Abstract
Identification of novel molecular targets is critical in development of new and efficient cancer therapies. Kinases are one of the most common drug targets with a potential for cancer therapy. Cell cycle progression is regulated by a number of kinases, some of which are being developed to treat cancer. Cdc7 is a serine-threonine kinase originally discovered in budding yeast, which has been shown to be necessary to initiate the S phase. Inhibition of Cdc7 in cancer cells retards the progression of the S phase, accumulates DNA damage, and induces p53-independent cell death, but the same treatment in normal cells does not significantly affect of less than viability. Low-molecular-weight compounds that inhibit Cdc7 kinase with an IC50 10 nM have been identified, and shown to be effective in the inhibition of tumor growth in animal models. Thus Cdc7 kinase can be recognized as a novel molecular target for cancer therapy.
Keywords: Cdc7 kinase, cell cycle, replication fork, genome stability, DNA damages, ATP-binding pocket, kinase inhibitor
Introduction
Recent studies on the molecular basis of various diseases have revealed that the deregulation of protein phosphorylation reactions by kinases is clearly associated with many disease states, especially cancer (Noble et al 2004). Thus modulating aberrant kinase activities in cancer cells may form an effective cancer therapy, with fewer side effects than traditional cytotoxic drugs. A number of kinases have been studied as potential targets for cancer therapy (Collins and Workman 2006). Kinases regulating the cell cycle, including Cdk (cyclin-dependent kinase), Aurora kinases, and Chk1 kinase, have also been the focus of many studies on cancer-treating agents (Fischer et al 2003; Collins and Garrett, 2005; Warner et al 2006; de Carcer et al 2007) in part because these cell cycle-regulating kinases are overproduced in cancer cells.
Cdc7 is a conserved serine-threonine kinase, which was originally identified in budding yeast and shown to be required for the initiation of DNA replication (Hartwell 1971; Patterson et al 1986; Sclafani 2000). Subsequent studies showed that Cdc7 is required not only for initiation but also for stable maintenance of replication forks (Snaith et al 2000; Takeda et al 2001; Fung et al 2002; Matsumoto et al 2005; Sommariva et al 2005). This brief article summarizes the latest knowledge on the functions of Cdc7 kinase, and current efforts on the design of a drug based on this interesting molecule.
Roles of Cdc7 in initiation of DNA replication and checkpoint regulation
Initial characterization of a budding yeast cdc7 temperature-sensitive mutant indicated that Cdc7 is required to initiate DNA synthesis. Cdc7 is a serine-threonine kinase, which belongs to a unique group in the kinase family (Figure 1). Cdc7 forms a complex with Dbf4 (Johnston and Thomas 1982; Kitada et al 1992; Jackson et al 1993), an activation subunit. Both cdc7 and dbf4 are temperature-sensitive mutants of budding yeast and arrest with 1C DNA content at a non-permissive temperature, suggesting a defect in the initiation of DNA replication. In 1995, the first functional homologue of Cdc7 was identified in fission yeast (Masai et al 1995). After this discovery, conserved presence of both Cdc7 and Dbf4 subunits was demonstrated across the species (Masai et al 1999; Masai and Arai 2002).
Figure 1.
Homology analyses of human Cdc7 kinase. A)The human kinome tree. Kinase family members were classified into the following eight subgroups on the basis of their primary structures: CAMK (calcium/calmodulin-dependent kinase group), TK (tyrosine kinase group), RGC (receptor guanylyl cyclase group), TKL (tyrosine kinase-like group), STE (sterile phenotype kinase group), CK1 (cell kinase 1/casein kinase 1 group), AGC (protein kinases A, G and C group), CMGC (cyclin-dependent-kinase [CDK], mitogen-activated-kinase [MAPK], glycogen-synthase-kinase [GSK] and CDK-like kinase group). B) A phylogenetic tree of Cdc7 and some human protein kinases which are most similar to Cdc7.
Cdc7 is essential for viability in yeasts. Knockout of Cdc7 genes in mice leads to early embryo death; the mutant embryos die between E3.5 and 6.5. Conditional knockout of Cdc7 genes in mouse embryonic stem cells resulted in the arrest of DNA synthesis, accumulation of nuclear DNA damage, and eventual p53-dependent cell death (Kim et al 2002). These results suggest that Cdc7 kinase has critical roles in DNA replication, which may be conserved across species.
The conserved targets for Cdc7 kinase are MCM subunits, and phosphorylation of the N-terminal non-conserved tails of MCM2, 4, and 6 proteins has been shown to facilitate the association of Cdc45 and other replisome factors with pre-replicative complex (pre-RC) (Masai et al 2000, 2006; Sheu and Stillman 2006). This step is crucial for the generation of active and efficient replication fork structures (Figure 2).
