Summary
The notochord is a defining feature of the chordate body plan. Experiments in ascidian, frog and mouse embryos have shown that co-expression of Brachyury and FoxA class transcription factors is required for notochord development. However, studies on the cis-regulatory sequences mediating the synergistic effects of these transcription factors are complicated by the limited knowledge of notochord genes and cis-regulatory modules (CRMs) that are directly targeted by both. We have identified an easily testable model for such investigations in a 155-bp notochord-specific CRM from the ascidian Ciona intestinalis. This CRM contains functional binding sites for both Ciona Brachyury (Ci-Bra) and FoxA (Ci-FoxA-a). By combining point mutation analysis and misexpression experiments, we demonstrate that binding of both transcription factors to this CRM is necessary and sufficient to activate transcription. To gain insights into the cis-regulatory criteria controlling its activity, we investigated the organization of the transcription factor binding sites within the 155-bp CRM. The 155-bp sequence contains two Ci-Bra binding sites with identical core sequences but opposite orientations, only one of which is required for enhancer activity. Changes in both orientation and spacing of these sites substantially affect the activity of the CRM, as clusters of identical sites found in the Ciona genome with different arrangements are unable to activate transcription in notochord cells. This work presents the first evidence of a synergistic interaction between Brachyury and FoxA in the activation of an individual notochord CRM, and highlights the importance of transcription factor binding site arrangement for its function.
Keywords: Ciona, Brachyury, FoxA, Notochord, Muscle, Ascidian, T-box, Cis-regulatory module, Transcription
INTRODUCTION
The T-box transcription factor Brachyury and members of the FoxA winged-helix/forkhead box transcription factor orthology group have been demonstrated to play pivotal roles in notochord development (e.g. Friedman and Kaestner, 2006; Herrmann and Kispert, 1994). Evidence for the genetic interaction of Brachyury and FoxA orthologs in notochord formation derives from studies in various model systems, including mouse and ascidian embryos. In mouse, a detailed analysis of the truncate (tc) mutation has shown that this genetic defect, which is characterized by impaired development of the posterior notochord, is due to a point mutation in the Noto gene, which is downregulated in both brachyury and Foxa2 mutant embryos (Abdelkhalek et al., 2004).
In the ascidian Ciona intestinalis (hereinafter referred to as Ciona), the Foxa2 ortholog Ci-FoxA-a (formerly known as forkhead or HNF3-beta) is expressed in the notochord, as well as in central nervous system, endoderm and epidermal precursors (Di Gregorio et al., 2001; Imai et al., 2004). In Ciona and other ascidians, Brachyury is expressed exclusively in notochord cells, which is in contrast to the pan-mesodermal expression of this gene during the early development of vertebrates and amphioxus (Wilkinson et al., 1990; Smith et al., 1991; Yasuo and Satoh, 1993; Holland et al., 1995; Corbo et al., 1997; Takada et al., 2002). In Ciona, expression of Ci-FoxA-a in notochord precursors precedes that of Ci-Brachyury (Ci-Bra), suggesting that Ci-FoxA-a might be acting upstream of Ci-Bra (Di Gregorio et al., 2001). This hypothesis is supported by evidence showing that Ci-Bra expression is abolished in embryos in which Ci-FoxA-a has been knocked down via morpholino oligonucleotides (Imai et al., 2006). A synergistic interaction between Foxa2 and Brachyury orthologs in ascidian notochord formation was first suggested by studies in Halocynthia roretzi, in which it has been shown that co-injection of their respective mRNAs into precursors of non-notochord lineages is sufficient to switch the fates of these blastomeres to notochord (Yasuo and Satoh, 1998; Shimauchi et al., 2001).
Related findings have originated from studies in Xenopus, in which animal caps from embryos co-injected with pintallavis (a Foxa4 class gene) and Xbra mRNAs gave rise to dorsal mesoderm and notochord in a dose-dependent fashion (O'Reilly et al., 1995). Interestingly, a similar synergy has likewise been shown in the non-chordate invertebrate Drosophila, in which the Brachyury and Foxa2 orthologs, brachyenteron (byn) and forkhead (fkh), respectively, cooperate in the specification and formation of the caudal visceral mesoderm, which is absent in byn/fkh double mutant flies and which is ectopically formed in the anterior mesoderm when both genes are overexpressed (Kusch and Reuter, 1999). Together, these reports from phylogenetically distant taxa indicate that the synergistic interactions between Foxa2 and Brachyury orthologs are a key step in the formation of specialized mesodermal compartments.
Despite this large body of evidence on the cooperative function of Foxa2 and Brachyury orthologs in the notochord, there is still no report of a cis-regulatory module (CRM) directly controlled by both of these transcription factors that could be used to explore these interactions at the single-nucleotide level. Therefore, the information on the structure, number and arrangement of the binding sites for these transcription factors, and on possible architectural constraints that might be regulating their interplay, is still very limited. To gather such information, which is crucial for elucidating the molecular mechanisms by which Foxa2 and Brachyury orthologs coordinately control notochord development, we have investigated a 155-bp notochord CRM isolated from Ciona. Rapid embryonic development, together with a compact, fully sequenced genome and the ease of transgenesis, make this invertebrate chordate amenable for the rapid identification and characterization of CRMs controlling gene expression (e.g. Kusakabe, 2005; Shi et al., 2005; Passamaneck and Di Gregorio, 2005).
The notochord CRM described here directs robust gene expression in notochord cells and is physically associated with a novel notochord gene, Ci-tune. The notochord activity of the 155-bp Ci-tune CRM relies upon two distinct cis-regulatory sequences, which are Ci-Bra and Ci-FoxA-a binding sites, respectively. We show that the simultaneous misexpression of Ci-Bra and Ci-FoxA-a is sufficient to trigger ectopic activation of the CRM in developing muscle cells. By combining manipulations of the spatial organization of these sites with genome-wide searches for related sequences, we found evidence that within the Ci-tune CRM, as well as within unrelated but similarly organized genomic sequences, both the orientation and the spacing of Ci-Bra and Ci-FoxA-a binding sites are limiting factors for notochord activity.
MATERIALS AND METHODS
Embryo rearing and cytochalasin B treatment
Adult Ciona intestinalis and Ciona savignyi were purchased from Marine Research and Educational Products (M-REP; Carlsbad, CA, USA) and kept in an aquarium in recirculating artificial sea water at ∼18°C. For cleavage-arrest experiments, fertilized, dechorionated embryos were electroporated at the one-cell stage and reared in filtered seawater until the 110-cell or the early gastrula stage, at which point cytochalasin B was added to a final concentration of 2 μg/ml. Treated embryos were maintained in cytochalasin B solution until untreated control embryos from the same clutch, cultured in parallel, had reached the late-tailbud stage.
Plasmid construction and electroporation
The XPA16474 plasmid consists of a 1655-bp genomic fragment cloned into the pCES vector (Harafuji et al., 2002). All subsequent inserts used for deletion and mutation analysis were generated by PCR amplification using this plasmid as a template, and were cloned into the pFBΔSP6 vector (Oda-Ishii and Di Gregorio, 2007). A list of the oligonucleotides employed for PCR amplifications is provided in Table 1.
Table 1.
Primer sequences for Ci-tune CRM truncations
| Ci-tune CRM truncation* | Forward primer sequence (5′-3′) | Reverse primer sequence (5′-3′) |
|---|---|---|
| B | GTAAACTAGCACGAGGTGTATG | CTGCAGCAGTGATTCATCATGA |
| C | CTCTTGCGAGATGGAGGCGACA | ACAAGGGCATGGCCTCGCAATA |
| D | CTCTTGCGAGATGGAGGCGACA | GATCACGTATAGTGTATGCTA |
| E | GCGTGCAAAATATACCAACTAT | ACAAGGGCATGGCCTCGCAATA |
| F | GCGTGCAAAATATACCAACTAT | AAGTCCGCATTAAAAGCAGGGC |
| G | TGTACTGAATAGTTGCTGATTC | ACAAGGGCATGGCCTCGCAATA |
| H | GCGTGCAAAATATACCAACTAT | GCTCGCTTATTGTTCTCATTTG |
| I | CGCAATCTGTACCTTACAAATG | AAGTCCGCATTAAAAGCAGGGC |
| J | GCGTGCAAAATATACCAACTAT | CTGACTTTGAGTGTGTACGCAA |
| K | TAAATATTGGTTCGTGGCTGAT | AAGTCCGCATTAAAAGCAGGGC |
| L | CGCAATCTGTACCTTACAAATG | GCTCGCTTATTGTTCTCATTTG |
| M | CAAATGAGAACAATAAGCGAGC | CATTTGTAAGGTACAGATTGCG |
Truncation clone names correspond to those in Fig. S1 in the supplementary material.
