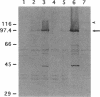
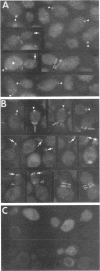
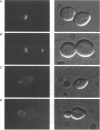
Abstract
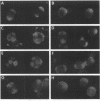
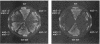
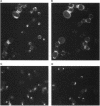
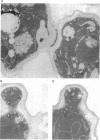
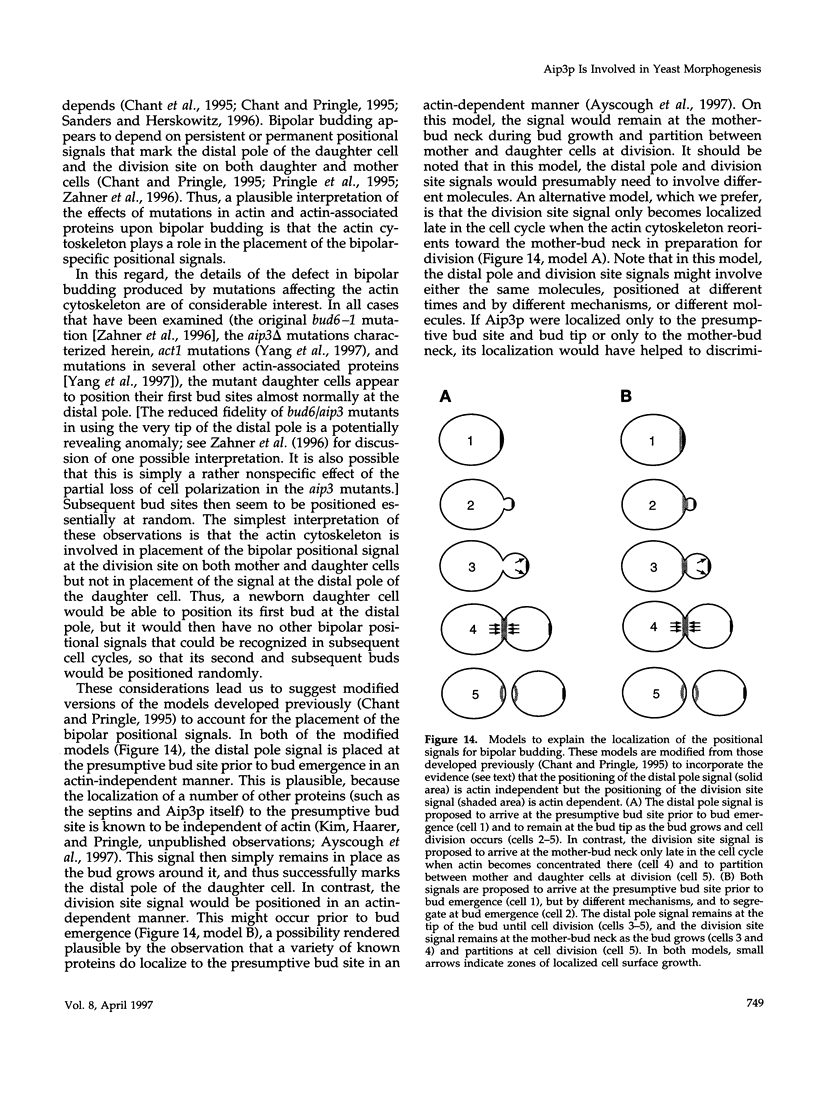
A search for Saccharomyces cerevisiae proteins that interact with actin in the two-hybrid system and a screen for mutants that affect the bipolar budding pattern identified the same gene, AIP3/BUD6. This gene is not essential for mitotic growth but is necessary for normal morphogenesis. MATa/alpha daughter cells lacking Aip3p place their first buds normally at their distal poles but choose random sites for budding in subsequent cell cycles. This suggests that actin and associated proteins are involved in placing the bipolar positional marker at the division site but not at the distal tip of the daughter cell. In addition, although aip3 mutant cells are not obviously defective in the initial polarization of the cytoskeleton at the time of bud emergence, they appear to lose cytoskeletal polarity as the bud enlarges, resulting in the formation of cells that are larger and rounder than normal. aip3 mutant cells also show inefficient nuclear migration and nuclear division, defects in the organization of the secretory system, and abnormal septation, all defects that presumably reflect the involvement of Aip3p in the organization and/or function of the actin cytoskeleton. The sequence of Aip3p is novel but contains a predicted coiled-coil domain near its C terminus that may mediate the observed homo-oligomerization of the protein. Aip3p shows a distinctive localization pattern that correlates well with its likely sites of action: it appears at the presumptive bud site prior to bud emergence, remains near the tips of small bund, and forms a ring (or pair of rings) in the mother-bud neck that is detectable early in the cell cycle but becomes more prominent prior to cytokinesis. Surprisingly, the localization of Aip3p does not appear to require either polarized actin or the septin proteins of the neck filaments.