Figure 2.
Scheme of initiation of eukaryotic DNA replication and action of Cdc7 kinase. Eukaryotic DNA replication is initiated by binding of ORC (origin recognition complex) at a replication origin. With the aid of Cdc6 and Cdt1 proteins, Mcm (minichromosome maintenance) is delivered at the origin, generating pre-RC (pre-replicative complex). Cdc45 associates with the pre-RC, followed by GINS complex. Phosphorylation by Cdk and Cdc7 is required for this step. It was reported that phosphorylation of the N-terminal tails of Mcm2, Mcm4, and Mcm6 proteins facilitates association of Cdc45 and other proteins with Mcm (Masai et al 2006; Sheu et al 2006). Active replication forks are generated by association of three DNA polymerases at the origin.
The replication fork is under continuous attack both internally and externally, clearly indicated by the fact that a recombinational repair system is essential for the viability of vertebrate cells (Sonoda et al 1998). Proper processing of stalled replication forks and the resumption of DNA replication are essential for completion of the entire genome duplication within the given S phase. Cellular responses to stalled replication forks are regulated by checkpoint reactions. A defect in checkpoint regulation poses serious threats to the stable maintenance of the genome. Indeed mutations in checkpoint regulators have been demonstrated to be responsible for various tumors or diseases (Michelson and Weinert 2000; D’Andrea and Grompe 2003;Narek and Lukas 2003). Checkpoint responses are composed of two phases; the mediator and effector phases. The former is involved in activating checkpoint kinases, while the latter is involved in executing the checkpoint effects (Niida and Nakanishi 2006). Accumulating evidence indicates the critical roles of Cdc7 kinase in both phases of DNA replication checkpoint responses. Chk1 is activated in response to replication fork arrest, and Cdc7 appears to be required for this activation (Kim et al 2008). Claspin is a mediator of checkpoint responses and is hyper-phosphorylated in response to stalled replication forks. Cdc7 is required for this step (Kim et al 2008). In fission yeast, the activation of Cds1, a checkpoint effector kinase, and hyperphosphorylation of Mrc1 (a fission yeast counterpart of Claspin) in response to fork arrest, depend on Hsk1, the fission yeast homologue of Cdc7 kinase (Takeda et al 2001; Shimmoto et al unpublished data). Claspin/Mrc1 is efficiently phosphorylated by Cdc7/Hsk1 in vitro, suggesting that possibly Cdc7 directly regulates the checkpoint activation (Kim et al 2008; Kakusho et al unpublished data). The requirement of Cdc7 for continuous activation of the Rad53 checkpoint kinase in response to HU has also been reported in budding yeast (Ogi et al 2008). On the other hand, the role of Cdc7 in the effector phase was suggested by the fact that Cdc7 is inactivated in Xenopus egg extracts in response to etopside, a DNA-damaging agent introducing single-stranded nicks (Costanzo et al 2003). Cdc7 was also shown to be down-regulated by etopside in human cells (Dierov et al 2004). Evidence showing that the fork block inhibits Cdc7 kinase was also reported in budding yeast (Weinreich and Stillman 1999). These results strongly suggest the dual roles of Cdc7 kinase in the mediator and effector phases of checkpoint responses. However researchers report that Cdc7 activity is unaltered by replication fork blocks in human cells (Tenca et al 2007). Furthermore, we cannot rule out the possibility that defects in the DNA replication fork caused by Cdc7 depletion contributes to the apparent defect in checkpoint activation. Whatever the mechanisms are, loss of Cdc7 functions in actively proliferating cells causes replication fork arrest which results in DNA damage, and simultaneously leads to defective checkpoint activation.
Structure and activation of Cdc7 kinase complex
Unique features of Cdc7 include the presence of two or three amino acid segments which interrupt the kinase domains at conserved locations (Figure 3). Although the functional significance of these kinase insert sequences has not been defined, association of nuclear localization and nuclear export activities with the second and third inserts, respectively, was reported for human Cdc7 (Kim et al 2007). Cdc7 associates with chromatin during the S through G2 phase, and dissociates from chromatin during the M phase (Masai et al 2006) when it is phosphorylated, presumably by Cdc2 kinase. Kinase activity may not be regulated by Cdk-mediated phosphorylation, since Escherichia coli-generated Cdc7 kinase is highly active and the activity is not affected by Cdk (Kakusho et al unpublished data).
Figure 3.
Schematic drawing of Cdc7 kinase. Human Cdc7 (574 amino acids) are composed of kinase conserved domains (white segments) and three kinase insert sequences (KI-1, -2, and -3; gray segments). Roman numerals indicate the kinase subdomains defined previously (Hanks et al 1988). KI-1 is not obvious in human Cdc7, but is longer in Cdc7 from other species. Although the length and sequences of kinase inserts are less conserved between species, they are located at the conserved positions among the kinase subdomains. The gatekeeper residue for the ATP-binding pocket (methionine 134) is also indicated.
