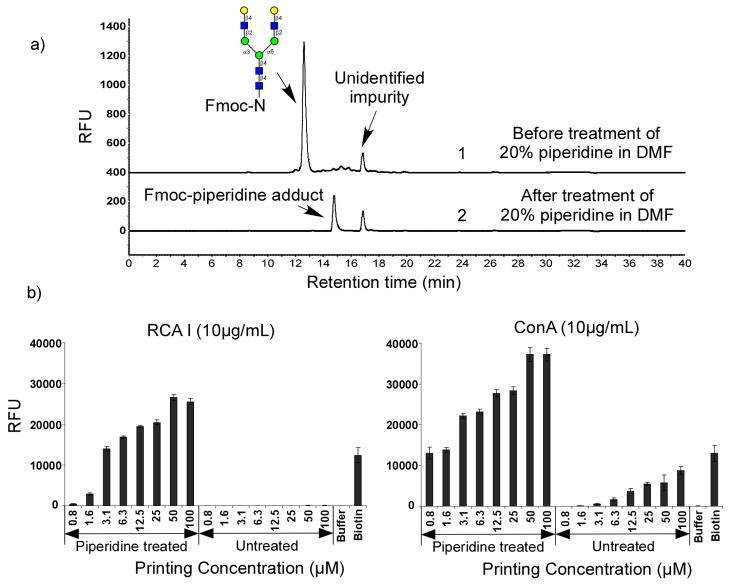
The practicability of Fmoc-protected glycoamino acids and glycopeptides for microarray application was evaluated by applying a simple deprotection procedure to the Fmoc-derivatives before the standard printing procedure. Fig. 3a shows the HPLC profiles of (Hex)5(HexNAc)4-Asn-Fmoc before and after standard Fmoc-deprotection by treatment with 20% piperidine in dimethylformamide (DMF) for 30 minutes. Full deprotection was confirmed by the complete disappearance of the starting material and generation of an Fmoc-piperidine adduct (Fig. 3a). The regenerated glycoamino acid (Hex)5(HexNAc)4-Asn is not fluorescent and is not detected. Thus, the loss of fluorescence can be used to monitor the completion of the reaction.
Figure 3.
The printing of N-NA2-Fmoc: a) HPLC profiles of N-NA2-Fmoc (top) and after 20% piperidine treatment (bottom); b) The printed N-NA2-Fmoc before and after deprotection validated by RCA I and ConA binding.
After evaporation of all the solvents, the deprotected glycoamino acid was printed on NHS-activated glass slides without any further purification, along with other compounds, including Fmoc-protected glycoamino acid, buffer control, and biotin-hydrazine. The microarray slide was then assayed with the biotinylated plant lectins RCA I and ConA [31, 32] followed by incubation with cyanine5-streptavidin (Fig. 3b). For deprotected (Hex)5(HexNAc)4-sn, both RCA I and ConA (each at 10 μ) showed binding with increasing printing concentrations of glycopeptide up to 100 μThe binding of the lectins corresponded to the terminal βlactosylated N-glycan structure recognized by RCA I [33, 34] and the hybrid-type N-glycan structure recognized by ConA [31, 32, 35]. The lowest detectable printing concentration at ~1 μM, shows the suitability of this strategy for glycan microarray development. RCA I did not show any binding to Fmoc-protected (Hex)5(HexNAc)4-Asn, however, ConA did show significant binding signals to Fmoc-protected (Hex)5(HexNAc)4-Asn. This is presumably due to trace amounts of deprotected NA2-N during the printing process at pH 8.5, which is detected by ConA. Nevertheless, the lowest detectable concentration (~3 μM) for NA2-N-Fmoc suggests an extremely high sensitivity of NA2-N to ConA binding on the microarray platform.
Generation of an Fmoc-derivatized TGL from HRP
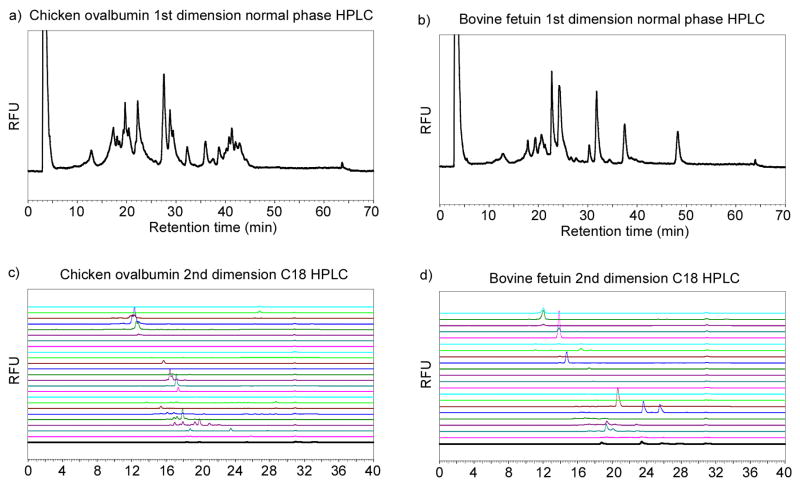
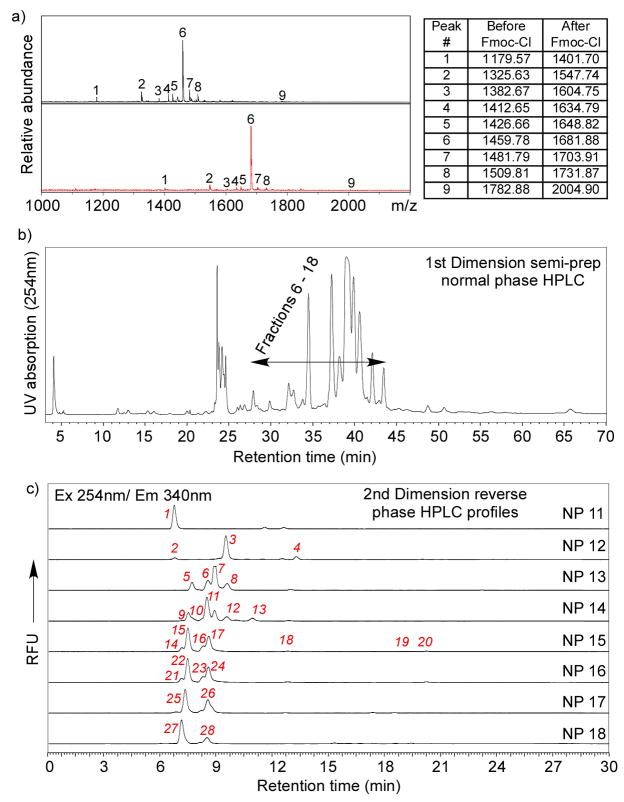
To evaluate whether Fmoc-derivatized glycopeptides from pronase digestion of glycoproteins can be separated by HPLC to near-homogeneous fractions, we tested a 2D-HPLC (normal phase followed by reverse phase) strategy on chicken ovalbumin and bovine fetuin. The profiles of 1st dimension normal phase HPLC are shown in Figs. 4a and b. Fractions collected from normal phase HPLC were subjected to 2nd dimension reverse phase (C18) HPLC (Figs. 4c and d). Although it is usually challenging to separate oligosaccharides or glycopeptides efficiently only by RP-HPLC [36], the reasonably well-separated peaks in Fig. 4 indicated that 2D-HPLC is sufficient to resolve most Fmoc-glycopeptides, which is essential for generation of a TGL. Horseradish peroxidase (HRP) has multiple N-glycosylation sites with