Full text
PDF






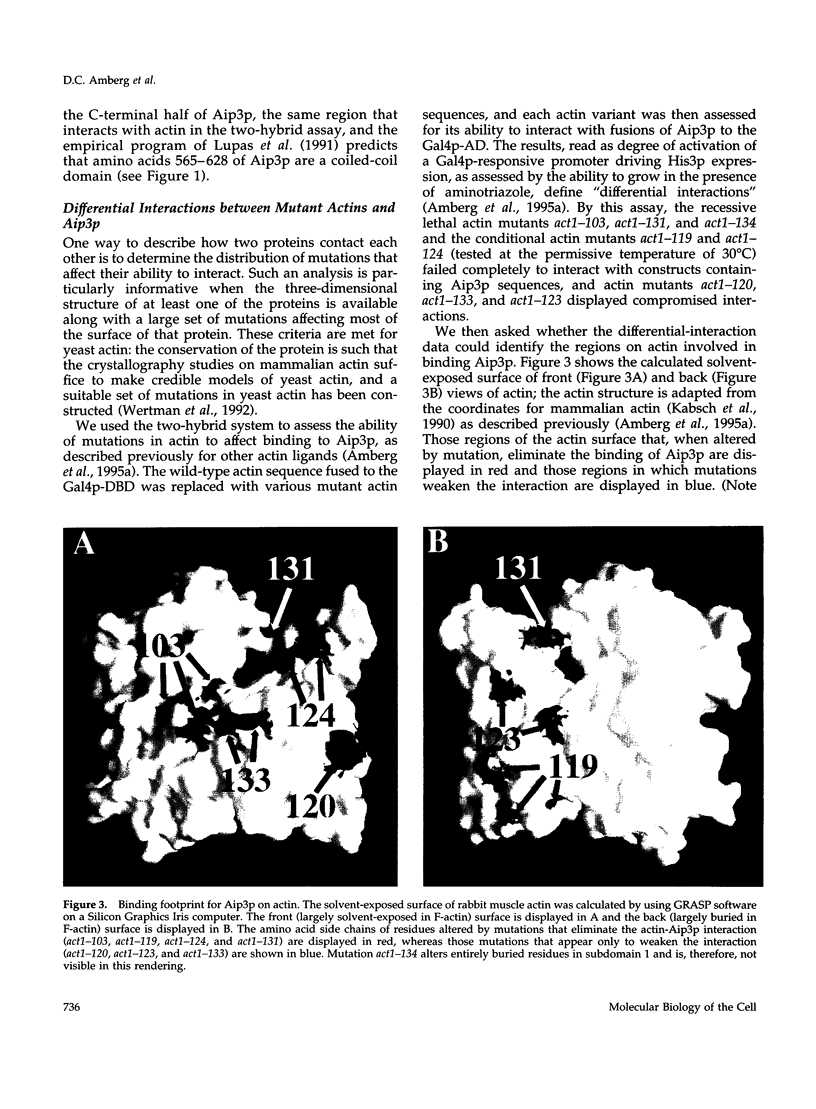








Images in this article
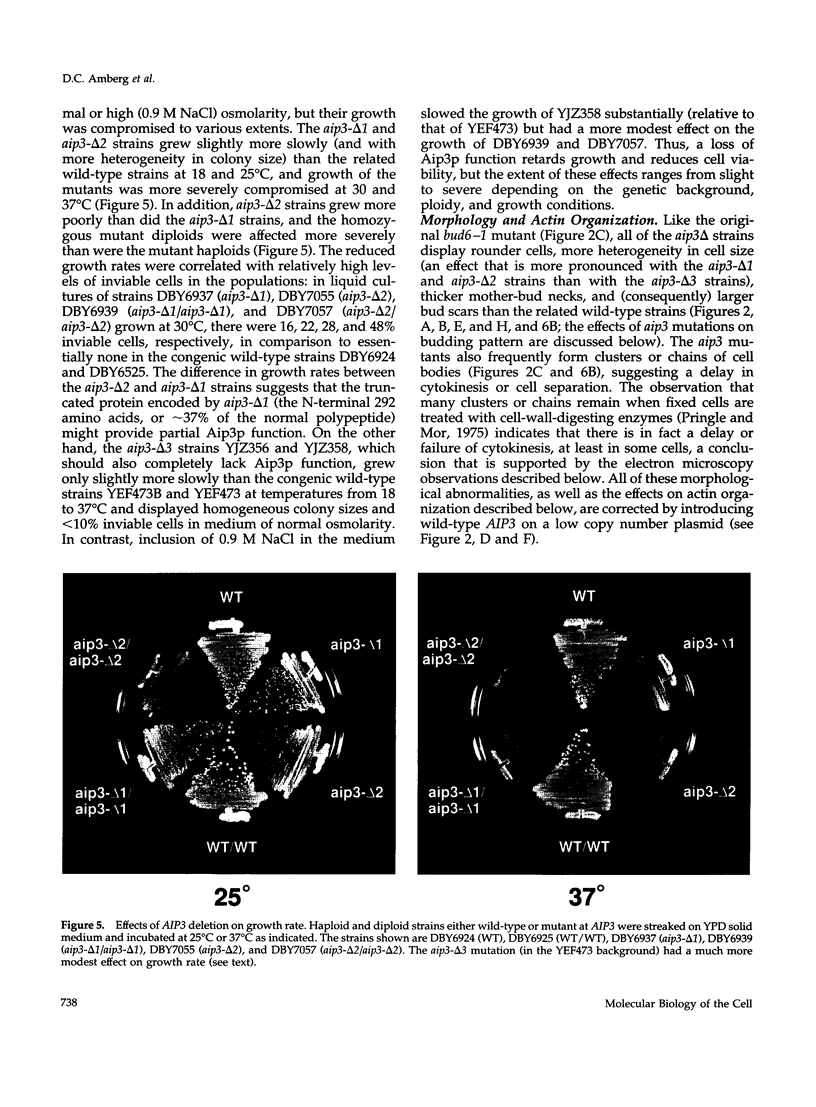
Selected References
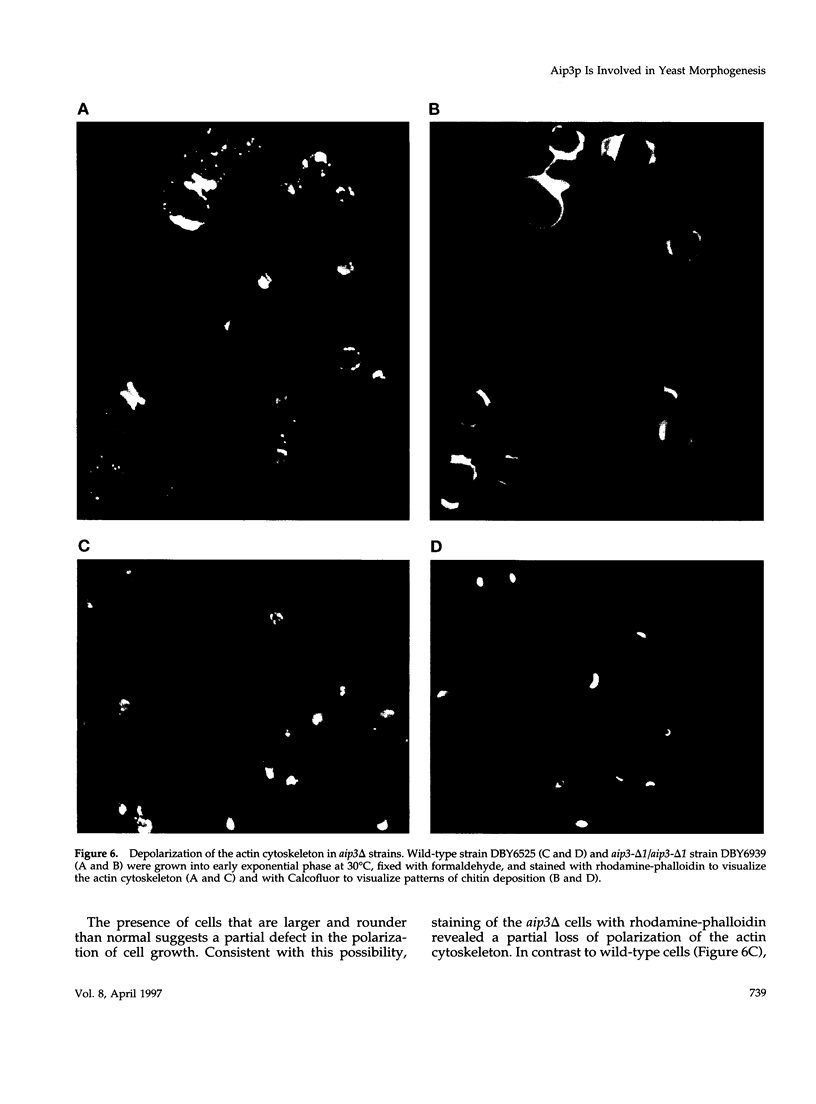
These references are in PubMed. This may not be the complete list of references from this article.
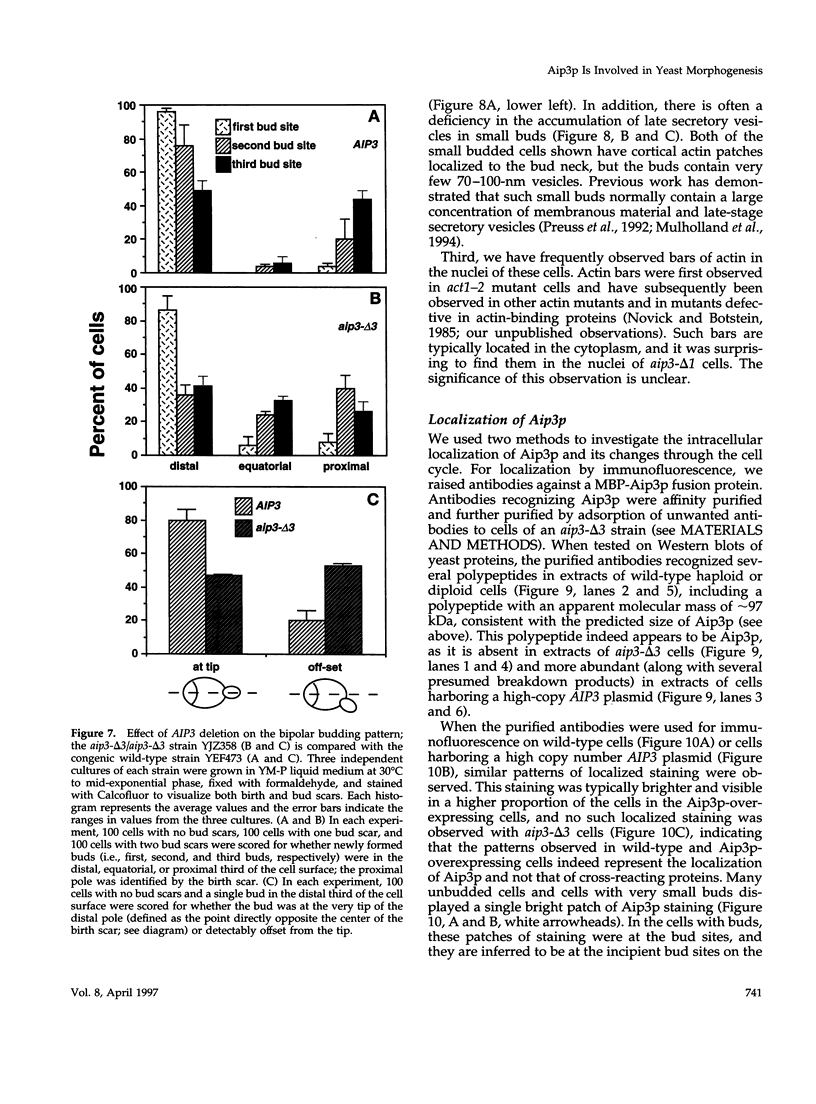
- Adams A. E., Botstein D., Drubin D. G. Requirement of yeast fimbrin for actin organization and morphogenesis in vivo. Nature. 1991 Dec 5;354(6352):404–408. doi: 10.1038/354404a0. [DOI] [PubMed] [Google Scholar]
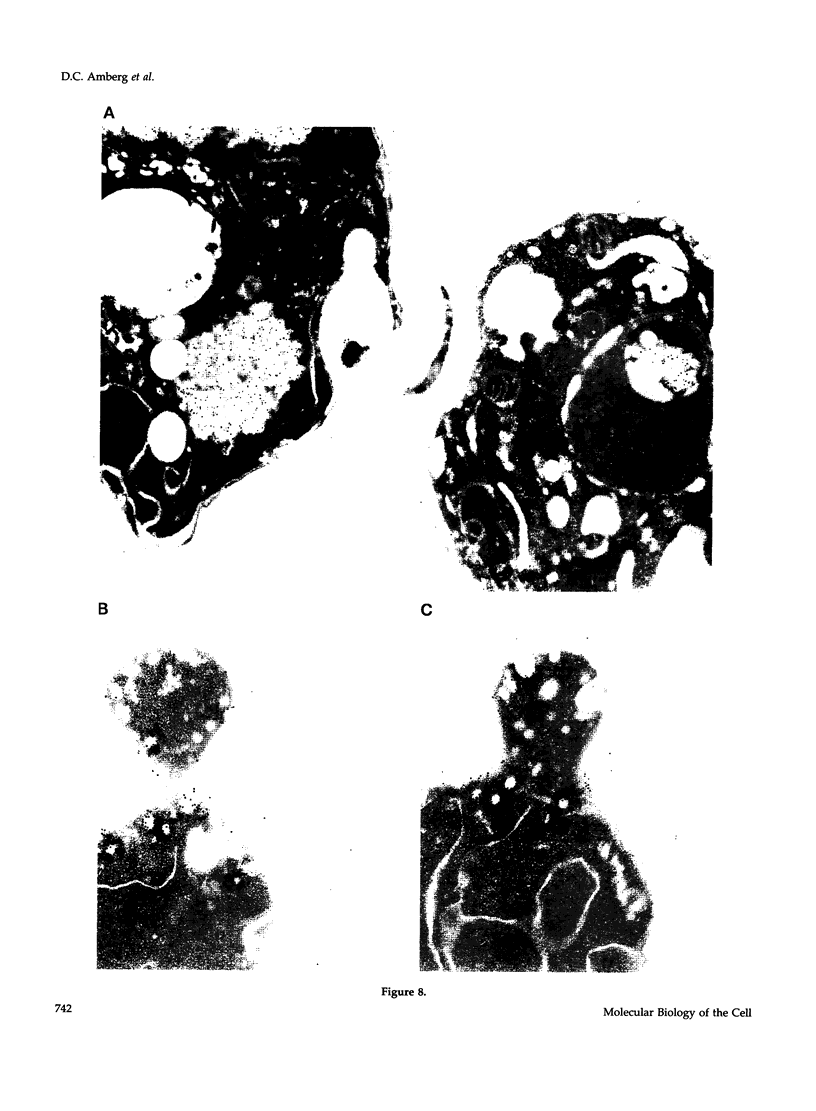
- Adams A. E., Pringle J. R. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984 Mar;98(3):934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
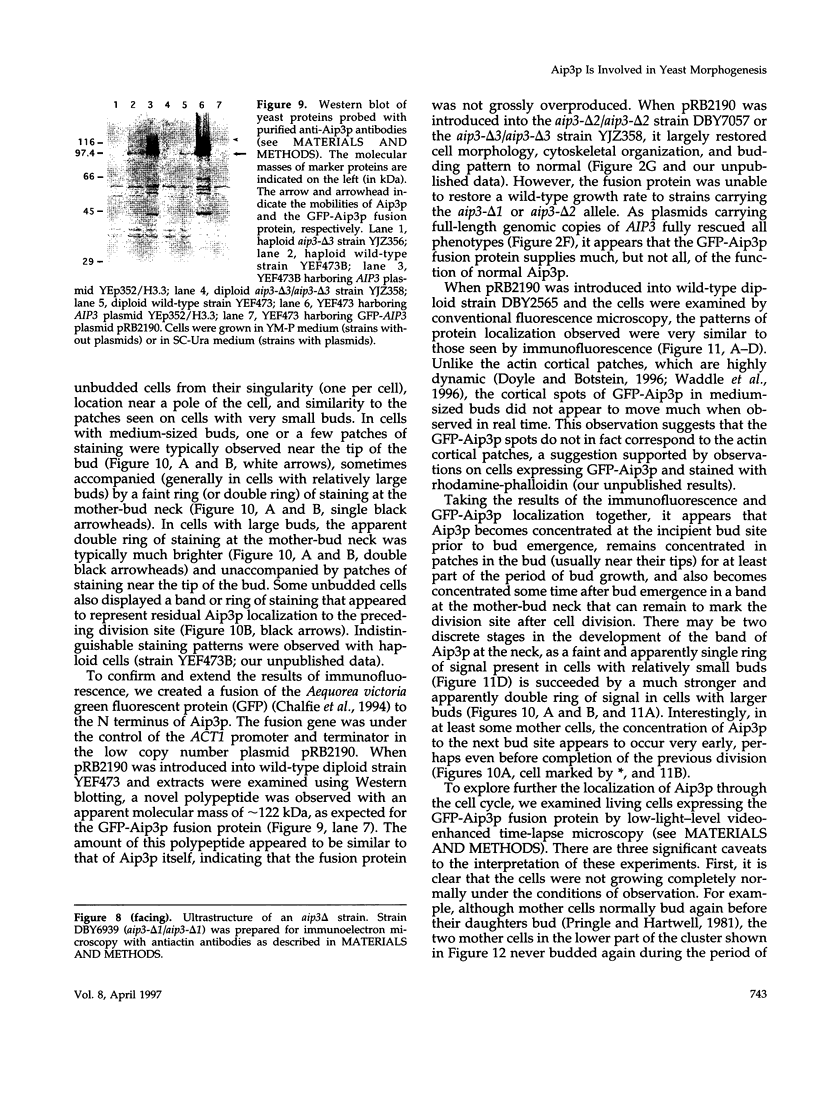
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
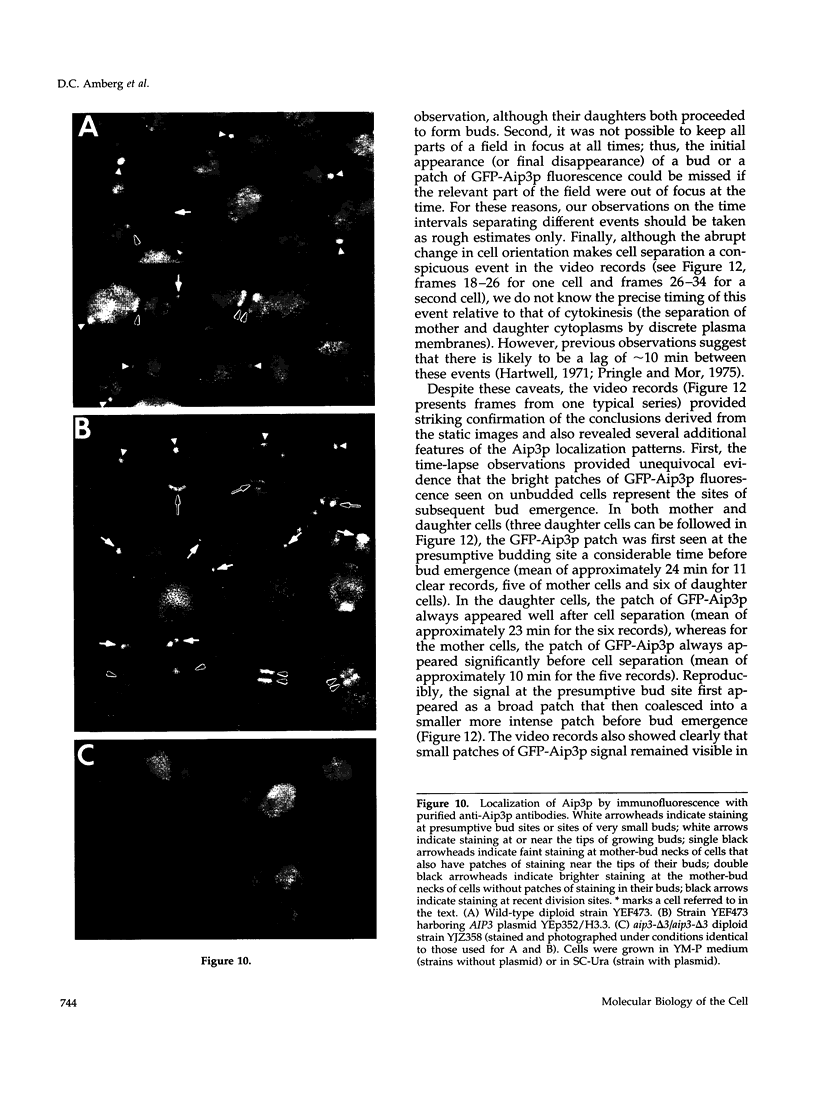
- Amatruda J. F., Cooper J. A. Purification, characterization, and immunofluorescence localization of Saccharomyces cerevisiae capping protein. J Cell Biol. 1992 Jun;117(5):1067–1076. doi: 10.1083/jcb.117.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
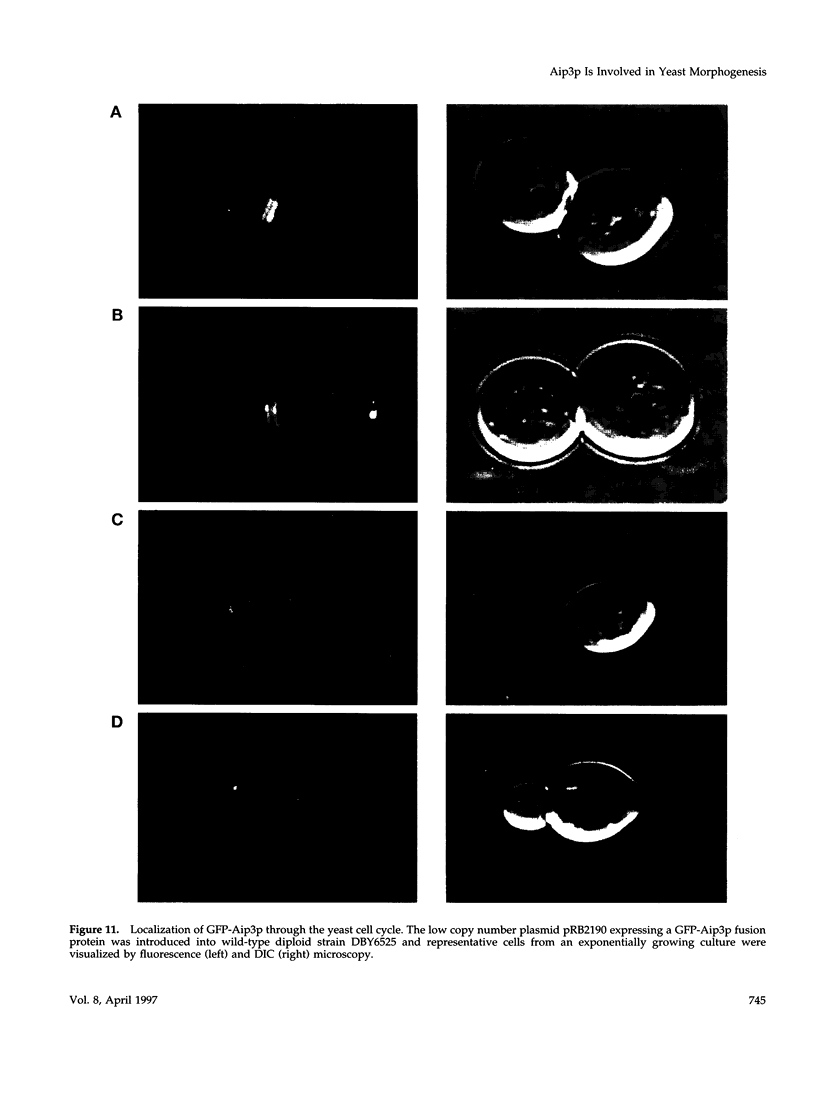
- Amberg D. C., Basart E., Botstein D. Defining protein interactions with yeast actin in vivo. Nat Struct Biol. 1995 Jan;2(1):28–35. doi: 10.1038/nsb0195-28. [DOI] [PubMed] [Google Scholar]
- Amberg D. C., Botstein D., Beasley E. M. Precise gene disruption in Saccharomyces cerevisiae by double fusion polymerase chain reaction. Yeast. 1995 Oct;11(13):1275–1280. doi: 10.1002/yea.320111307. [DOI] [PubMed] [Google Scholar]
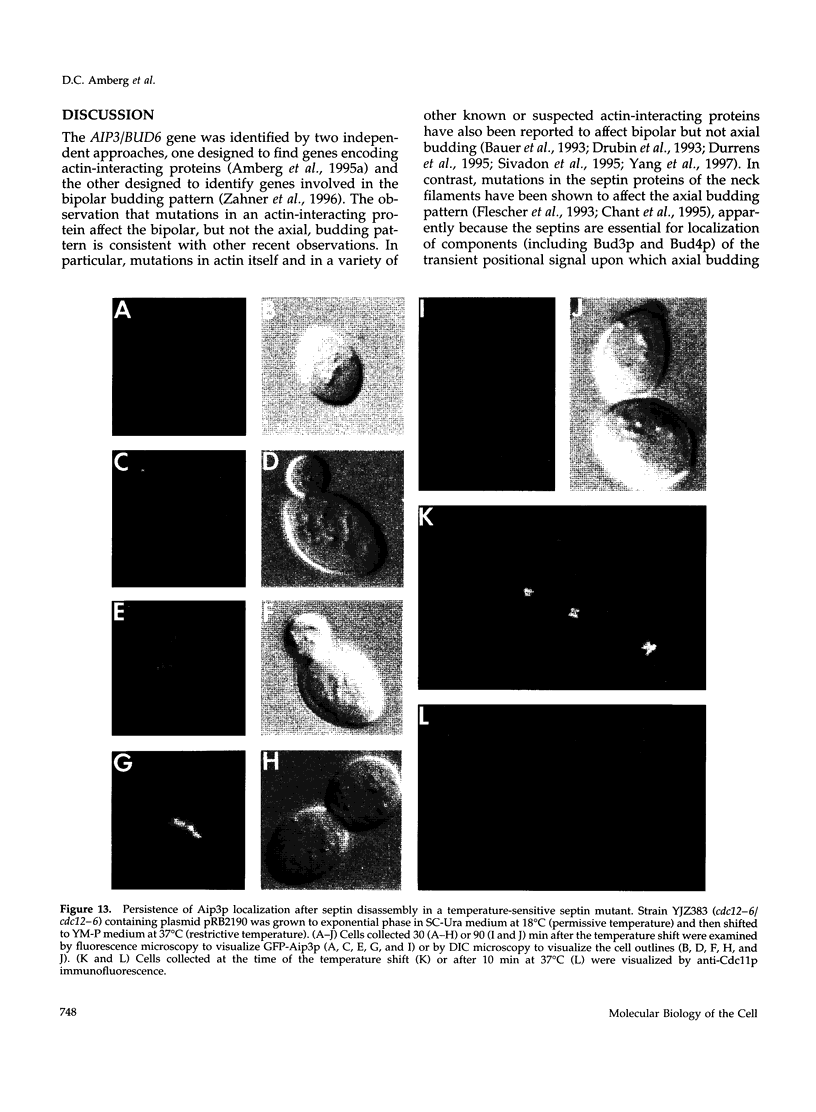
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993 Jul 11;21(14):3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer F., Urdaci M., Aigle M., Crouzet M. Alteration of a yeast SH3 protein leads to conditional viability with defects in cytoskeletal and budding patterns. Mol Cell Biol. 1993 Aug;13(8):5070–5084. doi: 10.1128/mcb.13.8.5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D. M., Guarente L. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 1991;194:182–187. doi: 10.1016/0076-6879(91)94015-5. [DOI] [PubMed] [Google Scholar]