Cdc7 kinase is generally active only when it associates with an activation subunit (Johstone et al 1999; Masai et al 1999, 2000; Masai and Arai 2002). The Dbf4-like activation subunits share three conserved motifs, namely Dbf4-motif-N, -M and -C (Masai and Arai 2000). The Dbf4-motif-N, a BRCT-like motif, is not essential for kinase activation, nor for cell viability in yeasts, although mutations in motif-N result in sensitivity to replication fork blockages induced by HU and other agents in yeasts (Ogino et al 2001; Varrin et al 2005; Gabrielse et al 2006). The Dbf4-motif-M (proline-rich novel motif) or motif-C (C2H2-type zinc-finger motif) is capable of binding to Cdc7 on its own, but the maximum activation, and viability in fission yeast cells through meiosis, require the presence of both motifs connected by a non-conserved linker segment (Ogino et al 2001). On the other hand, survival through mitosis in fission yeast can be maintained only by the motif-M, albeit with reduced growth rate, whereas motif-C is required for resistance to the alkylating reagent, methyl methanesulfonate (Fung et al 2002). In mammalian cells, in addition to motif-M and motif-C, motif-N is also required for viability (Yamashita et al 2005).
Members of kinases structurally close to Cdc7 include casein kinase (Figure 1B). Although the precise substrate recognition code is not known for Cdc7 kinase, it is clear that Cdc7 is an acidophylic kinase, preferring the presence of acidic residues near the phosphorylation site. This feature is shared by casein kinase. It was reported recently that positively charged polymers such as polyamines strongly stimulate Cdc7 kinase activity, whereas the negatively charged polymers such as nucleic acids severely inhibit Cdc7 kinase activity (Kakusho et al 2008). These characters of Cdc7 are also shared by casein kinase (Hathaway and Traugh 1984), reinforcing the structural similarity between Cdc7 and casein kinase.
Downregulation of Cdc7 causes cell death preferentially in cancer cells
siRNA-mediated downregulation of Cdc7 expression in various cancer cells induced retardation of S phase progression, followed by accumulation of nuclear damage, presumably induced by a stalled replication fork which was not properly stabilized and collapsed (Montagnoli et al 2004; Yoshizaw-Sugata et al 2005; Im and Lee 2008; Kim et al 2008). Cells then undergo chromosome fragmentation and die, which occurs in both p53-positive and -minus cells, indicating that cell death is p53-independent (Montagnoli et al 2004). Cells appear to die mostly by entering aberrant mitosis. In contrast, normal fibroblast cells do not die in significant quantities under the same conditions. S phase cells slow down DNA synthesis and some DNA damage appears in nuclei, but is eventually repaired. Cells in other cell cycles may be arrested in the G1 phase in a p53-dependent manner (Montagnoli et al 2004; Ishii et al unpublished data).
The precise mechanisms of cancer-specific cell death are not clear. Depletion of Cdc7 would cause defects in firing of the replication origins as well as acute destabilization of replication forks which would eventually collapse. Furthermore, checkpoint defects may trigger uncoupling of the M phase with the S phase. Therefore we suggest that the combined effect of replication fork arrest and deficient checkpoint responses might augment cell death by inducing aberrant cell cycle progression. Whereas normal cells could cope with these situations through backup checkpoint mechanisms, cancer cells, in which checkpoint mechanisms are very often partially abrogated, would suffer from aberrant M phase progression caused by checkpoint abrogation. Researchers reported recently that ATR-induced activation of p38 MAP kinase causes caspase activation in Cdc7-depleted cancer cells (Im and Lee 2008).
Structural features of ATP-binding pocket of Cdc7 kinase
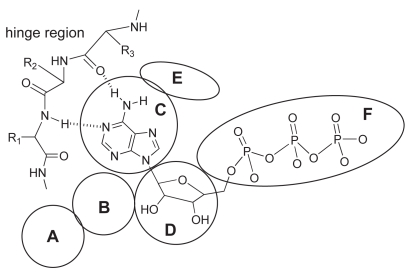
A ligand ATP binds to the ATP-binding pocket through hydrogen bonds with the hinge region connecting the N- and C-terminal lobes of the kinase catalytic domain (Figure 4). Most kinase inhibitors reported so far also bind to this hinge region with additional interactions with two hydrophobic regions (sites B and E in Figure 4) adjacent to the ATP-binding pocket. The hydrophobic region I (E in Figure 4) is commonly referred to as the selectivity pocket, because the size of a single residue in the interior of this region, termed the gatekeeper, has been shown to be a critical determinant of inhibitor sensitivity (Cherry and Williams 2004; Vieth et al 2004). Based on our in-house human kinome database, approximately 90 kinases possess a small threonine gatekeeper, and 190 kinases have a large methionine residue. According to a homology model of Cdc7 we built in-house, the large gatekeeper residue (Met134) appears to make the entrance to the hydrophobic region I very narrow, and the back pocket of this site is occupied by residues Val120 and Ile132 (Figure 5). The central portion of the adenine-binding region is formed by a hydrophobic sandwich with two residues, Met118 and Ile64, suggesting that a flat hydrophobic core may be preferred as an inhibitor scaffold. In the same region, typical hydrogen bonds may be formed with the backbone of the hinge region. Two ionic residues, Lys90 and Asp196, could provide opportunities for the design of potent inhibitors with appropriate substituent targeting these residues.