To create the trop>FoxA::enRD construct, the lacZ sequence was excised from a construct containing a 1.8-kb fragment of the Ci-tropomyosin-like cis-regulatory region (Di Gregorio and Levine, 1999) by digestion with NotI and BlpI, and the plasmid religated in the presence of the annealed oligos Not.Apa.Spe.linker.F (5′-GGCCGCGGAGGAGGGCCCGGAGGAACTAGTGGAGGAGC-3′) and Not.Apa.Spe.linker.R (5′-TCAGCTCCTCCACTAGTTCCTCCGGGCCCTCCTCCG-3′) to generate the vector pFB1.8trop.link. The GFPci (Ciona codon-optimized) coding region (Zeller et al., 2006) was amplified with the primers GFPci.F.Spe (5′-AGGAACTAGTGGAGGAGGAGGAATGTCCAAAGGTGAAGAACT-3′) and GFPci.R.Blp (5′-TTTATTGCTCAGCTTATTTATACAGCTCATCCAT-3′) and cloned into the SpeI/BlpI sites of pFB1.8trop.link to make the pFB1.8trop.GFP intermediate vector. The sequence encoding the DNA-binding domain of Ci-FoxA-a was amplified with the primers FoxDBD.F.Apa (5′-GGGCCCATGGCGCTCATGTTAAACAGAAGGACCG-3′) and FoxDBD.R.Spe (5′-TCCTCCACTAGTTCCAGCGCCCTTGGATTTC-3′) and cloned into the ApaI/SpeI sites of pFB1.8trop.GFP to make the pFB1.8trop.FoxA.GFP cloning intermediate. The sequence encoding the repression domain (RD) of the Drosophila engrailed gene was amplified with the primers En.F.Not (5′-AGCTGCGGCCGCTATGGCCCTGGAGGATCGCTGCAG-3′) and En.R.Apa (5′-GAGCGCCATGGGCCCTCCTCCTCCTCCCAGGGA-3′). The resultant fragment was cloned into the NotI/ApaI sites of pFB1.8trop.FoxA.GFP to produce the trop>FoxA::enRD construct.
To generate the Sna>FoxA-a misexpression construct, the entire open reading frame of Ci-FoxA-a was amplified with the primers CiFoxA-a.F.Nco (5′-GTGGCCATGGTGTTGTCGTCTCCACCGTC-3′) and CiFoxA-a.R.EcoRI (5′-AGCTGGAATTCTTACTTGTACATTAGCTTGC-3′). The resultant fragment was cloned into the NcoI/EcoRI sites of pSP72-1.27-GFP vector (Corbo et al., 1997) to generate the plasmid pSP72-1.27-FoxAa. A 737-bp fragment of the Ci-Snail promoter was amplified from the Sna>eGFP vector (Rhee et al., 2005) with the primers CiSna737.F.Xho (5′-GAACTCGAGCAGCTGAAGCTTGCATGCCTGC-3′) and CiSna737.R.Nco (5′-CAACACCATGGGTACCCGGGGATCCGATGTAA-3′) and cloned into the XhoI/NcoI sites of pSP72-1.27-FoxAa to generate the Sna>FoxA-a construct.
All constructs carrying mutations or alterations in spacing and orientation were generated by PCR amplification. Genomic fragments containing the sites found in the Ci-tune 155-bp CRM, which were identified using the GUFEE program (see below), were PCR-amplified from Ciona genomic DNA as 131-bp fragments and cloned into pFBΔSP6. The sequences of the oligonucleotides employed are shown in Table 2. Plasmid purifications and electroporations were carried out as previously described (Oda-Ishii and Di Gregorio, 2007).
Table 2.
Primer sequences for the GUFEE constructs
| GUFEE construct | Forward primer sequence (5′-3′) | Reverse primer sequence (5′-3′) |
|---|---|---|
| G3 | GTTACCACATTTAAATATATTA | GTGGTTGAAAAAATGTTAGGC |
| G2 | AGGTCCGTATCGGAA | TATAGCAATCAACCAATAAG |
| G8 | GTAGCAATATACCTTGTTGT | CATAAGTCATAAGTGAAC |
| G9 | TTGCATGAGTAAAGCTTAT | CAAATCGACGAATATCATT |
| G7 | TCACTATCATCAGCACA | CAGCTTCACTTTGATTGTGAT |
| G1 | TTTTGGTCAAACCAACATAA | TACCTGTATAAAATTGACTGT |
| G6 | TTGATCAGTTTGTAAACTAC | CACTACTTAAATAAATAATCC |
| G4 | AGCAAATTGTTAAGACACGA | ACACGTGTTTCTGATTGGTTAA |
| G5 | ACACAACTCACTCAAAAATAAT | GTAAAGGCAGATTTTGTTTAT |
| G12 | TACGTTAATTCCAGATGAGCCGC | ATGTGCAATCTAGTTGCACAAAA |
Whole-mount in situ hybridization (WMISH)
A digoxigenin-labeled antisense RNA was synthesized in vitro from the linearized Ci-tune cDNA clone GC05a19. Probe labeling and WMISH experiments were performed as previously reported (Oda-Ishii and Di Gregorio, 2007).
Electrophoretic mobility shift assays (EMSA)
We employed the previously described GST-Ci-Bra (Di Gregorio and Levine, 1999) and GST-Ci-FoxA-a (Dunn and Di Gregorio, 2009) fusion proteins and the following radiolabeled double-stranded oligonucleotides (only the 5′-3′ strand is reported, mutations are underlined):
164T1, 5′-TGTTCTCATTTGCACTCAGACGAG-3′;
164T1m, 5′-TGTTCTCATTTGACATCAGACGAG-3′;
164Fox, 5′-CGAACCAATATTTACTGACTTTGAG-3′; and
164Foxm, 5′-CGAACCAATAGGGCAGGACTTTGAG-3′.
Protein-DNA complexes were formed at room temperature for 30 minutes, then resolved on polyacrylamide gels as previously described (Dunn and Di Gregorio, 2009). For the competition experiments, the concentrations of unlabeled double-stranded oligonucleotides were 1 μM and 5 μM for the low and high concentrations, respectively.
Genome-wide and database searches for related combinations of Ci-Bra and Ci-FoxA-a binding sites
We used the program GUFEE (Genomic Utility For finding Enhancer Elements), a command-line software written in PERL that we developed, to scan the latest available assembly of the Ciona intestinalis genome (version 2.0, October 2002; http://genome.jgi-psf.org/Cioin2/Cioin2.download.ftp.html) and to search a database of genomic sequences from bona fide Ci-Bra target genes that had been previously identified (e.g. Takahashi et al., 1999a).
RESULTS
The XPA16474 sequence is a bona fide transcriptional enhancer for the Ciona gene ci0100137819
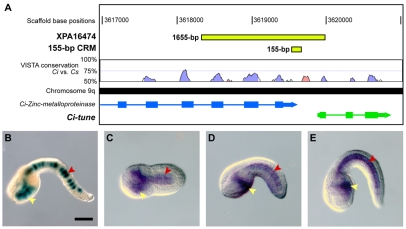
The 1655-bp XPA16474 sequence (Fig. 1A) was isolated through a screen of random genomic fragments for cis-regulatory activity, essentially as previously described (Harafuji et al., 2002). The XPA16474 plasmid was found to direct strong notochord and endoderm expression in the majority of the embryos transfected (Fig. 1B) and was therefore chosen for further analysis. BLAST searches (Altschul et al., 1990) of the Ciona intestinalis genome (http://genome.jgi-psf.org/Cioin2/Cioin2.home.html) (Dehal et al., 2002) showed that the XPA16474 sequence straddles two closely neighboring gene models, ci0100137797 and ci0100137819, on chromosome 9q (Shoguchi et al., 2006). The XPA16474 sequence encompasses the last three exons and intervening introns of ci0100137797, which encodes a putative Zinc metalloproteinase, as well as the 5′-flanking region and part of the first exon of ci0100137819, a short (934 bp in total) unannotated gene model (Fig. 1A).
Fig. 1.
The cis-regulatory activity of the XPA16474 sequence recapitulates the expression of the gene that it flanks. (A) Mapping of the XPA16474 sequence (green rectangle at the top) to the Ciona intestinalis genome shows that this fragment covers the last three exons (rectangles) and intervening introns (thin lines) of the Ci-Zinc-metalloproteinase gene (ci0100137797), as well as an intergenic region and the first exon of a neighboring gene, which we named Ci-tune (ci0100137819). The location of the minimal 155-bp CRM (detailed in Fig. S2 in the supplementary material) is also shown (smaller green rectangle). VISTA phylogenetic footprint (http://pipeline.lbl.gov/cgi-bin/gateway2) of this region in Ciona intestinalis and Ciona savignyi was performed employing the following parameters: calculation window, 100 bp; minimum conservation width, 100 bp; conservation identity, 50%. Conserved coding regions are depicted as blue peaks, conserved non-coding sequences as pink peaks. (B) X-Gal staining of a late-tailbud Ciona embryo electroporated at the one-cell stage with the 1655-bp XPA16474 sequence. (C-E) Detection of Ci-tune transcripts by WMISH reveals expression in the notochord and trunk endoderm at (C) early-tailbud, (D) mid-tailbud and (E) late-tailbud stages. In B, D and E, embryos are oriented with anterior to the left and dorsal up; C is a ventral view, anterior to the left. Yellow arrowheads indicate endodermal cells; red arrowheads, notochord cells. Ci, Ciona intestinalis; Cs, Ciona savignyi. Scale bar: 50 μm.