predominantly pausi-mannose-like structures [37, 38] (Fig. 1b). These N-glycans are resistant to widely used PNGase F and can only be released by treatment with PNGase A or harsh chemicals, such as sodium hydroxide and hydrazine. As an economic alternative, HRP was treated with Pronase so that the linkages between glycans and peptide chain remain intact. These glycopeptides were then derivatized with FmocCl. The MALDI-TOF spectra of the glycopeptides before and after FmocCl treatment are shown in Fig. 5a. Similar to egg yolk glycopeptides and ovalbumin glycopeptides (Fig. 2), all the glycopeptides showed a characteristic mass shift of 222 after FmocCl treatment, without affecting the general mass profile. It is interesting to note that the dominant peaks shown in both spectra (1459.78[M+H]+ before FmocCl and 1681.88[M+H]+ after FmocCl) have the composition of (Hex)3(HexNAc)2(Xyl)1(Fuc)1-Asn-Arg. This peak, however, is not the most abundant species as shown below by the 2D-HPLC analysis. The Fmoc-glycopeptides were purified by 2D-HPLC using a semi-preparative normal phase column for the first dimension (Fig. 5b) and a C18 reverse phase analytical column for the second dimension (Fig. 5c). Peaks were collected, characterized by MALDI-TOF/TOF, and re-quantified based on fluorescence. The selected fractions were saved as the TGL from HRP (Table 1a). Their glycan and peptide compositions are proposed based on MALDI/TOF-TOF experiments and known structures. Based on the fluorescence, it is possible to quantify the relative abundance of each glycopeptide. The HRP-glycans can be roughly categorized as fucosylated, xylosylated, both fucosylated and xylosylated, and high-mannose structures (Fig. 1b). Their relative abundance are listed in Table 1b, which shows that (Hex)3(HexNAc)2(Xyl)1(Fuc)1 is the dominant glycan structure. These results are largely consistent with previous reports [37, 38], although the identified structures and their abundance may vary due to the source of HRP and the analytical methods applied.
Figure 4.
2D HPLC separation of Fmoc-glycopeptide from pronase digested glycoproteins: a) 1st dimension normal phase HPLC profile of Fmoc-glycopeptides from pronase digested chicken ovalbumin; b) 1st dimension normal phase HPLC profile of Fmoc-glycopeptides from pronase digested bovine fetuin; c) 2nd dimension reverse phase (C18) HPLC profiles of fractions collected from normal phase HPLC of chicken ovalbumin Fmoc-glycopeptides; d) 2nd dimension reverse phase (C18) HPLC profiles of fractions collected from normal phase HPLC of bovine fetuin Fmoc-glycopeptides. Fluorescence detection at Ex 254 nm/Em 340 nm is shown.
Figure 5.
Generation of a tagged glycan library from horseradish peroxidase (HRP): a) MALDI-TOF spectra of Pronase treated HRP before (top) and after (bottom) FmocCl treatment, where the minor component (Hex)3(HexNAc)2(Xyl)1(Fuc)1-Asn-Arg was shown to be the dominant peak; b) The first dimensional normal phase HPLC; c) The second dimensional C18 reverse phase HPLC with all of the 2D fractions marked. The scale of the Y-axis of each profile was individually adjusted to show the minor peaks. The relative abundance was calculated based on re-quantification of each 2D fraction.