Figure 4.
Schematic representation of general binding mode of ATP; interactions with specific segments of protein kinases: A) solvent accessible region, B) hydrophobic region II, C) adenine-binding region, D) sugar pocket, E) hydrophobic region I, F) phosphate-binding region.
Figure 5.
A homology model of Cdc7 kinase. A) Mapping of important residues within the ATP-binding pocket of Cdc7 kinase based on a homology model. The dotted circle area marks the hydrophobic area (adenine-binding region) formed by a hydrophobic sandwich with two residues Met118 and Ile64. Possible hydrogen bonds between an inhibitor and the ATP-binding site of Cdc7 are indicated by the arrows. B) A model of staurosporine bound to the ATP-binding site of Cdc7 shown by Van der Waals surfaces. The negatively and positively charged regions are shown in pink and blue, respectively, on the Van der Waals surfaces. Oxygen and nitrogen are shown in red and blue, respectively. The carbon atoms in the ligand (staurosporine) and in the protein are shown in white and green, respectively.
Emerging Cdc7 kinase inhibitors
Only a handful of Cdc7 kinase inhibitors have been reported so far (Figures 6 and 7). A series of pyrrolopyridinone compounds with potent activity against Cdc7 has been reported by Nerviano Medical Sciences (Montagnoli et al 2008a; Vanotti et al 2008). PHA-767491 (#1) was identified from high throughput screening (HTS) and characterized as an ATP-competitive inhibitor of Cdc7 (IC50 = 10 nM). This compound recapitulates the most relevant molecular phenotypes that are expected from Cdc7 inhibition (Montagnoli et al 2008a). It inhibited Cdc7-directed phosphorylation of a substrate protein (Mcm2) as well as DNA synthesis, and induced cell death in multiple cancer cell lines, but did not affect the viability of normal cells (NHDF; normal human dermal fibroblasts). Treatment of NHDF with PHA-767491 indicates that Cdc7 inhibition may prevent the activation of replication origins but does not trigger a sustained DNA damage response.
Figure 6.
Cdc7 kinase inhibitors reported by Nerviano Medical Sciences. 1, PHA-767491 (IC50 =10 nM), 2, 3-aminopyrimidine analog (IC50 = 7 nM), 3, (S)-2-(2-aminopyrimidiny-4-yl)-7-(2-fluoroethyl)-1,5,6,7-tetrahydropyrrolo[3,2-c]pyridinone (IC50 =2 nM), and 4, the pyrrolopyridinone analog (IC50 = 3 nM).
Figure 7.
Structures of other Cdc7 inhibitors. 5, Tricyclic Cdc7 inhibitor (Roche); 6, indazolylpyrimidin-2(1H)-one inhibitor (Novartis); 7, thienopyrazole-base inhibitor (Pfizer); 8, 2-pyrimidyl-5-amidothiophene analog (Novartis Vaccines and Diagnostics); 9, imidazolone-based inhibitor (Sanofi-Aventis).
Subsequent optimization of this scaffold led to 3-aminopyrimidine analog (compound #2; IC50 = 7 nM). The Structure Activity Relationship (SAR) study suggested that the aminopyrimidine group probably interacts with the hinge. Although the compound #2 showed antiproliferative activity against the A-2780 ovarian cancer cell line, it exhibited poor membrane permeability in the PAMPA assay which would cause low oral absorption. Further optimization led to (S)-2-(2-aminopyrimidiny-4-yl)-7-(2-fluoroethyl)-1,5,6,7-tetrahydropyrrolo[3,2-c]pyridinone (compound #3; Menichincheri et al 2007). Compound #3, which had an IC50 value of 2 nM against Cdc7 with greater than 60-fold selectivity over other kinases, showed excellent inhibition of cell proliferation in various tumor lines with submicromolar IC50 values. In addition, compound #3 was found to be effective in an A2780 cell line mouse xenograft model, when administered twice daily at an oral dose of 60 mg/kg for 10 days. In the DMBA-induced mammary carcinoma rat model, compound #3 was also active at a dose of 20 mg/kg.