For the Zinc metalloproteinase gene, whole-mount in situ hybridization (WMISH) data available from the Ghost database (http://ghost.zool.kyoto-u.ac.jp) reported that weak expression was seen throughout the body of tailbud embryos, and particularly in notochord, papillae and mesenchyme cells (Satou et al., 2001). In the case of ci0100137819, for which no in situ data were available, our WMISH experiments on embryos at various developmental stages revealed strong expression in notochord and endoderm, starting from the neurula stage (Fig. 1C-E). This pattern was faithfully recapitulated by the XPA16474 transgene (Fig. 1B). We have named the gene corresponding to this locus Ci-tune (tunicate notochord and endoderm), because of its expression pattern. We performed 5′-RACE (rapid amplification of cDNA ends) experiments to validate the predicted gene model (Frohman, 1993), and no significant differences in the length of its coding region were observed (data not shown). The putative protein product of Ci-tune spans 116 amino acid residues and does not contain any apparent conserved domains, but it is tentatively indicated as a putative component of the secretory pathway by publicly available prediction software (SignalP 3.0; http://www.cbs.dtu.dk/services/SignalP/) (Emanuelsson et al., 2007).
TBLASTN searches (Altschul et al., 1990) of publicly available genomic and expressed sequence tags (ESTs) identified an ortholog of Ci-tune in the sister species Ciona savignyi (E-value=1×10-38) and a putative ortholog in the stolidobranch tunicate Molgula tectiformis (E-value=3×10-5). BLAST searches of datasets for amphioxus (Branchiostoma floridae), sea urchin (Strongylocentrotus purpuratus) and other deuterostomes failed to identify any sequence similarity outside of that of tunicates.
The XPA16474 enhancer is controlled by Ci-Bra and Ci-FoxA-a via a compact cluster of binding sites
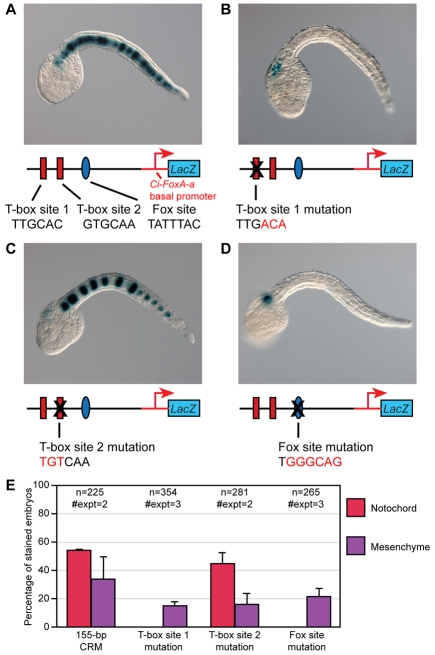
To identify the minimal cis-regulatory sequences required for notochord activity of the 1655-bp XPA16474 sequence, we created a series of 10 truncations and tested their activity following electroporation into Ciona embryos (see Fig. S1 in the supplementary material). These experiments narrowed down the active sequence to a 155-bp CRM, which still directed intense and consistent notochord staining regardless of its orientation (Fig. S1 in the supplementary material) and which was therefore selected for further analyses. Sequence inspection of the 155-bp CRM (Fig. 2A) revealed the presence of two identical T-box half-sites (TTGCAC) with opposite orientation and one putative Fox-binding site (TATTTAC). We focused on analyzing these T-box and Fox sites because of the well-known function of both Ci-Bra and Ci-FoxA-a in notochord formation in ascidians, and we mutated these sequences to assess their respective contributions to the CRM activity. Mutation of either the distal-most T-box site (`T-box site 1'; Fig. 2B) or the putative Fox site (Fig. 2D) caused a complete loss of notochord activity. By contrast, mutation of the proximal T-box site (`T-box site 2') did not have any effect (Fig. 2C). A quantification of the effects of these mutations is shown in Fig. 2E.
Fig. 2.
Requirement of T-box and Fox transcription factor binding sites for notochord activity. (A-D) Late-tailbud Ciona embryos electroporated at the one-cell stage with different variants of the 155-bp minimal notochord CRM and stained for β-galactosidase (blue). Not all 40 notochord cells are stained, owing to mosaic incorporation of the transgene(s). (A) Embryo electroporated with the 155-bp wild-type notochord CRM. The schematic underneath depicts the 155-bp sequence, with two predicted T-box transcription factor binding sites (red rectangles) and one putative Fox transcription factor binding site (blue oval), cloned upstream of the Ci-FoxA-a basal promoter (red line and arrow) and of the lacZ reporter gene (blue box; not drawn to scale). (B) Embryo electroporated with the 155-bp CRM carrying a mutation in the distal T-box site (T-box site 1). (C) Embryo electroporated with the 155-bp CRM in which the proximal T-box site (T-box site 2) has been mutated. (D) Embryo electroporated with the 155-bp CRM carrying a mutant Fox site. (E) Percentage of embryos with notochord and mesenchyme staining following electroporation with either wild-type or mutagenized plasmids. All properly developed embryos, with and without staining, were scored; embryos showing staining in both mesenchyme and notochord were scored separately for each tissue.
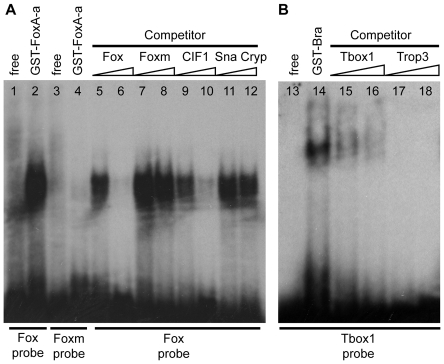
The binding of Ci-FoxA-a and Ci-Bra to their putative target sites was tested by electrophoretic mobility shift assays (EMSA). As shown in Fig. 3A, a radiolabeled, double-stranded oligonucleotide containing the putative Ci-FoxA-a site (the `Fox probe') was bound very efficiently by a GST-Ci-FoxA-a fusion protein, whereas a mutant probe (`Foxm') bearing the same mutation that abolishes notochord activity in vivo (shown in Fig. 2D) was not recognized by the same amount of protein. Similarly, in competition assays, only increasing amounts of the unlabeled wild-type Ci-FoxA-a site inhibited binding of the radiolabeled probe by Ci-FoxA-a, while the unlabeled mutant competitor Foxm did not interfere with the formation of the protein-DNA complex. A previously published high-affinity Ci-FoxA-a binding site, CIF1, competed efficiently the wild-type Fox probe, whereas an unlabeled, unrelated competitor, Sna Cryp, which is a known binding site for Ci-Snail (Di Gregorio et al., 2001), did not. As shown in Fig. 3B, the T-box1 probe was efficiently bound in vitro by a GST-Ci-Bra fusion protein. This binding was abolished in reactions containing increasing amounts either of the unlabeled T-box1 oligonucleotide or of an unlabeled oligonucleotide containing one of the previously published Ci-Bra binding sites found in the Ci-tropomyosin-like (Ci-trop) notochord CRM (Trop3, previously known as Ci-Bra #3) (Di Gregorio and Levine, 1999). A probe containing the dispensable T-box site 2 was likewise bound by the GST-Ci-Bra fusion protein (data not shown).
Fig. 3.
In vitro binding of the sequences necessary for notochord activity of the Ci-tune CRM by the Ci-Bra and Ci-FoxA-a proteins. (A) Electrophoretic mobility shift assays (EMSA) with a radiolabeled probe containing the predicted Ci-FoxA-a binding site and a GST-Ci-FoxA-a fusion protein. Lanes contained: (1,2) radiolabeled probe containing the predicted Ci-FoxA-a binding site, (1) free, and (2) incubated with the GST-Ci-FoxA-a fusion protein; (3,4) radiolabeled probe containing the mutated Ci-FoxA-a binding site, (3) free, and (4) incubated with GST-Ci-FoxA-a; (5-12) radiolabeled probe containing the predicted Ci-FoxA-a binding site and unlabeled competitors, at 1 μM (lanes 5,7,9,11) and 5 μM (lanes 6,8,10,12) concentrations, incubated with GST-Ci-FoxA-a. (B) EMSA with radiolabeled probe and a GST-Ci-Bra fusion protein. Lanes contained: (13,14) radiolabeled probe containing the predicted Ci-Bra binding site T-box site 1, (13) free, and (14) incubated with GST-Ci-Bra fusion protein; (15-18) radiolabeled T-box site 1 probe and unlabeled competitors, at 1 μM (lanes 15,17) and 5 μM (lanes 16,18) concentrations, incubated with GST-Ci-Bra.