Table 1.
a) The masses (Calculated and Found) for Fmoc-protected glycoamino acids and glycopeptides as 2D-HPLC fractions obtained from horseradish peroxidase, their proposed compositions and relative abundances. All the masses are sodium adducts ([M+Na]+) except fractions 1 & 2, which are proton adducts ([M+H]+). Green = fucosylated structures, Yellow = xylosylated structures, White = fucosylated and xylosylated structures, Blue = high mannose structures;b) The relative abundances of fucosylated, xylosylated, both fucosylated and xylosylated, and high-mannose glycan structures from HRP.
| a) | ||||
|---|---|---|---|---|
| 2D fraction No. | Mass (Found) | Mass (Calc.) | Proposed composition | Relative abundance (%) |
| 1 | 1681.75 | 1681.55 | Hex3HexNAc2Fuc1 Xyl1 -NR | 1.8 |
| 2 | 1681.68 | 1681.55 | Hex3HexNAc2Fuc1 Xyl1 -NR | 0.3 |
| 3 | 1401.54 | 1401.37 | Hex3HexNAc2Xyl1-N | 3.0 |
| 4 | 1646.72 | 1646.49 | Hex3HexNAc2Fuc1 Xyl1 -NV | 0.4 |
| 5 | 1458.60 | 1458.39 | Hex3HexNAc2Xyl1-GN | 1.2 |
| 6 | 1415.59 | 1415.43 | Hex3HexNAc2Fuc1-N | 0.9 |
| 7 | 1401.53 | 1401.37 | Hex3HexNAc2Xyl1-N | 3.9 |
| 8 | 1486.65 | 1486.46 | Hex3HexNAc2Fuc1-NA | 0.4 |
| 9 | 1634.63 | 1634.46 | Hex3HexNAc2Fuc1 Xyl1 -NS | 5.3 |
| 10 | 1648.67 | 1648.47 | Hex3HexNAc2Fuc1Xyl1-NT | 6.3 |
| 11 | 1547.63 | 1547.43 | Hex3HexNAc2Fuc1Xyl1-N | 32.0 |
| 12 | 1731.74 | 1731.55 | Hex3HexNAc2Fuc1Xyl1-LNA | 6.2 |
| 13 | 1849.82 | 1849.57 | Hex3HexNAc3Fuc1Xyl1-NV | 0.7 |
| 14 | 1634.53 | 1634.46 | Hex3HexNAc2Fuc1Xyl1-NS | 3.8 |
| 15 | 1603.86 | 1604.45 | Hex3HexNAc2Fuc1Xyl1-GN | 0.5 |
| 16 | 1648.63 | 1648.47 | Hex3HexNAc2Fuc1Xyl1-NT | 8.1 |
| 17 | 1547.59 | 1547.43 | Hex3HexNAc2Fuc1 Xyl1 -N | 4.4 |
| 18 | 1731.75 | 1731.55 | Hex3HexNAc2Fuc1Xyl1-LNA | 1.8 |
| 19 | 1804.71 | 1804.56 | Hex3HexNAc2Fuc1 Xyl 1 -SVNA | 0.4 |
| 20 | 1802.74 | 1802.55 | Hex3HexNAc2Fuc1Xyl1-SPNA | 1.2 |
| 21 | 1675.63 | 1675.49 | Hex3HexNAc2Fuc1Xyl1-NQ | 0.7 |
| 22 | 1634.62 | 1634.46 | Hex3HexNAc2Fuc1 Xyh -NS | 5.7 |
| 23 | 1807.69 | 1807.53 | Hex3HexNAc3Fuc1 Xyl1 -NG | 1.2 |
| 24 | 1750.68 | 1750.51 | Hex3HexNAc3Fuc1Xyl1-N | 4.6 |
| 25 | 1842.63 | 1842.56 | Hex6HexNAc2-NS | 1.2 |
| 26 | 1912.71 | 1912.56 | Hex4HexNAc3Fuc1 Xyl1 -N | 0.9 |
| 27 | 2004.70 | 2004.61 | Hex7HexNAc2-NS | 2.4 |
| 28 | 1917.72 | 1917.58 | Hex7HexNAc2-N | 1.0 |
| b) | |
|---|---|
| N-glycan Category | Molar Percentage(%) |
| Fucosylated only | 1.3 |
| Xylosylated only | 8.1 |
| Both fucosylated and xylosylated | 86.1 |
| High-mannose | 4.5 |
Glycan microarray of HRP TGL
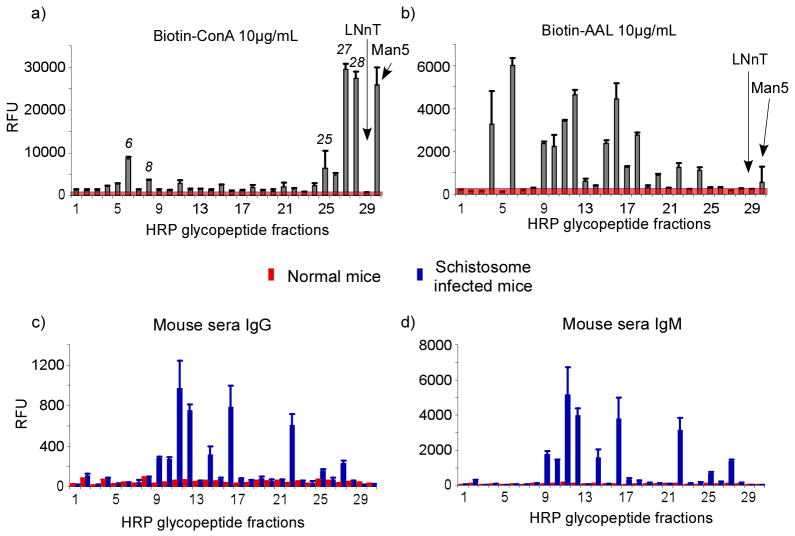
The Fmoc-derivatized glycopeptides from HRP were printed after simple deprotection by piperidine and interrogated with biotinylated lectins followed by incubation with cyanine5-streptavidin. ConA showed strong binding to several high-mannose-type structures (#25, #27, #28) (Fig. 6a). The presence of the core α3-linked fucose residue slightly decreased the binding (#6 and #8), whereas the presence of the β2-linked xylose residue largely abolished the binding. The elimination of ConA binding to core-xylosylated N-glycans is consistent with previous findings [39]. AAL, an α-fucose binding protein [40], showed binding to most of the glycans (Fig. 6b), in agreement with the fact that most of the N-glycans of HRP possess a core α3-linked fucose. AAL did not bind to glycans lacking fucosylation (#3, #5, #7, #25, #27, and #28). Several glycopeptide fractions, although confirmed by mass spectra to possess a fucose (#1, 2, 8, 21, 23), did not show significant binding by AAL. While the reason is not clear, it is possible that amino acids in proximity to the glycan as it is derivatized on the slide surface may interfere with lectin accessibility, since the core-fucose is very close to the peptide chain. This is a unique possibility that should be explored further using defined glycans within a large number of peptide sequences, since the compromised recognition by AAL could also be reflected in compromised recognition by anti-glycan antibodies that recognize the α3-linked fucose residue as a partial determinant. Thus, this is a not a limitation of the approach, but an advantage, since multiple peptide species are likely to arise from Pronase digestion, giving a range of glycopeptides with similar glycans to test.