The Nerviano group also examined other possible modifications to the pyrrolopyridinone scaffold and disclosed novel pyrrole compounds, such as compound #4 (Vanotti et al 2007). A clinical candidate, NMS-354, which is derived from the same chemical class as compound #4, was reported as an orally available Cdc7 inhibitor (IC50 = 3 nM; Montagnoli et al 2008b). NMS-354 is potent in blocking proliferation and inducing apoptosis in a broad panel of 120 cancer cell lines with IC50 values in the submicromolar range. In vivo, NMS-354 has a broad range of activities when administered singly (20 mg/kg) in different xenograft tumor models (ovarian, colon, mammary, leukemia) and also in a rat mammary carcinogenic-induced tumor model (DMBA) with tumor growth inhibition of more than 80% without toxic effects. NMS-354 is currently under preclinical evaluation.
Researchers at Hoffmann-La Roche explored a series of tricyclic Cdc7 inhibitors and identified potent and selective compounds (Zhao et al 2005). A pyridothienopyrimidine was identified from HTS and subsequent structural optimization resulted in the discovery of compound #5 which is a potent Cdc7 inhibitor with Ki of 2 nM (Figure 7). Compound #5 showed high selectivity against a panel of 11 kinases. However, no further progress on this compound has been reported.
Recently, Novartis reported a series of 4-indazolylpyrimidin-2(1H)-one as potent Cdc7 inhibitors (Shafer et al 2007, 2008). Compound #6 was identified as a potent inhibitor of Cdc7 with an IC50 = 5 nM (Figure 7). They predict the binding mode of this series of compounds on the basis of the co-crystal structure of the parent compound, phenol pyrimidin-2(1H)-one, with PIM-1 kinase. The structure shows that the phenol and pyrimidinone are coplanar, and this flat shape is expected to fit well with the Cdc7 ATP-binding site as described in the previous section. Although the details of the interactions may be different between PIM-1 and Cdc7, the structure suggested that the NH group in the indazole ring is likely to bind to the hinge region of Cdc7. Unfortunately, this series of compounds exhibits low membrane permeability as well as metabolic liability, and, in fact, showed weak cellular activity.
Various Cdc7 kinase inhibitors have also been disclosed in the patent literatures recently (Figure 7). Thienopyrazoles have been claimed as Cdc7 inhibitors by Pharmacia (now Pfizer) (Tonani et al 2004). In this application, compound #7 was described as possessing Cdc7 kinase-inhibiting activity with IC50 <1000 nM. However, no kinase selectivity data were presented.
The 2-pyrimidyl-5-amidothiophenes were identified as an AKT inhibitor by Chiron (now Novartis Vaccines and Diagnostics; Lin et al 2005). The patent application for this compound series claims that these analogs also have inhibitory activities against Cdc7 as exemplified by compound #8. However, no activity data were presented.
A series of imidazolones was recently disclosed in a patent application by Sanofi-Aventis (Leroy et al 2008). They claim that the members of this series, as represented in compound #9, have inhibitory activities against Cdc7. However, no data were presented to substantiate these claims.
Conclusions and perspectives
Although Cdc7 is essential for the viability of eukaryotic cells, its inhibition or depletion leads to cell death specifically in cancer cells. This is caused by acute genomic instability induced by the combined effects of replication fork arrest, its destabilization and aberrant M phase due to checkpoint defect. Cancer cells are generally associated with defects in checkpoint response pathways, and thus exhibit greater sensitivity to S phase perturbation than normal cells. Thus Cdc7 has emerged as a potentially effective target for cancer therapy.
Potent compounds capable of inhibiting Cdc7 kinase with nM IC50 were recently reported, and were shown to be effective in reducing tumor growth in mouse xenograft models. Only a few patent applications have been disclosed so far for Cdc7 kinase inhibitors, and it is not yet possible to determine which compounds will prove most promising, because detailed evaluation of the compounds at the cellular level or in animal models has not been conducted for most of these compounds. Nevertheless, because of the unique nature of its structure and action, inhibitors of Cdc7 kinase would be promising candidates for novel classes of cancer drugs.
Acknowledgments
We thank Carna Biosicences for production of the human kinome tree (Figure 1) and also Dr Eiji Nishiwaki and Mr Fumio Nakajima of Carna Biosciences for their in silico work. We thank Dr Yutaka Kanoh for his original artwork of Figure 2. We thank Dr Sayuri Ito, Ms Chika Tanaka-Taniyama and other members of our laboratory for sharing the unpublished data and helpful discussion. We also thank Mr Takayuki Irie of Carna Biosciences for stimulating discussion. We report no conflicts of interest in this work.