Interestingly, no significant phylogenetic conservation between Ciona intestinalis and Ciona savignyi was seen within the 155-bp CRM, for either the T-box or the Fox sites (see Fig. S2A in the supplementary material). However, electroporation of the 1655-bp XPA16474 sequence into C. savignyi produced staining in ∼30% of the embryos; the construct was active in both endoderm and notochord, although the number of notochord cells displaying staining in C. savignyi was considerably lower than that in C. intestinalis (see Fig. S2B-D in the supplementary material). In contrast to what is seen in C. intestinalis, where the 155-bp CRM functions as a notochord-specific enhancer, this CRM retains activity in both these tissues when electroporated into C. savignyi embryos (see Fig. S2E in the supplementary material). Notably, sequence inspection revealed that the C. savignyi sequence corresponding to the Ci-tune CRM also contains two T-box sites and one Fox-binding site, although with different cores and orientations (highlighted in Fig. S2 in the supplementary material). These observations suggest that even though the C. savignyi sequence does not contain the same Brachyury- and Fox-binding sites that are found in the C. intestinalis sequence, the cis-regulatory logic of the Ci-tune CRM is, to some extent, conserved.
Repressor forms of Ci-Bra and Ci-FoxA-a abolish notochord activity of the 155-bp Ci-tune CRM
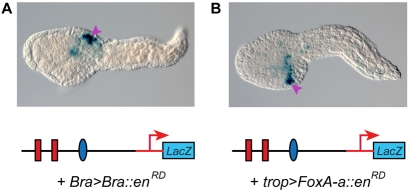
After specific binding of both Ci-FoxA-a and Ci-Bra to the sites found in the Ci-tune CRM was verified in vitro through EMSA, we tested the effects of synthetic repressor forms of each transcription factor on the activity of the Ci-tune CRM. When the 155-bp CRM was co-electroporated into Ciona zygotes along with the Bra>Bra::enRD plasmid, which expresses a repressor form of Ci-Bra in notochord cells (Kugler et al., 2008), staining was detected only in mesenchyme cells (Fig. 4A). This matched the background staining that was observed when the empty parent vector was transfected (data not shown).
Fig. 4.
Inhibition of notochord activity of the Ci-tune CRM by repressor forms of Ci-Bra and Ci-FoxA-a. (A) Late-tailbud embryo co-electroporated with the 155-bp notochord CRM and with the Bra>Bra::enRD construct, which directs notochord expression of Ci-Bra fused to the repression domain of Drosophila Engrailed (enRD); stained for β-galactosidase (blue). (B) Late-tailbud stage embryo co-electroporated with the 155-bp notochord CRM and a repressor form of Ci-FoxA-a under the control of the Ci-tropomyosin-like (Ci-trop) notochord promoter (trop>FoxA-a::enRD); stained for β-galactosidase (blue). Purple arrowheads indicate mesenchyme cells.
To assess the effects of a repressor form of Ci-FoxA-a, we fused the sequence encoding the Ci-FoxA-a DNA-binding domain to the sequence encoding the Drosophila Engrailed repression domain (enRD). Because early expression of this repressor under the control of the Ci-Bra promoter adversely affected overall morphogenesis (data not shown), we replaced the Ci-Bra promoter with the Ci-trop promoter. The Ci-trop promoter is a target of Ci-Bra and hence it is activated a few cell divisions later than the Ci-Bra promoter; the Ci-trop promoter has been employed in previous studies to delay and mitigate the effects of other transcription factors (Di Gregorio et al., 2002). As expected, the trop>FoxAa::enRD plasmid produced a milder phenotype and allowed a clear visualization of the repression of notochord activity of the 155-bp CRM (Fig. 4B). The notochord-specific effect of the repressor form of Ci-FoxA-a was confirmed by the fact that the background mesenchyme activity of the vector was still detectable (purple arrowhead in Fig. 4B).
Context-independent sufficiency of Ci-FoxA-a and Ci-Bra for the activation of the Ci-tune CRM
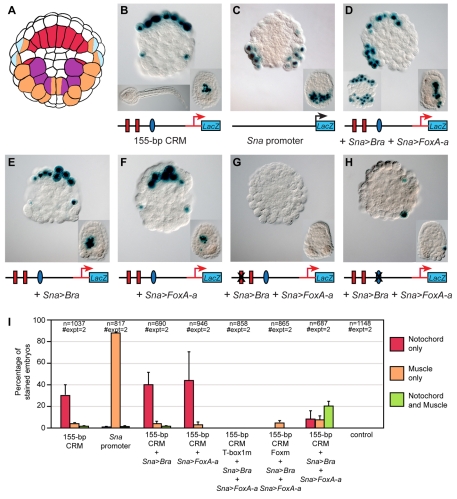
Having demonstrated the necessity of both Ci-Bra and Ci-FoxA-a for notochord activity of the 155-bp CRM, we next tested the sufficiency of these transcription factors for ectopic activation of the Ci-tune CRM by misexpression assays. We chose to misexpress Ci-Bra and Ci-FoxA-a in muscle cells because muscle represents a tractable territory where neither transcription factor is normally expressed. To this end, we cloned their coding regions downstream of the Ci-Snail (Ci-Sna) promoter region, which is active in muscle precursors by the onset of gastrulation (Erives et al., 1998). Because misexpression of Ci-Bra and Ci-FoxA-a in muscle cells caused morphogenetic perturbations that complicated their identification, we performed all misexpression experiments with embryos that were cleavage arrested at the 110-cell stage (Fig. 5A), or at the early gastrula stage, by treatment with cytochalasin B.
Fig. 5.
Requirement of both Ci-Bra and Ci-FoxA-a for ectopic activation of the 155-bp notochord CRM. (A) Simplified lineage map of the vegetal half of the Ciona embryo at the 110-cell stage. Notochord precursors are labeled in red, muscle precursors in orange, trunk mesenchyme in purple and neural precursors in light blue. Precursors of two different lineages are labeled with two colors. (B-H) Embryos electroporated at the one-cell stage with the constructs indicated underneath each panel. Binding sites that have been mutated are covered by an `X'. Embryos were cleavage arrested with cytochalasin B at the 110-cell stage and stained for β-galactosidase (blue). In all panels, the insets at the bottom right show embryos treated with cytochalasin B at the early gastrula stage. (B) Activity of the 155-bp Ci-tune CRM. Bottom left inset shows an untreated control embryo from the same clutch, grown in parallel with the cleavage-arrested embryos shown in all panels. (C) Activity of the Ci-Sna promoter, which was used to drive misexpression of Ci-Bra and Ci-FoxA-a. (D) Triple electroporation of the 155-bp CRM with misexpression constructs for Ci-Bra (Sna>Bra) and Ci-FoxA-a (Sna>FoxA-a) showing ectopic muscle staining only on one side, due to mosaic incorporation. Bottom left inset depicts another embryo from the same experiment, showing bilateral muscle staining. (E) Co-electroporation of the 155-bp CRM with the Sna>Bra misexpression construct. (F) Co-electroporation of the 155-bp CRM with the Sna>FoxA-a misexpression construct. (I) Graph showing the percentage of cleavage-arrested embryos with staining in notochord (red bars), muscle (orange bars), or in both tissues (green bars). The transgenes employed are indicated underneath the x-axis.
Embryos electroporated at the one-cell stage with the 155-bp Ci-tune CRM and cleavage arrested at the 110-cell stage displayed staining in notochord precursors (Fig. 5B), whereas embryos electroporated with the 737-bp Ci-Sna promoter upstream of lacZ (Erives et al., 1998) had staining only in the muscle and mesenchyme lineages (Fig. 5C). Triple electroporation of the 155-bp CRM with both the Sna>Bra (Oda-Ishii and Di Gregorio, 2007) and Sna>FoxA-a misexpression constructs resulted in ectopic staining in muscle precursors, in addition to the notochord and mesenchyme staining observed when the 155-bp CRM alone was electroporated (Fig. 5D). To validate that ectopic activation of the 155-bp CRM was dependent upon concomitant expression of Ci-Bra and Ci-FoxA-a, we co-electroporated it with each misexpression construct individually. In embryos co-electroporated with only the 155-bp CRM and the Sna>Bra misexpression construct, staining was present in the notochord, but no ectopic staining in muscle was observed (Fig. 5E). Likewise, when the 155-bp CRM was co-electroporated with the Sna>FoxA-a construct, staining was still restricted to the notochord (Fig. 5F).