Figure 6.
The lectin and sera binding to the 2D-HPLC separation of Fmoc-glycopeptides from HRP. The average RFU of 6 replicates on the microarray was plotted against each fraction and the standard deviation was incorporated as the error bar. a) Binding of biotinylated ConA (10 μg/mL) was detected by cyanine5-streptavidin; b) Binding of biotinylated AAL (10 μg/mL) was detected by cyanine5-streptavidin; c) Binding of normal (red) and Schistosome-infected (blue) mouse serum (1:20 dilution) was detected by Alexa568 labeled goat anti-mouse IgG; d) Binding of normal (red) and Schistosome-infected (blue) mouse serum (1:20 dilution) was detected by Alexa488 labeled goat anti-mouse IgM.
We then screened the HRP microarrays with sera from mice infected with Schistosoma mansoni. This vascular parasitic worm synthesizes N-glycans that contain core α3-linked fucose and core β2-linked xylose residues [41–43], and antibodies to those glycans are produced during infection [44, 45]. It is believed that glycoconjugates play important roles in host-parasite interactions [46–48]. To test recognition of glycopeptides on the microarray, sera from normal, uninfected mice and 20-week infected mice were tested, followed by fluorescently labeled anti-mouse IgG (Fig. 6c) and IgM (Fig. 6d). While there was no antibody binding observed with normal mouse serum, strong reactivity was observed with 20-week infected sera for both IgG and IgM. Both IgG and IgM showed significant binding to most core-fucosylated and xylosylated glycans to different degrees. When glycopeptides are immobilized onto glass slides through the N-terminus, the peptide segments adjacent to the glycans could significantly affect the presentation of the glycans to antibodies, and therefore change the binding pattern. The existence of antibodies recognizing the structures close to the glycan-peptide linkages is also possible. The 20-week infected mouse sera also showed antibody recognition of high-mannose structures, which has also been observed previously [49]. These results demonstrate the presence of antibodies from the sera of S. mansoni-infected mice to core-fucosylated and xylosylated glycans, and confirm the utility of the microarray presentation in order to identify these glycan antigens [41, 43]. These results also show the value of preparing glycopeptides with multiple peptide moieties to enhance glycan presentation, since clearly the linking peptide and other potential linking strategies could influence the recognition of antigenic glycan determinants that are near the reducing end of the glycan.
Discussion
Glycan microarrays have become powerful tools for exploring and defining protein-carbohydrate interactions, which are important in numerous biological processes, including cell-cell adhesion, cell signaling, glycoprotein turnover and biosynthesis, and genetic disorders and infectious diseases. The success of a glycan microarray essentially depends on the size, diversity, and representative nature of the glycan library included in the microarray. By developing a novel heterobifunctional linker, 2-amino-N-(2-amino-ethyl)-benzamide (AEAB), we have demonstrated that natural tagged glycan libraries (TGLs) provide a feasible and efficient way to expand glycan microarrays for functional glycomics studies [24]. Although this strategy is generally useful for most studies, the reducing end structures are lost due to the reductive amination. As a supplementary study, we developed this new strategy for glycanase-resistant N-glycans, O-glycans, and other situations where reductive amination is not favored and glycan release from peptides would compromise their structural integrity and recognition.
The 9-fluorenylmethyl chloroformate (Fmoc)-based solid phase peptide synthesis (SPPS) is the most popular peptide synthesis method, since it involves the easy installation of Fmoc to amino groups and its removal by mild conditions [50, 51]. Fmoc-protected glycoamino acids have also been widely used for assembly of glycopeptides [52–57]. In our study, we have explored the application of Fmoc as a fluorescent tag for generating TGLs. This approach has several advantages. The fluorescent property of the Fmoc is essential for sensitive detection and quantification for microscale glycopeptides. The hydrophobic nature of Fmoc can also act as an affinity tag during the separation, since it imparts a unique hydrophobic group to the highly hydrophilic glycopeptides. In addition, the active amino group on the peptide that is required for covalent linking in glycan microarrays can be easily regenerated without interference of other structures. Preliminary study using glycoamino acid and glycopeptide mixtures from chicken egg yolk and ovalbumin showed successful Fmoc-derivatization of both glycoamino acids and glycopeptides, but not free reducing glycans (Fig. 2). The hydrophobicity of Fmoc group provide an easy clean-up of the material away from salts, free reducing glycans, and trace amounts of underivatized glycopeptides.