References
- Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–9. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- Cherry M, Williams DH. Recent kinase and kinase inhibitor X-ray structures: mechanisms of inhibition and selectivity insights. Curr Med Chem. 2004;11:663–73. doi: 10.2174/0929867043455792. [DOI] [PubMed] [Google Scholar]
- Collins I, Garrett MD. Targeting the cell division cycle in cancer: CDK and cell cycle checkpoint kinase inhibitors. Curr Opin Pharmacol. 2005;5:366–73. doi: 10.1016/j.coph.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Collins I, Workman P. New approaches to molecular cancer therapeutics. Nat Chem Biol. 2006;2:689–700. doi: 10.1038/nchembio840. [DOI] [PubMed] [Google Scholar]
- Costanzo V, Shechter D, Lupardus PJ, et al. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell. 2003;11:203–13. doi: 10.1016/s1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- D’Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- de Cárcer G, de Castro IP, Malumbres M. Targeting cell cycle kinases for cancer therapy. Curr Med Chem. 2007;14:969–85. doi: 10.2174/092986707780362925. [DOI] [PubMed] [Google Scholar]
- Dierov J, Dierova R, Carroll M. BCR/ABL translocates to the nucleus and disrupts an ATR-dependent intra-S phase checkpoint. Cancer Cell. 2004;5:275–85. doi: 10.1016/s1535-6108(04)00056-x. [DOI] [PubMed] [Google Scholar]
- Fischer PM, Endicott J, Meijer L. Cyclin-dependent kinase inhibitors. Prog Cell Cycle Res. 2003;5:235–48. [PubMed] [Google Scholar]
- Fung AD, Ou J, Bueler S, et al. A conserved domain of Schizosaccharomyces pombe dfp1(+) is uniquely required for chromosome stability following alkylation damage during S phase. Mol Cell Biol. 2002;22:4477–90. doi: 10.1128/MCB.22.13.4477-4490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielse C, Miller CT, McConnell KH, et al. A Dbf4p BRCA1 C-terminal-like domain required for the response to replication fork arrest in budding yeast. Genetics. 2006;173:541–55. doi: 10.1534/genetics.106.057521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Hartwell LH. Genetic control of the cell division cycle in yeast. II. Genes controlling DNA replication and its initiation. J Mol Biol. 1971;59:183–94. doi: 10.1016/0022-2836(71)90420-7. [DOI] [PubMed] [Google Scholar]
- Hathaway GM, Traugh JA. Kinetics of activation of casein kinase II by polyamines and reversal of 2,3-bisphosphoglycerate inhibition. J Biol Chem. 1984;259:7011–15. [PubMed] [Google Scholar]
- Im JS, Lee JK. ATR-dependent activation of p38 MAP kinase is responsible for apoptotic cell death in cells depleted of Cdc7. J Biol Chem. 2008;283:25171–7. doi: 10.1074/jbc.M802851200. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Pahl PM, Harrison K, et al. Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol Cell Biol. 1993;13:2899–908. doi: 10.1128/mcb.13.5.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LH, Masai H, Sugino A. First the CDKs, now the DDKs. Trends Cell Biol. 1999;9:249–52. doi: 10.1016/s0962-8924(99)01586-x. [DOI] [PubMed] [Google Scholar]
- Johnston LH, Thomas AP. A further two mutants defective in initiation of the S phase in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1982;186:445–8. doi: 10.1007/BF00729467. [DOI] [PubMed] [Google Scholar]
- Kakusho N, Taniyama C, Masai H. Identification of stimulators and inhibitors of CDC7 kinase in vitro. J Biol Chem. 2008;283:19211–18. doi: 10.1074/jbc.M803113200. [DOI] [PubMed] [Google Scholar]
- Kim BJ, Kim SY, Lee H. Identification and characterization of human cdc7 nuclear retention and export sequences in the context of chromatin binding. J Biol Chem. 2007;282:30029–38. doi: 10.1074/jbc.M703705200. [DOI] [PubMed] [Google Scholar]
- Kim JM, Kakusho N, Yamada M, et al. Cdc7 kinase mediates Claspin phosphorylation in DNA replication checkpoint. Oncogene. 2008;27:3475–82. doi: 10.1038/sj.onc.1210994. [DOI] [PubMed] [Google Scholar]