Lastly, we tested the dependence of the ectopic CRM activation upon the previously identified Ci-Bra and Ci-FoxA-a binding sites by co-electroporating Sna>Bra and Sna>FoxA-a with mutant versions of the 155-bp CRM lacking either the T-box site 1 or the Fox site. The triple electroporation of the T-box site 1 mutant produced no staining (Fig. 5G); the triple electroporation of the Fox site mutant resulted in sporadic activity in one or two notochord and/or muscle cells (Fig. 5H). For all misexpression experiments, comparable results were obtained in embryos treated with cytochalasin B at the early gastrula stage, which showed a larger number of stained notochord and/or muscle precursors (insets in Fig. 5B-H). Quantitative data for these experiments are shown in the graph in Fig. 5I and low-magnification group micrographs are presented in Fig. S3 in the supplementary material.
Together with the data obtained from the mutation analyses, these results provide evidence that Ci-Bra and Ci-FoxA-a are both necessary and sufficient to activate transcription driven by the Ci-tune CRM.
Effects of changes in orientation of the Ci-Bra and Ci-FoxA-a binding sites on the notochord activity of the Ci-tune CRM
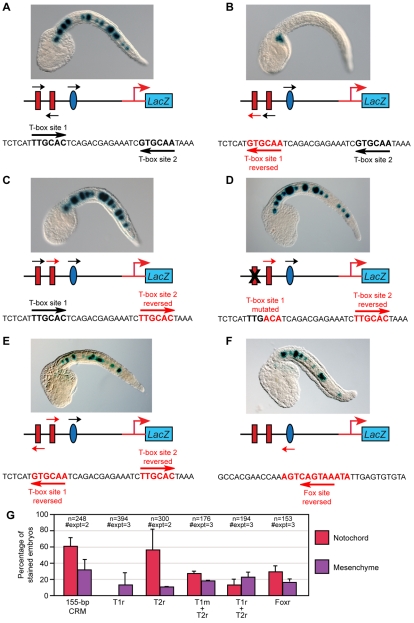
The observation that only one of the two T-box sites in the 155-bp minimal notochord CRM, namely T-box site 1 (Fig. 2B), was required for notochord activity prompted us to investigate the effects of changes in the organization of these sites on cis-regulatory activity. Because the two T-box sites have identical core sequences but are arranged in opposite orientations (Fig. 6A), we first tested the effects of changes in their palindromic arrangement. Similarly to orthologous proteins from other model systems, Ci-Bra is able to bind palindromic sites, possibly in the form of a dimer (Di Gregorio and Levine, 1999), with dimerization likely being mediated by the evolutionarily conserved PDSPNF amino acid motif within its T-domain (Kusch et al., 2002). A mutation that generated the reverse complement of the 6-bp core sequence of the T-box site 1, thus destroying the palindromic arrangement of the T-box sites, completely eliminated notochord staining (Fig. 6B,G). By contrast, reversal of the T-box site 2, which is not required for CRM function (Fig. 2C), did not decrease notochord activity (Fig. 6C,G). However, reversal of T-box site 2 was sufficient to compensate for the mutation of T-box site 1 and to partially restore notochord activity (Fig. 6D,G). A mutant Ci-tune CRM carrying both T-box sites in a reversed orientation retained notochord activity, although both the percentage of stained embryos and the qualitative intensity of the staining were below the levels driven by the wild-type CRM (Fig. 6E,G; see also Fig. S4E in the supplementary material). These results show that the palindromic arrangement of the T-box sites is not required.
Fig. 6.
Effects of changes in the orientation of the Ci-Bra and Ci-FoxA-a binding sites on notochord activity of the Ci-tune CRM. Late-tailbud embryos electroporated at the one-cell stage with either the wild-type 155-bp Ci-tune CRM or with one of its mutant versions, schematized underneath each microphotograph. Black arrows indicate the native orientation of the sites, red arrows indicate the new orientation created by mutagenesis. Embryos are stained for β-galactosidase (blue). (A) In the wild-type Ci-tune CRM, the cores of T-box site 1 and T-box site 2 are identical, but are oriented in opposite directions. (B) Reversal of T-box site 1. (C) Reversal of T-box site 2. (D) Reversal of T-box site 2, combined with a mutation of T-box site 1. (E) Reversal of both T-box sites. (F) Reversal of the Ci-FoxA-a site. (G) Quantification of notochord and mesenchyme staining following electroporation with the wild-type 155-bp CRM and with its derivatives in which the Ci-Bra and Ci-FoxA-a sites are reversed and/or mutated. Embryos were scored as in Fig. 2. T1r, T-box site 1 reversed; T1m, T-box site 1 mutated; T2r, T-box 2 site reversed; Foxr, Fox site reversed.
The inversion of the Fox site (Fig. 6F,G) also reduced, but did not abolish, notochord activity, and was unable to rescue either the mutation or the inversion of the T-box site 1 (see Fig. S4A,B in the supplementary material). Likewise, the inversion of the Fox site did not have any visible effect on either the mutation or the inversion of the T-box site 2 (Fig. S4C,D in the supplementary material); however, the percentage of embryos showing notochord staining was reduced when the Fox site was reversed in a construct where both T-box sites were reversed (see Fig. S4E,F in the supplementary material). Constructs containing the CRM in either syn (5′→3′) or anti (3′→5′) orientation with respect to the basal promoter, produced equivalent notochord staining (Fig. S1 in the supplementary material).
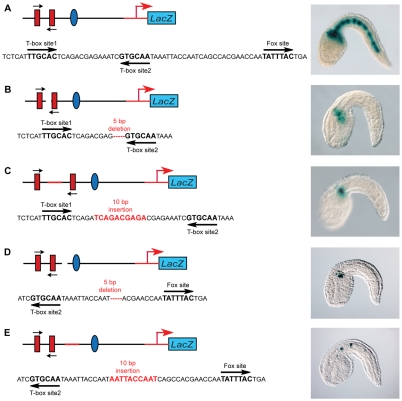
To evaluate the role of binding site spacing on CRM function, we introduced a series of insertions or deletions in the 155-bp sequence to alter the distance between the Ci-Bra and Ci-FoxA-a sites required for CRM function. In the wild-type 155-bp CRM, the core T-box binding sites are spaced by 14 bp and T-box site 2 is separated from the Fox site by 26 bp (Fig. 7A). Deletion of a 5-bp fragment from the sequence in between the two Ci-Bra sites completely eliminated notochord activity (Fig. 7B). CRM function was also lost when 5 bp (data not shown) or 10 bp were inserted between these sites (Fig. 7C). Similarly, the deletion of 5 bp between Ci-Bra and Ci-FoxA-a ablated notochord activity (Fig. 7D), and the insertion of 10 bp in this region drastically reduced activity (Fig. 7E).
Fig. 7.
Effects of changes in the spacing of the binding sites on notochord activity of the Ci-tune CRM. Late-tailbud embryos electroporated at the one-cell stage with either the wild-type 155-bp Ci-tune CRM or with versions containing insertions or deletions, schematized to the left of each micrograph. Embryos are stained for β-galactosidase (blue). (A) Wild-type Ci-tune CRM with the two Ci-Bra binding sites with identical core sequences spaced by 14 bp, and the Ci-FoxA-a binding site separated from T-box site 2 by 26 bp. (B) Deletion of 5 bp within the 14-bp spacer between the Ci-Bra binding sites. (C) Insertion of 10 bp in the 14-bp spacer. (D) Deletion of 5 bp between the T-box site 2 and the Ci-FoxA-a binding sites. (E) Insertion of 10 bp between the T-box site 2 and the Ci-FoxA-a binding sites nearly abolishes notochord activity, which is detected in a few cells in ∼9% of the transgenic embryos.
To determine whether the presence of Ci-Bra and Ci-FoxA-a binding sites is a recurring feature of a class of co-regulated notochord CRMs, we sought to identify genomic regions that contained clustered binding sites matching the Ci-Bra and Ci-FoxA-a site core sequences identified in the 155-bp Ci-tune CRM. To narrow down the results, we included in our query an additional nucleotide from the 5′-end of each of the T-box sites, leaving the Ci-FoxA-a site unchanged, and we narrowed the width of the targeted intervals to 70 bp. This search retrieved 14 hits, four of which were duplicates, and one of which was, as expected, the Ci-tune CRM, which therefore provided an internal control for our software. The sequences containing the remaining nine hits were separately PCR-amplified, cloned and tested in vivo. Of these nine constructs, none was found to have notochord activity (see Fig. S5 in the supplementary material). Additionally, a construct containing the Ci-Bra sites in the same orientation and spacing as the Ci-tune CRM but with a differently spaced Ci-FoxA-a site did not have notochord activity (G12 in Fig. S5 in the supplementary material). However, searches focused to the genomic loci of known Ci-Bra target genes showed that putative Ci-Bra and Ci-FoxA-a binding sites were often associated within short sequence intervals (data not shown), suggesting that some of these genes might be controlled by both transcription factors.