The printing of deprotected Fmoc-glycoamino acids was validated by lectin binding (Fig. 3), which shows that this array is a good platform for screening glycan-protein interactions. The printed glycans retained their natural linkages between the glycans and the peptides. Although in most cases protein-carbohydrate interactions occur at the non-reducing termini, there are many examples of the importance of the reducing termini and the close-ring structure of the reducing end, along with the adjacent aglycon. It is apparent that the easiest way to retain all such information is not to break the glycosidic linkages. It is noteworthy that although the deprotection of Fmoc by 20% piperidine in DMF at room temperature is generally considered safe for most base-catalyzed reactions including β-elimination, this has not been validated for O-glycopeptides. Extra caution may be needed when these systems are being studied.
Digestion of glycoproteins with Pronase, which is a protease mixture capable of cleaving virtually all peptide linkages [29], represents an inexpensive and convenient way to release glycoamino acids and glycopeptides of both N- and O-glycan related structures from glycoproteins. While PNGase F has been established as an efficient way to release most N-glycans from glycoproteins, it is too expensive to be applied to relatively large scale reactions, and more importantly, some N-glycans are resistant to PNGase F cleavage. Such is the case for N-glycans with core α3-linked fucose attached to the reducing GlcNAc residue, for which PNGase A has to be used [58]. The most established chemical method, hydrazinolysis, is known to be too harsh for labile groups, such as sialic acids and O- and N-acetylation, and requires chemical re-N-acetylation of glycans. There are no enzymatic methods to release large-sized O-glycans containing more than 2 sugar residues, and the only methods to release large O-glycans from Ser/Thr residues is base-catalyzed release, which may compromise the integrity of the glycans [59–61]. In most cases release of O-glycans by chemical treatment is accompanied by inclusion of sodium borohydride to reduce the glycan or the production of glycosylamines that have to be converted to reducing free glycans, with loss of efficiency. Thus, there are many advantages in avoiding chemical release methods for O-glycans and using protease treatments. Pronase digestion is a commonly used method to extract glycoamino acids and glycopeptides from glycoproteins. Other non-glycan components could be easily separated out based on the highly hydrophilic property of glycoamino acids and glycopeptides. Solid phase extraction (SPE) including C18 Sep-pak and Carbograph has been employed in the enrichment/purification of glycopeptides. One critical issue is to avoid over-saturation of the capacities of SPE columns. This can be easily confirmed by the traditional phenol-sulfuric assay for hexose, in which >95% hexose should be observed in the final elution from the Carbograph column.
It is also interesting that Pronase digestion often does not result in complete digestion to a glycan linked to a single Asn residue. While the heterogeneity of the peptide portion of glycopeptides could increase the complexity of the product mixture, making the separation and characterization of individual glycans difficult, this heterogeneity in peptide regions could offer an advantage, since it allows the exploration of the glycan recognition in the context of several types of peptide presentations. Extensive Pronase digestion can reduce the heterogeneity of the glycopeptide mixture, as in the case of chicken ovalbumin (Fig. 2b, c), which has only a single N-glycosylation site. While digestion of all glycopeptides from all sources to single asparagine-glycans may not be practical, the HPLC profiles of Pronase-digested and Fmoc-derivatized glycopeptide mixture from HRP suggests that chromatographic separation of such mixtures is achievable (Fig. 5a, b), especially by 2D-HPLC. It is noteworthy that although the glycan attached to a single asparagine was the most dominant molecular species (Table 1a, fraction 11), it did not appear as the major peak in MALDI-TOF analysis of the glycopeptide mixture (Fig. 5a) either before or after Fmoc-treatment. This suggests that regular MALDI-TOF of a glycan mixture is not reliable for quantification where tagging is necessary.
Horseradish peroxidase (HRP) is known to include pausi-mannose glycan structures, which are resistant to PNGase F digestion. These glycan structures are known to be allergens and showed cross-reactivity towards Schistosome-specific antibodies [41–45]. We therefore applied the Pronase-Fmoc strategy to generate a TGL of HRP for microarray studies. The Fmoc-glycoamino acids and glycopeptides could be efficiently separated by 2D-HPLC separation (Fig. 5) and the fractions can be characterized by MALDI-TOF/TOF. The interrogation of this natural glycan microarray from HRP showed expected lectin interactions. It also showed significant response to Schistosome-infected mouse sera, confirming the cross-reactivity between typical plant glycans and Schistosome related antibodies. It is interesting that the results show that glycopeptides containing glycans with both core-fucose and xylose are best recognized by anti-sera from schistosome-infected mice, whereas the individual epitopes as either core xylose or core fucose alone are weakly recognized. Future studies should be aimed at further defining these important immunodeterminants using immobilized glycopeptides of varying peptide moieties. These data support the general applicability of this strategy for analyzing various glycan binding proteins and antibodies, and should make this approach especially useful for exploring glycan determinants in association with their natural peptide linkages.
Acknowledgments
This work was supported by in part by a Bridge Grant to R.D.C from the Consortium for Functional Glycomics under NIGMS through NIH Grant GM62116 and by NIH Grant RO1AI47214 to R.D.C. We thank Dr. Jamie Heimburg-Molinaro (Emory University School of Medicine) for manuscript editing and review. The authors declare they have no financial interest.