- Bi E., Pringle J. R. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol Cell Biol. 1996 Oct;16(10):5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilbe G., Delabie J., Brüggen J., Richener H., Asselbergs F. A., Cerletti N., Sorg C., Odink K., Tarcsay L., Wiesendanger W. Restin: a novel intermediate filament-associated protein highly expressed in the Reed-Sternberg cells of Hodgkin's disease. EMBO J. 1992 Jun;11(6):2103–2113. doi: 10.1002/j.1460-2075.1992.tb05269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A., Drees B., Harsay E., Schott D., Wang T. What are the basic functions of microfilaments? Insights from studies in budding yeast. J Cell Biol. 1994 Aug;126(4):821–825. doi: 10.1083/jcb.126.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Goetsch L. A highly ordered ring of membrane-associated filaments in budding yeast. J Cell Biol. 1976 Jun;69(3):717–721. doi: 10.1083/jcb.69.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bénédetti H., Raths S., Crausaz F., Riezman H. The END3 gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol Biol Cell. 1994 Sep;5(9):1023–1037. doi: 10.1091/mbc.5.9.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M., Tu Y., Euskirchen G., Ward W. W., Prasher D. C. Green fluorescent protein as a marker for gene expression. Science. 1994 Feb 11;263(5148):802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Chant J., Herskowitz I. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell. 1991 Jun 28;65(7):1203–1212. doi: 10.1016/0092-8674(91)90015-q. [DOI] [PubMed] [Google Scholar]
- Chant J., Mischke M., Mitchell E., Herskowitz I., Pringle J. R. Role of Bud3p in producing the axial budding pattern of yeast. J Cell Biol. 1995 May;129(3):767–778. doi: 10.1083/jcb.129.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J., Pringle J. R. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J Cell Biol. 1995 May;129(3):751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Sullivan D. S., Huffaker T. C. Two yeast genes with similarity to TCP-1 are required for microtubule and actin function in vivo. Proc Natl Acad Sci U S A. 1994 Sep 13;91(19):9111–9115. doi: 10.1073/pnas.91.19.9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S., Smith K. W., Gustin M. C. Osmotic stress and the yeast cytoskeleton: phenotype-specific suppression of an actin mutation. J Cell Biol. 1992 Aug;118(3):561–571. doi: 10.1083/jcb.118.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle T., Botstein D. Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc Natl Acad Sci U S A. 1996 Apr 30;93(9):3886–3891. doi: 10.1073/pnas.93.9.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees B., Brown C., Barrell B. G., Bretscher A. Tropomyosin is essential in yeast, yet the TPM1 and TPM2 products perform distinct functions. J Cell Biol. 1995 Feb;128(3):383–392. doi: 10.1083/jcb.128.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin D. G., Jones H. D., Wertman K. F. Actin structure and function: roles in mitochondrial organization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol Biol Cell. 1993 Dec;4(12):1277–1294. doi: 10.1091/mbc.4.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin D. G., Nelson W. J. Origins of cell polarity. Cell. 1996 Feb 9;84(3):335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Durrens P., Revardel E., Bonneu M., Aigle M. Evidence for a branched pathway in the polarized cell division of Saccharomyces cerevisiae. Curr Genet. 1995 Feb;27(3):213–216. doi: 10.1007/BF00326151. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fields S., Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989 Jul 20;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Fishkind D. J., Wang Y. L. New horizons for cytokinesis. Curr Opin Cell Biol. 1995 Feb;7(1):23–31. doi: 10.1016/0955-0674(95)80041-7. [DOI] [PubMed] [Google Scholar]