DISCUSSION
Combinatorial control of a Ciona notochord CRM by FoxA-a and Brachyury
The Ci-tune notochord CRM that we have described was identified through a position-unbiased genome-wide screen for Ciona CRMs (e.g. Harafuji et al., 2002). To verify that this sequence acted as a bona fide transcriptional enhancer within a genomic context, we analyzed the similarities between its territories of activity and the expression patterns of its neighboring genes. Our data show that the expression pattern of the 3′ neighboring gene, which we named Ci-tune, is faithfully recapitulated by this CRM, which in its `full-length' form is active in notochord and endoderm, although we do not rule out the possibility that this CRM might also control notochord expression of its 5′ neighbor, Ci-Zinc metalloproteinase, possibly as a long-range regulatory module.
A 155-bp CRM was found to be the minimal functional sequence with notochord activity. This sequence was subjected to mutation analysis and was found to require for its activity one T-box site and one putative Fox-binding site. Specific binding of these sites by Ci-Bra and Ci-FoxA-a, respectively, was verified in vitro by EMSA, and the binding of Ci-Bra to this sequence was confirmed by preliminary chromatin immunoprecipitation assays (data not shown); the requirement of both transcription factors for CRM notochord activity was confirmed in vivo by co-electroporation with dominant-negative forms of each transcription factor. Lastly, we used a misexpression assay to validate that the co-expression of Ci-FoxA-a and Ci-Bra is necessary and sufficient for activation of the 155-bp CRM. Together, these results demonstrate that Ci-FoxA-a and Ci-Bra both bind sites in the 155-bp CRM, and that a combinatorial effect of these two transcription factors is required for notochord activity.
Our observations of the ectopic activation of a notochord CRM in a non-notochord lineage complement previously published results, which showed that co-injection of mRNAs for Foxa2 and Brachyury ascidian orthologs into non-notochord blastomeres of the ascidian H. roretzi is sufficient to downregulate non-notochord differentiation markers and to ectopically induce notochord-like features (Yasuo and Satoh, 1998; Shimauchi et al., 2001).
A working model for the synergistic activation of notochord gene expression by Ci-FoxA-a and Ci-Bra
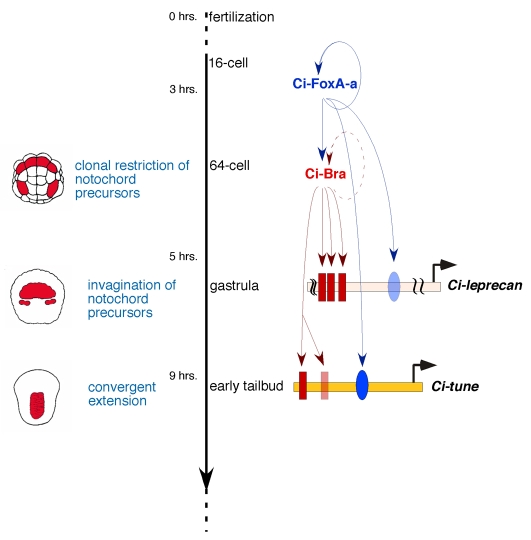
A simplified model that recapitulates the synergistic interactions between Ci-FoxA-a and Ci-Bra at the level of individual CRMs during ascidian notochord development is shown in Fig. 8. Ci-FoxA-a transcripts become visible in the cytoplasm starting from the 16-cell stage (Di Gregorio et al., 2001); as the mature Ci-FoxA-a protein accumulates, it positively regulates its own transcription by binding to autoregulatory sites found in the Ci-FoxA-a cis-regulatory region (Di Gregorio et al., 2001). In addition, in notochord cells, Ci-FoxA-a positively contributes to the transcription of Ci-Bra, as shown by the downregulation of Ci-Bra in embryos in which expression of Ci-FoxA-a has been knocked down (Imai et al., 2006), and by the presence of sites bound in vitro by Ci-FoxA-a in the Ci-Bra 5′-flanking region (A.D.G., unpublished).
Fig. 8.
Model for the synergistic function of Ci-FoxA-a and Ci-Bra during ascidian notochord development. Model for the interactions between Ci-FoxA-a and Ci-Bra at the level of individual notochord CRMs during ascidian development. On the left side are schematic representations of embryos at the 64-cell (top), gastrula (middle) and early tailbud (bottom) stages. Notochord cells and their precursors are colored in red. The main events in notochord formation (blue font) are plotted against the developmental timeline (black vertical arrow) for Ciona embryos at the temperature of 18°C. Two CRMs controlled by Ci-FoxA-a and Ci-Bra, Ci-leprecan (Dunn and Di Gregorio, 2009) and Ci-tune, are depicted as horizontal colored bars. Red rectangles symbolize sites bound in vitro by Ci-Bra, blue ovals indicate sites bound in vitro by Ci-FoxA-a. Faded colors are used to indicate sites that play minor or no role in notochord activity. Circular arrows indicate autoregulatory loops on the transcription factor promoters, either demonstrated (continuous line) or inferred (dotted line). For simplicity, the Ci-FoxA-a and Ci-Bra CRMs are omitted.
In the ascidian H. roretzi, Brachyury has been shown to regulate its own transcription through a binding site found in its minimal promoter sequence (Takahashi et al., 1999b). The presence of putative T-box binding sites in the minimal Ci-Bra CRM (data not shown) indicates that autoregulation of Brachyury transcription might occur also in Ciona. Ci-Bra in turn activates transcription of at least 50 notochord genes (Takahashi et al., 1999a; Di Gregorio and Levine, 1999; Hotta et al., 2000; Hotta et al., 2008; Oda-Ishii and Di Gregorio, 2007; Kugler et al., 2008), among which are Ci-leprecan (Dunn and Di Gregorio, 2009) and Ci-tune (presented here). Both the Ci-leprecan and the Ci-tune notochord CRMs contain sites that are efficiently bound in vitro by Ci-FoxA-a and by Ci-Bra (Fig. 8, indicated by blue ovals and red rectangles, respectively). However, in the case of Ci-leprecan, the Ci-FoxA-a site contributes only minimally to notochord activity (Fig. 8, faded blue oval) (Dunn and Di Gregorio, 2009); in the case of Ci-tune, the Ci-FoxA-a site is indispensable (Fig. 8, dark blue oval). The Ci-Bra binding sites in the Ci-leprecan CRM are all involved, although to different extents, in notochord activity (Fig. 8, dark red rectangles), whereas one of the Ci-Bra binding sites in the Ci-tune notochord CRM is dispensable (Fig. 8, faded red rectangle). Notably, the core sequences of the Ci-Bra binding sites in the Ci-leprecan CRM all differ from each other (i.e. are various permutations of the TNNCAC generic core consensus sequence) (Dunn and Di Gregorio, 2009) and also differ from the core sequence of the Ci-Bra binding sites in the Ci-tune CRM. Similarly, the sequence of the Ci-FoxA-a binding site found in the region of the Ci-leprecan CRM depicted in Fig. 8 has seven out of 10 matches with the extended consensus sequence that we used as a reference (WTRTTKRYTY) (Qian and Costa, 1995) (our unpublished observations), whereas the Ci-FoxA-a binding site found in the Ci-tune CRM fits the consensus more stringently (9/10 matches). These observations suggest that the variable number and combinations of Ci-FoxA-a and Ci-Bra binding sites with slightly different core sequences and binding affinities might ensure a fine-tuned control of the transcriptional onset and overall expression levels of the genes targeted by these factors.
Effects of architectural alterations on the Ci-tune cis-regulatory information
The two Ci-Bra binding sites found in the Ci-tune CRM have identical cores (TTGCAC) but opposite orientation. Despite their identical core sequence, the functional requirements for these sites are widely different, with the distal Ci-Bra binding site (T-box site 1) being necessary for activity, and the proximal Ci-Bra binding site (T-box site 2) being dispensable. This observation suggested that their orientation might be a determinant for their activity.
To test this hypothesis, we prepared a variant of the 155-bp Ci-tune CRM, in which we simultaneously mutated the T-box site 1 and reversed the orientation of the T-box site 2. We found that, in this case, the mutation of the T-box site 1 was partially compensated by the reversal of the T-box site 2, as was the reversal of T-box site 1 in a construct in which the T-box site 2 is also reversed. These results seem to suggest that as long as one of the two T-box sites is in the 5′→3′ orientation, the CRM is able to function. Based on the results of both the reversal and the mutation experiments, this orientation is likely to be independent from that of the other T-box site; this latter observation, in turn, rules out the possibility that the palindromic arrangement of T-box site 1 and T-box site 2 is required for their activity and binding by Ci-Bra.