- Kim JM, Nakao K, Nakamura K, et al. Inactivation of Cdc7 kinase in mouse ES cells results in S-phase arrest and p53-dependent cell death. EMBO J. 2002;21:2168–79. doi: 10.1093/emboj/21.9.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada K, Johnston LH, Sugino T, et al. Temperature-sensitive cdc7 mutations of Saccharomyces cerevisiae are suppressed by the DBF4 gene, which is required for the G1/S cell cycle transition. Genetics. 1992;131:21–9. doi: 10.1093/genetics/131.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy V, Bacque E, Conseille E, et al. New imidazolone derivatives, preparation thereof as drugs, pharmaceutical compositions, and use thereof as protein kinase inhibitors, in particular CDC7. 2008 Patent: WO2008/046982. [Google Scholar]
- Lin X, Rico A, Zhou Y, et al. Substitued thiophene derivatives as anti-cancer agents. 2005 Patent: WO2005/095386. [Google Scholar]
- Masai H, Miyake T, Arai K. hsk1+, a Schizosaccharomyces pombe gene related to Saccharomyces cerevisiae CDC7, is required for chromosomal replication. EMBO J. 1995;14:3094–104. doi: 10.1002/j.1460-2075.1995.tb07312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Sato N, Takeda T, et al. CDC7 kinase complex as a molecular switch for DNA replication. Front Biosci. 1999;4:D834–40. doi: 10.2741/masai. [DOI] [PubMed] [Google Scholar]
- Masai H, Arai K. Dbf4 motifs: conserved motifs in activation subunits for Cdc7 kinases essential for S-phase. Biochem Biophys Res Commun. 2000;275:228–32. doi: 10.1006/bbrc.2000.3281. [DOI] [PubMed] [Google Scholar]
- Masai H, Matsui E, You Z, et al. Human Cdc7-related kinase complex. In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a criticial threonine residue of Cdc7 by Cdks. J Biol Chem. 2000;275:29042–52. doi: 10.1074/jbc.M002713200. [DOI] [PubMed] [Google Scholar]
- Masai H, Arai K. Cdc7 kinase complex: a key regulator in the initiation of DNA replication. J Cell Physiol. 2002;190:287–96. doi: 10.1002/jcp.10070. [DOI] [PubMed] [Google Scholar]
- Masai H, Taniyama C, Ogino K, et al. Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J Biol Chem. 2006;281:39249–61. doi: 10.1074/jbc.M608935200. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Ogino K, Noguchi E, et al. Hsk1-Dfp1/Him1, the Cdc7-Dbf4 kinase in Schizosaccharomyces pombe, associates with Swi1, a component of the replication fork protection complex. J Biol Chem. 2005;280:42536–42. doi: 10.1074/jbc.M510575200. [DOI] [PubMed] [Google Scholar]
- Menichincheri M, Bargiotti A, Berthelsen J, et al. Cdc7 kinase inhibitors: 7-Substituted pyrrolopyridinones as potent and orally active antitumor agents [Abstract]. 19th EORTC-NCI-AACR Symposium; October 22–26; San Franscisco, CA, USA. 2007. p. A224. [Google Scholar]
- Michelson RJ, Weinert T. Closing the gaps among a web of DNA repair disorders. Bioessays. 2000;22:966–9. doi: 10.1002/1521-1878(200011)22:11<966::AID-BIES2>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Montagnoli A, Santocanale C, Valsasina B, et al. A new and potent orally available class of Cdc7 inhibitors with anti-tumor activity [abstract]. AACR 99th Annual Meeting; April 12–16; San Diego, CA, USA. 2008b. #755. [Google Scholar]
- Montagnoli A, Tenca P, Sola F, et al. Cdc7 inhibition reveals a p53-dependent replication checkpoint that is defective in cancer cells. Cancer Res. 2004;64:7110–16. doi: 10.1158/0008-5472.CAN-04-1547. [DOI] [PubMed] [Google Scholar]
- Montagnoli A, Valsasina B, Croci V, et al. A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat Chem Biol. 2008a;4:357–65. doi: 10.1038/nchembio.90. [DOI] [PubMed] [Google Scholar]
- Niida H, Nakanishi M. DNA damage checkpoints in mammals. Mutagenesis. 2006;21:3–9. doi: 10.1093/mutage/gei063. [DOI] [PubMed] [Google Scholar]
- Noble ME, Endicott JA, Johnson LN. Protein kinase inhibitors: insights into drug design from structure. Science. 2004;303:1800–5. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- Ogi H, Wang CZ, Nakai W, et al. The role of the Saccharomyces cerevisiae Cdc7-Dbf4 complex in the replication checkpoint. Gene. 2008;414:32–40. doi: 10.1016/j.gene.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Ogino K, Takeda T, Matsui E, et al. Bipartite binding of a kinase activator activates Cdc7-related kinase essential for S phase. J Biol Chem. 2001;276:31376–87. doi: 10.1074/jbc.M102197200. [DOI] [PubMed] [Google Scholar]