- Flescher E. G., Madden K., Snyder M. Components required for cytokinesis are important for bud site selection in yeast. J Cell Biol. 1993 Jul;122(2):373–386. doi: 10.1083/jcb.122.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford S. K., Pringle J. R. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC11 gene product and the timing of events at the budding site. Dev Genet. 1991;12(4):281–292. doi: 10.1002/dvg.1020120405. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988 Dec 30;74(2):527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Goodson H. V., Anderson B. L., Warrick H. M., Pon L. A., Spudich J. A. Synthetic lethality screen identifies a novel yeast myosin I gene (MYO5): myosin I proteins are required for polarization of the actin cytoskeleton. J Cell Biol. 1996 Jun;133(6):1277–1291. doi: 10.1083/jcb.133.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson H. V., Spudich J. A. Identification and molecular characterization of a yeast myosin I. Cell Motil Cytoskeleton. 1995;30(1):73–84. doi: 10.1002/cm.970300109. [DOI] [PubMed] [Google Scholar]
- Haarer B. K., Petzold A., Lillie S. H., Brown S. S. Identification of MYO4, a second class V myosin gene in yeast. J Cell Sci. 1994 Apr;107(Pt 4):1055–1064. doi: 10.1242/jcs.107.4.1055. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971 Dec;69(2):265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Holmes K. C., Popp D., Gebhard W., Kabsch W. Atomic model of the actin filament. Nature. 1990 Sep 6;347(6288):44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- Holtzman D. A., Wertman K. F., Drubin D. G. Mapping actin surfaces required for functional interactions in vivo. J Cell Biol. 1994 Jul;126(2):423–432. doi: 10.1083/jcb.126.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honts J. E., Sandrock T. S., Brower S. M., O'Dell J. L., Adams A. E. Actin mutations that show suppression with fimbrin mutations identify a likely fimbrin-binding site on actin. J Cell Biol. 1994 Jul;126(2):413–422. doi: 10.1083/jcb.126.