The orientation of transcription factor binding sites has been proven to be required for the full activity of CRMs identified in other model systems, particularly in Drosophila melanogaster, in which the orientation of REL and GATA binding sites has been studied in various CRMs controlling the expression of immunity genes in the fat body (Senger et al., 2004). In this particular case, the inversion of either binding site caused a 2- or 3-fold reduction in activity in most of the CRMs studied; these effects were abolished by the inversion of both sites (Senger et al., 2004). Analogous results were obtained when the orientations of linked binding sites for Dorsal, Twist and Snail were reverted within CRMs active in the ventral neurogenic ectoderm of the early fly embryo (Zinzen et al., 2006). The results reported here show that similar syntactic rules apply to the Ci-tune CRM.
When we tested the effects induced on notochord activity by alterations of the spacing between the binding sites found in the 155-bp CRM, we found that when the spacing between the two Ci-Bra sites was altered by either insertion of 5 or 10 bp, or by deletion of 5 bp, the notochord activity was lost. Similar results were obtained when 11 bp were inserted between the two sites (data not shown). Likewise, the deletion of 5 bp and the insertion of 10 bp between the Ci-Bra sites and the Ci-FoxA-a site both eliminated notochord activity. These findings suggest that, in addition to the helical phasing of the binding sites, which is perturbed by the 5-bp insertion/deletion and is expected to be restored by the insertion of a full helical turn of 10-11 bp (Makeev et al., 2003), additional architectural constraints might be modulating the assembly of transcription factors and nucleoproteins responsible for the function of the Ci-tune CRM.
Genome-wide validation of the geometric constraints identified for the Ci-tune CRM
It is generally acknowledged that at least subsets of CRMs active in the same tissue(s) contain related constellations of binding sites for transcriptional activators and/or repressors (e.g. Levine and Tjian, 2003; Markstein et al., 2002; Berman et al., 2002). This assumption has led to the formulation of the concept of a `cis-regulatory code' (e.g. Istrail and Davidson, 2005), or grammar, that could be extrapolated from co-regulated CRMs and used to predict related modules with similar cis-regulatory properties. We employed the syntactic rules that we had learned from the Ci-tune CRM to scan the Ciona genome in an attempt to identify novel, related notochord CRMs. However, we found that none of the combinations of Ci-FoxA-a and Ci-Bra binding sites that we tested was able to direct notochord activity under the experimental conditions employed. These results are not surprising if we consider that generic T-box and Fox binding sites have short core sequences and therefore occur very frequently along the genome, even within short (<160 bp) regions. Nevertheless, these findings show that related combinations of binding sites for the activators analyzed here, as well as sites with identical core sequences, can exhibit widely different behaviors. This in turn suggests that additional regulatory mechanisms, most likely epigenetic modifications and/or the action of transcriptional repressors, must be modulating the behavior of these binding sites in a context-dependent fashion, thus acting as `external operators' (Istrail and Davidson, 2005) and adding further layers of transcriptional control. Along these lines, it is notable that in other systems the Ci-FoxA-a ortholog Foxa2 has been shown to possess a chromatin-opening activity that is necessary to initiate activation of its target genes (Lee et al., 2005). In conclusion, the results of the orientation, spacing and genome-wide scanning experiments suggest that the Ci-tune CRM functions as a rather unique `enhanceosome' with relatively fixed geometric requirements (Arnosti and Kulkarni, 2005).
The genetic circuitry presided by FoxA and Brachyury in animal development and evolution
The repeated occurrence of developmentally relevant interactions between FoxA and Brachyury in phylogenetically distant animals is strongly suggestive of the reliability of their synergy in ensuring adequate control over a wide gene network, such as the one controlling mesoderm formation and regionalization. In the Ciona notochord, the positive regulation of Ci-Bra transcription by Ci-FoxA-a can be envisioned as an efficient way of establishing a robust feed-forward regulatory mechanism from the early stages of embryonic development. This mechanism can provide the rapid mobilization, through combinatorial transcriptional activation, of a large gene battery of effectors able to perform widely diverse functions and to coordinately execute complex morphogenetic movements within a relatively short amount of time.
Although a cooperative interaction between FoxA and Brachyury at the level of individual CRMs had been postulated from previously published observations coming from ascidians as well as from different model systems (e.g. O'Reilly et al., 1995; Kusch and Reuter, 1999; Shimauchi et al., 2001), the detailed molecular mechanisms through which it is achieved at the cis-regulatory level had yet to be analyzed. The work presented here provides a first case study of the cis-regulatory mechanisms underlying this synergy. Interestingly, while this manuscript was being prepared for submission, work carried out in zebrafish has shown that several CRMs targeted by the Brachyury ortholog No tail (Ntl) are enriched in a motif containing the TGTTT core consensus sequence found in Fox-binding sites (Morley et al., 2009). These exciting results suggest the applicability of our findings in Ciona to vertebrate systems, and open new perspectives for future studies.
The Ci-tune CRM should thus provide an easily manipulatable system for future investigations of CRM structure and function, and, potentially, for crystallographic analyses of the interactions that Foxa2 and Brachyury proteins establish with their target CRMs, and possibly with each other. In a broader perspective, the insights provided by this study should be applicable to other tissues and model systems in which Brachyury and FoxA orthologs have been shown to cooperatively regulate gene expression.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/136/21/3679/DC1
Supplementary Material
We thank Dr Yutaka Nibu and the members of the Di Gregorio and Nibu labs for helpful discussion and critical comments on the manuscript. We are indebted to two anonymous reviewers for their valuable comments and suggestions. We thank Dr David N. Keys for the XPA16474 construct, Ms Irina Pyatigorskaya for technical help and Dr Nori Satoh for the cDNA collection. A.D.G. is an Irma T. Hirschl Scholar. This work was supported by grants NIH/NICHD R01HD050704 and 1-FY08-430 from the March of Dimes Birth Defects Foundation to A.D.G. Deposited in PMC for release after 12 months.
References
- Abdelkhalek, H. B., Beckers, A., Schuster-Gossler, K., Pavlova, M. N., Burkhardt, H., Lickert, H., Rossant, J., Reinhardt, R., Schalkwyk, L. C., Muller, I. et al. (2004). The mouse homeobox gene Not is required for caudal notochord development and affected by the truncate mutation. Genes Dev. 18, 1725-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W. and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- Arnosti, D. N. and Kulkarni, M. M. (2005). Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J. Cell. Biochem. 94, 890-898. [DOI] [PubMed] [Google Scholar]
- Berman, B. P., Nibu, Y., Pfeiffer, B. D., Tomancak, P., Celniker, S. E., Levine, M., Rubin, G. M. and Eisen, M. B. (2002). Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. USA 99, 757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo, J. C., Levine, M. and Zeller, R. W. (1997). Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development 124, 589-602. [DOI] [PubMed] [Google Scholar]
- Dehal, P., Satou, Y., Campbell, R. K., Chapman, J., Degnan, B., De Tomaso, A., Davidson, B., Di Gregorio, A., Gelpke, M., Goodstein, D. M. et al. (2002). The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298, 2157-2167. [DOI] [PubMed] [Google Scholar]
- Di Gregorio, A. and Levine, M. (1999). Regulation of Ci-tropomyosin-like, a Brachyury target gene in the ascidian, Ciona intestinalis. Development 126, 5599-5609. [DOI] [PubMed] [Google Scholar]
- Di Gregorio, A., Corbo, J. C. and Levine, M. (2001). The regulation of forkhead/HNF-3beta expression in the Ciona embryo. Dev. Biol. 229, 31-43. [DOI] [PubMed] [Google Scholar]