Abbreviations
- AAL
Aleuria aurantia Lectin
- AEAB
2-amino-N-(2-aminoethyl)-benzamide
- ConA
Concanavalin A
- c.v
column volumes
- DAP
2,6-diaminopyridine
- DMF
dimethylformamide
- FmocCL
9-fluorenylmethyl chloroformate
- GBP
glycan binding protein
- HPLC
high performance liquid chromatography
- HRP
horseradish peroxidase
- NHS
N-hydroxysuccinimide
- RCA I
Ricinus communis Agglutinin I
- RFU
relative fluorescence unit
- SPPS
solid phase peptide synthesis
- TFA
trifluoroacetic acid
- TGL
Tagged Glycan Library
- THF
tetrahydrofuran
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 2009. [PubMed] [Google Scholar]
- 2.Taniguchi N, Hancock W, Lubman DM, Rudd PM. The second golden age of glycomics: from functional glycomics to clinical applications. J Proteome Res. 2009;8:425–6. doi: 10.1021/pr801057j. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez RA, Blixt O. Identification of ligand specificities for glycan-binding proteins using glycan arrays. Methods Enzymol. 2006;415:292–310. doi: 10.1016/S0076-6879(06)15018-1. [DOI] [PubMed] [Google Scholar]
- 4.Paulson JC, Blixt O, Collins BE. Sweet spots in functional glycomics. Nat Chem Biol. 2006;2:238–48. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- 5.Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci U S A. 2004;101:17033–8. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drickamer K, Taylor ME. Glycan arrays for functional glycomics. Genome Biol. 2002;3:REVIEWS1034. doi: 10.1186/gb-2002-3-12-reviews1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feizi T, Chai W. Oligosaccharide microarrays to decipher the glyco code. Nat Rev Mol Cell Biol. 2004;5:582–8. doi: 10.1038/nrm1428. [DOI] [PubMed] [Google Scholar]
- 8.Park S, Lee MR, Shin I. Chemical tools for functional studies of glycans. Chem Soc Rev. 2008;37:1579–91. doi: 10.1039/b713011m. [DOI] [PubMed] [Google Scholar]
- 9.Lopez PH, Schnaar RL. Determination of glycolipid-protein interaction specificity. Methods Enzymol. 2006;417:205–20. doi: 10.1016/S0076-6879(06)17015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magnani JL, Brockhaus M, Smith DF, Ginsburg V. Detection of glycolipid ligands by direct binding of carbohydrate-binding proteins to thin-layer chromatograms. Methods Enzymol. 1982;83:235–41. doi: 10.1016/0076-6879(82)83016-4. [DOI] [PubMed] [Google Scholar]
- 11.Feizi T, Fazio F, Chai W, Wong CH. Carbohydrate microarrays - a new set of technologies at the frontiers of glycomics. Curr Opin Struct Biol. 2003;13:637–45. doi: 10.1016/j.sbi.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Feizi T, Campanero-Rhodes MA, Childs RA, Zhang Y, Mulloy B, Evans PG, Osborn HM, Otto D, Crocker PR, Chai W. Neoglycolipid probes prepared via oxime ligation for microarray analysis of oligosaccharide-protein interactions. Chem Biol. 2007;14:847–59. doi: 10.1016/j.chembiol.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Disney MD, Seeberger PH. The use of carbohydrate microarrays to study carbohydrate-cell interactions and to detect pathogens. Chem Biol. 2004;11:1701–7. doi: 10.1016/j.chembiol.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 14.de Boer AR, Hokke CH, Deelder AM, Wuhrer M. General microarray technique for immobilization and screening of natural glycans. Anal Chem. 2007;79:8107–13. doi: 10.1021/ac071187g. [DOI] [PubMed] [Google Scholar]
- 15.Song X, Xia B, Lasanajak Y, Smith DF, Cummings RD. Quantifiable fluorescent glycan microarrays. Glycoconj J. 2008;25:15–25. doi: 10.1007/s10719-007-9066-8. [DOI] [PubMed] [Google Scholar]
- 16.Park S, Lee MR, Shin I. Fabrication of carbohydrate chips and their use to probe protein-carbohydrate interactions. Nat Protoc. 2007;2:2747–58. doi: 10.1038/nprot.2007.373. [DOI] [PubMed] [Google Scholar]
- 17.Park S, Lee MR, Pyo SJ, Shin I. Carbohydrate chips for studying high-throughput carbohydrate-protein interactions. J Am Chem Soc. 2004;126:4812–9. doi: 10.1021/ja0391661. [DOI] [PubMed] [Google Scholar]
- 18.Park S, Shin I. Fabrication of carbohydrate chips for studying protein-carbohydrate interactions. Angew Chem Int Ed Engl. 2002;41:3180–2. doi: 10.1002/1521-3773(20020902)41:17<3180::AID-ANIE3180>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 19.Ratner DM, Adams EW, Su J, O’Keefe BR, Mrksich M, Seeberger PH. Probing protein-carbohydrate interactions with microarrays of synthetic oligosaccharides. Chembiochem. 2004;5:379–82. doi: 10.1002/cbic.200300804. [DOI] [PubMed] [Google Scholar]
- 20.Bryan MC, Fazio F, Lee HK, Huang CY, Chang A, Best MD, Calarese DA, Blixt O, Paulson JC, Burton D, Wilson IA, Wong CH. Covalent display of oligosaccharide arrays in microtiter plates. J Am Chem Soc. 2004;126:8640–1. doi: 10.1021/ja048433f. [DOI] [PubMed] [Google Scholar]