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K., Moriyama K., Matsumoto S., Kawasaki H., Nishida E., Yahara I. Isolation of a yeast essential gene, COF1, that encodes a homologue of mammalian cofilin, a low-M(r) actin-binding and depolymerizing protein. Gene. 1993 Feb 14;124(1):115–120. doi: 10.1016/0378-1119(93)90770-4. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston G. C., Prendergast J. A., Singer R. A. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J Cell Biol. 1991 May;113(3):539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. S., Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990 Sep-Oct;6(5):363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Mannherz H. G., Suck D., Pai E. F., Holmes K. C. Atomic structure of the actin:DNase I complex. Nature. 1990 Sep 6;347(6288):37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- Kilmartin J. V., Adams A. E. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984 Mar;98(3):922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. B., Haarer B. K., Pringle J. R. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J Cell Biol. 1991 Feb;112(4):535–544. doi: 10.1083/jcb.112.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölling R., Nguyen T., Chen E. Y., Botstein D. A new yeast gene with a myosin-like heptad repeat structure. Mol Gen Genet. 1993 Mar;237(3):359–369. doi: 10.1007/BF00279439. [DOI] [PubMed] [Google Scholar]
- Kübler E., Riezman H. Actin and fimbrin are required for the internalization step of endocytosis in yeast. EMBO J. 1993 Jul;12(7):2855–2862. doi: 10.1002/j.1460-2075.1993.tb05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lew D. J., Reed S. I. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J Cell Biol. 1995 May;129(3):739–749. doi: 10.1083/jcb.129.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D. J., Reed S. I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993 Mar;120(6):1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie S. H., Pringle J. R. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol. 1980 Sep;143(3):1384–1394. doi: 10.1128/jb.143.3.1384-1394.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. P., Bretscher A. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell. 1989 Apr 21;57(2):233–242. doi: 10.1016/0092-8674(89)90961-6. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., DeMarini D. J., Valencik M. L., Al-Awar O. S., Fares H., De Virgilio C., Pringle J. R. The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996 Feb;8(1):106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- Magdolen V., Oechsner U., Müller G., Bandlow W. The intron-containing gene for yeast profilin (PFY) encodes a vital function. Mol Cell Biol. 1988 Dec;8(12):5108–5115. doi: 10.1128/mcb.8.12.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklos D., Caplan S., Mertens D., Hynes G., Pitluk Z., Kashi Y., Harrison-Lavoie K., Stevenson S., Brown C., Barrell B. Primary structure and function of a second essential member of the heterooligomeric TCP1 chaperonin complex of yeast, TCP1 beta. Proc Natl Acad Sci U S A. 1994 Mar 29;91(7):2743–2747. doi: 10.1073/pnas.91.7.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon A. L., Janmey P. A., Louie K. A., Drubin D. G. Cofilin is an essential component of the yeast cortical cytoskeleton. J Cell Biol. 1993 Jan;120(2):421–435. doi: 10.1083/jcb.120.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland J., Preuss D., Moon A., Wong A., Drubin D., Botstein D. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J Cell Biol. 1994 Apr;125(2):381–391. doi: 10.1083/jcb.125.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P., Botstein D. Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell. 1985 Feb;40(2):405–416. doi: 10.1016/0092-8674(85)90154-0. [DOI] [PubMed] [Google Scholar]
- Preuss D., Mulholland J., Franzusoff A., Segev N., Botstein D. Characterization of the Saccharomyces Golgi complex through the cell cycle by immunoelectron microscopy. Mol Biol Cell. 1992 Jul;3(7):789–803. doi: 10.1091/mbc.3.7.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J. R., Bi E., Harkins H. A., Zahner J. E., De Virgilio C., Chant J., Corrado K., Fares H. Establishment of cell polarity in yeast. Cold Spring Harb Symp Quant Biol. 1995;60:729–744. doi: 10.1101/sqb.1995.060.01.079. [DOI] [PubMed] [Google Scholar]