- Di Gregorio, A., Harland, R. M., Levine, M. and Casey, E. S. (2002). Tail morphogenesis in the ascidian, Ciona intestinalis, requires cooperation between notochord and muscle. Dev. Biol. 244, 385-395. [DOI] [PubMed] [Google Scholar]
- Dunn, M. P. and Di Gregorio, A. (2009). The evolutionarily conserved leprecan gene: its regulation by Brachyury and its role in the developing Ciona notochord. Dev. Biol. 328, 561-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Brunak, S., von Heijne, G. and Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953-971. [DOI] [PubMed] [Google Scholar]
- Erives, A., Corbo, J. C. and Levine, M. (1998). Lineage-specific regulation of the Ciona snail gene in the embryonic mesoderm and neuroectoderm. Dev. Biol. 194, 213-225. [DOI] [PubMed] [Google Scholar]
- Friedman, J. R. and Kaestner, K. H. (2006). The Foxa family of transcription factors in development and metabolism. Cell Mol. Life Sci. 63, 2317-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman, M. A. (1993). Rapid amplification of complementary DNA ends for generation of full-length complementary DNAs: thermal RACE. Methods Enzymol. 218, 340-356. [DOI] [PubMed] [Google Scholar]
- Harafuji, N., Keys, D. N. and Levine, M. (2002). Genome-wide identification of tissue-specific enhancers in the Ciona tadpole. Proc. Natl. Acad. Sci. USA 99, 6802-6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann, B. G. and Kispert, A. (1994). The T genes in embryogenesis. Trends Genet. 10, 280-286. [DOI] [PubMed] [Google Scholar]
- Holland, P. W., Koschorz, B., Holland, L. Z. and Herrmann, B. G. (1995). Conservation of Brachyury (T) genes in amphioxus and vertebrates: developmental and evolutionary implications. Development 121, 4283-4291. [DOI] [PubMed] [Google Scholar]
- Hotta, K., Takahashi, H., Asakura, T., Saitoh, B., Takatori, N., Satou, Y. and Satoh, N. (2000). Characterization of Brachyury-downstream notochord genes in the Ciona intestinalis embryo. Dev. Biol. 224, 69-80. [DOI] [PubMed] [Google Scholar]
- Hotta, K., Takahashi, H., Satoh, N. and Gojobori, T. (2008). Brachyury-downstream gene sets in a chordate, Ciona intestinalis: integrating notochord specification, morphogenesis and chordate evolution. Evol. Dev. 10, 37-51. [DOI] [PubMed] [Google Scholar]
- Imai, K. S., Hino, K., Yagi, K., Satoh, N. and Satou, Y. (2004). Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development 131, 4047-4058. [DOI] [PubMed] [Google Scholar]
- Imai, K. S., Levine, M., Satoh, N. and Satou, Y. (2006). Regulatory blueprint for a chordate embryo. Science 312, 1183-1187. [DOI] [PubMed] [Google Scholar]
- Istrail, S. and Davidson, E. H. (2005). Logic functions of the genomic cis-regulatory code. Proc. Natl. Acad. Sci. USA 102, 4954-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler, J. E., Passamaneck, Y. J., Feldman, T. G., Regnier, T. W. and Di Gregorio, A. (2008). Evolutionary conservation of vertebrate notochord genes in the ascidian Ciona intestinalis. Genesis 46, 697-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakabe, T. (2005). Decoding cis-regulatory systems in ascidians. Zool. Sci. 22, 129-146. [DOI] [PubMed] [Google Scholar]
- Kusch, T. and Reuter, R. (1999). Functions for Drosophila brachyenteron and forkhead in mesoderm specification and cell signalling. Development 126, 3991-4003. [DOI] [PubMed] [Google Scholar]
- Kusch, T., Storck, T., Walldorf, U. and Reuter, R. (2002). Brachyury proteins regulate target genes through modular binding sites in a cooperative fashion. Genes Dev. 16, 518-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. S., Friedman, J. R., Fulmer, J. T., and Kaestner, K. H. (2005). The initiation of liver development is dependent on Foxa transcription factors. Nature 435, 944-947. [DOI] [PubMed] [Google Scholar]
- Levine, M. and Tjian, R. (2003). Transcription regulation and animal diversity. Nature 424, 147-151. [DOI] [PubMed] [Google Scholar]
- Makeev, V. J., Lifanov, A. P., Nazina, A. G. and Papatsenko, D. A. (2003). Distance preferences in the arrangement of binding motifs and hierarchical levels in organization of transcription regulatory information. Nucleic Acids Res. 31, 6016-6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein, M., Markstein, P., Markstein, V. and Levine, M. S. (2002). Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 99, 763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley, R. H., Lachani, K., Keefe, D., Gilchrist, M. J., Flicek, P., Smith, J. C. and Wardle, F. C. (2009). A gene regulatory network directed by zebrafish No tail accounts for its roles in mesoderm formation. Proc. Natl. Acad. Sci. USA 106, 3829-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda-Ishii, I. and Di Gregorio, A. (2007). Lineage-independent mosaic expression and regulation of the Ciona multidom gene in the ancestral notochord. Dev. Dyn. 236, 1806-1819. [DOI] [PubMed] [Google Scholar]
- O'Reilly, M. A., Smith, J. C. and Cunliffe, V. (1995). Patterning of the mesoderm in Xenopus: dose-dependent and synergistic effects of Brachyury and Pintallavis. Development 121, 1351-1359. [DOI] [PubMed] [Google Scholar]
- Passamaneck, Y. J. and Di Gregorio, A. (2005). Ciona intestinalis: chordate development made simple. Dev. Dyn. 233, 1-19. [DOI] [PubMed] [Google Scholar]
- Qian, X. and Costa, R. H. (1995). Analysis of hepatocyte nuclear factor-3 beta protein domains required for transcriptional activation and nuclear targeting. Nucleic Acids Res. 23, 1184-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee, J. M., Oda-Ishii, I., Passamaneck, Y. J., Hadjantonakis, A. K. and Di Gregorio, A. (2005). Live imaging and morphometric analysis of embryonic development in the ascidian Ciona intestinalis. Genesis 43, 136-147. [DOI] [PubMed] [Google Scholar]
- Satou, Y., Takatori, N., Yamada, L., Mochizuki, Y., Hamaguchi, M., Ishikawa, H., Chiba, S., Imai, K., Kano, S., Murakami, S. D. et al. (2001). Gene expression profiles in Ciona intestinalis tailbud embryos. Development 128, 2893-2904. [DOI] [PubMed] [Google Scholar]
- Senger, K., Armstrong, G. W., Rowell, W. J., Kwan, J. M., Markstein, M. and Levine, M. (2004). Immunity regulatory DNAs share common organizational features in Drosophila. Mol. Cell 13, 19-32. [DOI] [PubMed] [Google Scholar]
- Shi, W., Levine, M. and Davidson, B. (2005). Unraveling genomic regulatory networks in the simple chordate, Ciona intestinalis. Genome Res. 15, 1668-1674. [DOI] [PubMed] [Google Scholar]
- Shimauchi, Y., Chiba, S. and Satoh, N. (2001). Synergistic action of HNF-3 and Brachyury in the notochord differentiation of ascidian embryos. Int. J. Dev. Biol. 45, 643-652. [PubMed] [Google Scholar]
- Shoguchi, E., Kawashima, T., Satou, Y., Hamaguchi, M., Sin-I, T., Kohara, Y., Putnam, N., Rokhsar, D. S. and Satoh, N. (2006). Chromosomal mapping of 170 BAC clones in the ascidian Ciona intestinalis. Genome Res. 16, 297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. C., Price, B. M., Green, J. B., Weigel, D. and Herrmann, B. G. (1991). Expression of a Xenopus homolog of Brachyury (T) is an immediate-early response to mesoderm induction. Cell 67, 79-87. [DOI] [PubMed] [Google Scholar]
- Takada, N., York, J., Davis, J. M., Schumpert, B., Yasuo, H., Satoh, N. and Swalla, B. J. (2002). Brachyury expression in tailless Molgulid ascidian embryos. Evol. Dev. 4, 205-211. [DOI] [PubMed] [Google Scholar]
- Takahashi, H., Hotta, K., Erives, A., Di Gregorio, A., Zeller, R. W., Levine, M. and Satoh, N. (1999a). Brachyury downstream notochord differentiation in the ascidian embryo. Genes Dev. 13, 1519-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, H., Mitani, Y., Satoh, G. and Satoh, N. (1999b). Evolutionary alterations of the minimal promoter for notochord-specific Brachyury expression in ascidian embryos. Development 126, 3725-3734. [DOI] [PubMed] [Google Scholar]
- Wilkinson, D. G., Bhatt, S. and Herrmann, B. G. (1990). Expression pattern of the mouse T gene and its role in mesoderm formation. Nature 343, 657-659. [DOI] [PubMed] [Google Scholar]
- Yasuo, H. and Satoh, N. (1993). Function of vertebrate T gene. Nature 364, 582-583. [DOI] [PubMed] [Google Scholar]
- Yasuo, H. and Satoh, N. (1998). Conservation of the developmental role of Brachyury in notochord formation in a urochordate, the ascidian Halocynthia roretzi. Dev. Biol. 200, 158-170. [DOI] [PubMed] [Google Scholar]
- Zeller, R. W., Weldon, D. S., Pellatiro, M. A. and Cone, A. C. (2006). Optimized green fluorescent protein variants provide improved single cell resolution of transgene expression in ascidian embryos. Dev. Dyn. 235, 456-467. [DOI] [PubMed] [Google Scholar]
- Zinzen, R. P., Senger, K., Levine, M. and Papatsenko, D. (2006). Computational models for neurogenic gene expression in the Drosophila embryo. Curr. Biol. 16, 1358-1365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.