- 21.Bigge JC, Patel TP, Bruce JA, Goulding PN, Charles SM, Parekh RB. Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal Biochem. 1995;230:229–38. doi: 10.1006/abio.1995.1468. [DOI] [PubMed] [Google Scholar]
- 22.Bohorov O, Andersson-Sand H, Hoffmann J, Blixt O. Arraying glycomics: a novel bi-functional spacer for one-step microscale derivatization of free reducing glycans. Glycobiology. 2006;16:21C–27C. doi: 10.1093/glycob/cwl044. [DOI] [PubMed] [Google Scholar]
- 23.Xia B, Kawar ZS, Ju T, Alvarez RA, Sachdev GP, Cummings RD. Versatile fluorescent derivatization of glycans for glycomic analysis. Nat Methods. 2005;2:845–50. doi: 10.1038/nmeth808. [DOI] [PubMed] [Google Scholar]
- 24.Song X, Xia B, Stowell SR, Lasanajak Y, Smith DF, Cummings RD. Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem Biol. 2009;16:36–47. doi: 10.1016/j.chembiol.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arsequell G, Dwek RA, Wong SY. 9-Fluorenylmethoxycarbonyl (Fmoc)-glycine coupling of saccharide beta-glycosylamines for the fractionation of oligosaccharides and the formation of neoglycoconjugates. Anal Biochem. 1994;216:165–70. doi: 10.1006/abio.1994.1021. [DOI] [PubMed] [Google Scholar]
- 26.Seko A, Koketsu M, Nishizono M, Enoki Y, Ibrahim HR, Juneja LR, Kim M, Yamamoto T. Occurence of a sialylglycopeptide and free sialylglycans in hen’s egg yolk. Biochim Biophys Acta. 1997;1335:23–32. doi: 10.1016/s0304-4165(96)00118-3. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Li X, Chan K, Zou W, Pribil P, Li XF, Sawyer MB, Li J. “One-pot” methylation in glycomics application: esterification of sialic acids and permanent charge construction. Anal Chem. 2007;79:3894–900. doi: 10.1021/ac070091j. [DOI] [PubMed] [Google Scholar]
- 28.Liu X, McNally DJ, Nothaft H, Szymanski CM, Brisson JR, Li J. Mass spectrometry-based glycomics strategy for exploring N-linked glycosylation in eukaryotes and bacteria. Anal Chem. 2006;78:6081–7. doi: 10.1021/ac060516m. [DOI] [PubMed] [Google Scholar]
- 29.Narahashi Y. Pronase. Methods Enzymol. 1970;19:651–664. [Google Scholar]
- 30.Nisbet AD, Saundry RH, Moir AJ, Fothergill LA, Fothergill JE. The complete amino-acid sequence of hen ovalbumin. Eur J Biochem. 1981;115:335–45. doi: 10.1111/j.1432-1033.1981.tb05243.x. [DOI] [PubMed] [Google Scholar]
- 31.Ogata S, Muramatsu T, Kobata A. Fractionation of glycopeptides by affinity column chromatography on concanavalin A-sepharose. J Biochem. 1975;78:687–96. doi: 10.1093/oxfordjournals.jbchem.a130956. [DOI] [PubMed] [Google Scholar]
- 32.Krusius T, Finne J, Rauvala H. The structural basis of the different affinities of two types of acidic N-glycosidic glycopeptides for concanavalin A--sepharose. FEBS Lett. 1976;72:117–20. doi: 10.1016/0014-5793(76)80911-8. [DOI] [PubMed] [Google Scholar]
- 33.Baenziger JU, Fiete D. Structural determinants of Ricinus communis agglutinin and toxin specificity for oligosaccharides. J Biol Chem. 1979;254:9795–9. [PubMed] [Google Scholar]
- 34.Narasimhan S, Freed JC, Schachter H. The effect of a “bisecting” N-acetylglucosaminyl group on the binding of biantennary, complex oligosaccharides to concanavalin A, Phaseolus vulgaris erythroagglutinin (E-PHA), and Ricinus communis agglutinin (RCA-120) immobilized on agarose. Carbohydr Res. 1986;149:65–83. doi: 10.1016/s0008-6215(00)90370-7. [DOI] [PubMed] [Google Scholar]
- 35.Kornfeld K, Reitman ML, Kornfeld R. The carbohydrate-binding specificity of pea and lentil lectins. Fucose is an important determinant. J Biol Chem. 1981;256:6633–40. [PubMed] [Google Scholar]
- 36.Corradi Da Silva ML, Stubbs HJ, Tamura T, Rice KG. 1H NMR characterization of a hen ovalbumin tyrosinamide N-linked oligosaccharide library. Arch Biochem Biophys. 1995;318:465–75. doi: 10.1006/abbi.1995.1255. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi N, Lee KB, Nakagawa H, Tsukamoto Y, Masuda K, Lee YC. New N-glycans in horseradish peroxidase. Anal Biochem. 1998;255:183–7. doi: 10.1006/abio.1997.2463. [DOI] [PubMed] [Google Scholar]
- 38.Yang BY, Gray JS, Montgomery R. The glycans of horseradish peroxidase. Carbohydr Res. 1996;287:203–12. doi: 10.1016/0008-6215(96)00073-0. [DOI] [PubMed] [Google Scholar]
- 39.Wilson IB, Altmann F. Concanavalin A binding and endoglycosidase D resistance of beta1,2-xylosylated and alpha1,3-fucosylated plant and insect oligosaccharides. Glycoconj J. 1998;15:203–6. doi: 10.1023/a:1006932725821. [DOI] [PubMed] [Google Scholar]
- 40.Yamashita K, Kochibe N, Ohkura T, Ueda I, Kobata A. Fractionation of L-fucose-containing oligosaccharides on immobilized Aleuria aurantia lectin. J Biol Chem. 1985;260:4688–93. [PubMed] [Google Scholar]