- Patterson M, Sclafani RA, Fangman WL, et al. Molecular characterization of cell cycle gene CDC7 from Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:1590–8. doi: 10.1128/mcb.6.5.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani RA. Cdc7p-Dbf4p becomes famous in the cell cycle. J Cell Sci. 2000;13:2111–17. doi: 10.1242/jcs.113.12.2111. [DOI] [PubMed] [Google Scholar]
- Shafer C, Walter A, Lindvall MK, et al. Indazole compounds and methods for inhibition of CDC7. 2007 Patent: WO2007/124288. [Google Scholar]
- Shafer C, Lindvall M, Bellamacina C, et al. 4-(1H-Indazol-5-yl)-6-phenylpyrimidin-2(1H)-one analogs as potent CDC7 inhibitors. Bioorg Med Chem Lett. 2008;18:4482–5. doi: 10.1016/j.bmcl.2008.07.061. [DOI] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell. 2006;24:101–13. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaith HA, Brown GW, Forsburg SL. Schizosaccharomyces pombe Hsk1p is a potential cds1p target required for genome integrity. Mol Cell Biol. 2000;20:7922–32. doi: 10.1128/mcb.20.21.7922-7932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommariva E, Pellny TK, Karahan N, et al. Schizosaccharomyces pombe Swi1, Swi3, and Hsk1 are components of a novel S-phase response pathway to alkylation damage. Mol Cell Biol. 2005;25:2770–84. doi: 10.1128/MCB.25.7.2770-2784.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E, Sasaki MS, Buerstedde JM, et al. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T, Ogino K, Tatebayashi K, et al. Regulation of initiation of S phase, replication checkpoint signaling, and maintenance of mitotic chromosome structures during S phase by Hsk1 kinase in the fission yeast. Mol Biol Cell. 2001;12:1257–74. doi: 10.1091/mbc.12.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenca P, Brotherton D, Montagnoli A, et al. Cdc7 is an active kinase in human cancer cells undergoing replication stress. J Biol Chem. 2007;282:208–15. doi: 10.1074/jbc.M604457200. [DOI] [PubMed] [Google Scholar]
- Tonani R, Bindi S, Fancelli D, et al. Heterobicyclic pyrazole derivatives as kinase inhibitors. 2004 Patent: WO2004/007504. [Google Scholar]
- Vanotti E, Amici R, Bargiotti A, et al. Cdc7 kinase inhibitors: pyrrolopyridinones as potential antitumor agents. 1. Synthesis and structure-activity relationships. J Med Chem. 2008;51:487–501. doi: 10.1021/jm700956r. [DOI] [PubMed] [Google Scholar]
- Vanotti E, Caldarelli M, Cirla A, et al. Pyridyl- and pyrimidinyl-substitued pyrrole-, thiophene- and furane-derivatives as kinase inhibitors. 2007 Patent: WO2007/110344. [Google Scholar]
- Varrin AE, Prasad AA, Scholz RP, et al. A mutation in Dbf4 motif M impairs interactions with DNA replication factors and confers increased resistance to genotoxic agents. Mol Cell Biol. 2005;25:7494–504. doi: 10.1128/MCB.25.17.7494-7504.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieth M, Higgs RE, Robertson DH, et al. Kinomics-structural biology and chemogenomics of kinase inhibitors and targets. Biochim Biophys Acta. 2004;1697:243–57. doi: 10.1016/j.bbapap.2003.11.028. [DOI] [PubMed] [Google Scholar]
- Warner SL, Gray PJ, Von Hoff DD. Tubulin-associated drug targets: Aurora kinases, Polo-like kinases, and others. Semin Oncol. 2006;33:436–48. doi: 10.1053/j.seminoncol.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Weinreich M, Stillman B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 1999;18:5334–46. doi: 10.1093/emboj/18.19.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita N, Kim JM, Koiwai O, et al. Functional analyses of mouse ASK, an activation subunit for Cdc7 kinase, using conditional ASK knockout ES cells. Genes Cells. 2005;10:551–63. doi: 10.1111/j.1365-2443.2005.00857.x. [DOI] [PubMed] [Google Scholar]
- Yoshizawa-Sugata N, Ishii A, Taniyama C, et al. A second human Dbf4/ASK-related protein, Drf1/ASKL1, is required for efficient progression of S and M phases. J Biol Chem. 2005;280:13062–70. doi: 10.1074/jbc.M411653200. [DOI] [PubMed] [Google Scholar]
- Zhao C, Chen L, Xu Q, et al. Synthesis and evaluation of pyridothieno-pyrrimidine as potent and selective CDC7 inhibitor [Abstract]. 229th ACS National Meeting; March 13–17; San Diego, CA, USA. 2005. MEDI 141. [Google Scholar]