- 41.Jang-Lee J, Curwen RS, Ashton PD, Tissot B, Mathieson W, Panico M, Dell A, Wilson RA, Haslam SM. Glycomics analysis of Schistosoma mansoni egg and cercarial secretions. Mol Cell Proteomics. 2007;6:1485–99. doi: 10.1074/mcp.M700004-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Khoo KH, Chatterjee D, Caulfield JP, Morris HR, Dell A. Structural mapping of the glycans from the egg glycoproteins of Schistosoma mansoni and Schistosoma japonicum: identification of novel core structures and terminal sequences. Glycobiology. 1997;7:663–77. doi: 10.1093/glycob/7.5.663. [DOI] [PubMed] [Google Scholar]
- 43.Khoo KH, Huang HH, Lee KM. Characteristic structural features of schistosome cercarial N-glycans: expression of Lewis X and core xylosylation. Glycobiology. 2001;11:149–63. doi: 10.1093/glycob/11.2.149. [DOI] [PubMed] [Google Scholar]
- 44.Nyame AK, Leppanen AM, Bogitsh BJ, Cummings RD. Antibody responses to the fucosylated LacdiNAc glycan antigen in Schistosoma mansoni-infected mice and expression of the glycan among schistosomes. Exp Parasitol. 2000;96:202–12. doi: 10.1006/expr.2000.4573. [DOI] [PubMed] [Google Scholar]
- 45.Simpson AJ, Ali PO, Yi XY, Smithers SR. Schistosome glycoconjugates as antigens and their relevance in experimental and human schistosomiasis. Biochem Soc Trans. 1988;16:264–5. doi: 10.1042/bst0160264. [DOI] [PubMed] [Google Scholar]
- 46.Hokke CH, Deelder AM. Schistosome glycoconjugates in host-parasite interplay. Glycoconj J. 2001;18:573–87. doi: 10.1023/a:1020634602161. [DOI] [PubMed] [Google Scholar]
- 47.Thomas PG, Harn DA., Jr Immune biasing by helminth glycans. Cell Microbiol. 2004;6:13–22. doi: 10.1046/j.1462-5822.2003.00337.x. [DOI] [PubMed] [Google Scholar]
- 48.van Die I, Cummings RD. Glycans modulate immune responses in helminth infections and allergy. Chem Immunol Allergy. 2006;90:91–112. doi: 10.1159/000088883. [DOI] [PubMed] [Google Scholar]
- 49.van Remoortere A, Bank CM, Nyame AK, Cummings RD, Deelder AM, van Die I. Schistosoma mansoni-infected mice produce antibodies that cross-react with plant, insect, and mammalian glycoproteins and recognize the truncated biantennaryN-glycan Man3GlcNAc2-R. Glycobiology. 2003;13:217–25. doi: 10.1093/glycob/cwg025. [DOI] [PubMed] [Google Scholar]
- 50.Fields GB, Noble RL. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int J Pept Protein Res. 1990;35:161–214. doi: 10.1111/j.1399-3011.1990.tb00939.x. [DOI] [PubMed] [Google Scholar]
- 51.Wellings DA, Atherton E. Standard Fmoc protocols. Methods Enzymol. 1997;289:44–67. doi: 10.1016/s0076-6879(97)89043-x. [DOI] [PubMed] [Google Scholar]
- 52.Cudic M, Burstein GD. Preparation of glycosylated amino acids suitable for Fmoc solid-phase assembly. Methods Mol Biol. 2008;494:187–208. doi: 10.1007/978-1-59745-419-3_11. [DOI] [PubMed] [Google Scholar]
- 53.Garg HG, von dem Bruch K, Kunz H. Developments in the synthesis of glycopeptides containing glycosyl L-asparagine, L-serine, and L-threonine. Adv Carbohydr Chem Biochem. 1994;50:277–310. doi: 10.1016/s0065-2318(08)60153-5. [DOI] [PubMed] [Google Scholar]
- 54.Kihlberg J, Elofsson M, Salvador LA. Direct synthesis of glycosylated amino acids from carbohydrate peracetates and Fmoc amino acids: solid-phase synthesis of biomedicinally interesting glycopeptides. Methods Enzymol. 1997;289:221–45. doi: 10.1016/s0076-6879(97)89050-7. [DOI] [PubMed] [Google Scholar]
- 55.Lavielle S, Ling NC, Guillemin RC. Solid-phase synthesis of two glycopeptides containing the amino acid sequence 5 to 9 of somatostatin. Carbohydr Res. 1981;89:221–8. doi: 10.1016/s0008-6215(00)85247-7. [DOI] [PubMed] [Google Scholar]
- 56.Liu M, Barany G, Live D. Parallel solid-phase synthesis of mucin-like glycopeptides. Carbohydr Res. 2005;340:2111–22. doi: 10.1016/j.carres.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 57.Norberg T, Luning B, Tejbrant J. Solid-phase synthesis of O-glycopeptides. Methods Enzymol. 1994;247:87–106. doi: 10.1016/s0076-6879(94)47008-1. [DOI] [PubMed] [Google Scholar]
- 58.Altmann F, Schweiszer S, Weber C. Kinetic comparison of peptide: N-glycosidases F and A reveals several differences in substrate specificity. Glycoconj J. 1995;12:84–93. doi: 10.1007/BF00731873. [DOI] [PubMed] [Google Scholar]
- 59.Chai W, Feizi T, Yuen CT, Lawson AM. Nonreductive release of O-linked oligosaccharides from mucin glycoproteins for structure/function assignments as neoglycolipids: application in the detection of novel ligands for E-selectin. Glycobiology. 1997;7:861–72. doi: 10.1093/glycob/7.6.861. [DOI] [PubMed] [Google Scholar]
- 60.Hounsell EF, Pickering NJ, Stoll MS, Lawson AM, Feizi T. The effect of mild alkali and alkaline borohydride on the carbohydrate and peptide moieties of fetuin. Biochem Soc Trans. 1984;12:607–10. doi: 10.1042/bst0120607. [DOI] [PubMed] [Google Scholar]
- 61.Huang Y, Mechref Y, Novotny MV. Microscale nonreductive release of O-linked glycans for subsequent analysis through MALDI mass spectrometry and capillary electrophoresis. Anal Chem. 2001;73:6063–9. doi: 10.1021/ac015534c. [DOI] [PubMed] [Google Scholar]