- Pringle J. R., Mor J. R. Methods for monitoring the growth of yeast cultures and for dealing with the clumping problem. Methods Cell Biol. 1975;11:131–168. doi: 10.1016/s0091-679x(08)60320-9. [DOI] [PubMed] [Google Scholar]
- Pringle J. R., Preston R. A., Adams A. E., Stearns T., Drubin D. G., Haarer B. K., Jones E. W. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Riles L., Dutchik J. E., Baktha A., McCauley B. K., Thayer E. C., Leckie M. P., Braden V. V., Depke J. E., Olson M. V. Physical maps of the six smallest chromosomes of Saccharomyces cerevisiae at a resolution of 2.6 kilobase pairs. Genetics. 1993 May;134(1):81–150. doi: 10.1093/genetics/134.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J. R., Paterson B. M. Yeast myosin heavy chain mutant: maintenance of the cell type specific budding pattern and the normal deposition of chitin and cell wall components requires an intact myosin heavy chain gene. Cell Motil Cytoskeleton. 1990;17(4):301–308. doi: 10.1002/cm.970170405. [DOI] [PubMed] [Google Scholar]
- Sanders S. L., Herskowitz I. The BUD4 protein of yeast, required for axial budding, is localized to the mother/BUD neck in a cell cycle-dependent manner. J Cell Biol. 1996 Jul;134(2):413–427. doi: 10.1083/jcb.134.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Heid H. W., Schäfer S., Nuber U. A., Zimbelmann R., Franke W. W. Desmosomes and cytoskeletal architecture in epithelial differentiation: cell type-specific plaque components and intermediate filament anchorage. Eur J Cell Biol. 1994 Dec;65(2):229–245. [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivadon P., Bauer F., Aigle M., Crouzet M. Actin cytoskeleton and budding pattern are altered in the yeast rvs161 mutant: the Rvs161 protein shares common domains with the brain protein amphiphysin. Mol Gen Genet. 1995 Feb 20;246(4):485–495. doi: 10.1007/BF00290452. [DOI] [PubMed] [Google Scholar]
- Snyder M., Gehrung S., Page B. D. Studies concerning the temporal and genetic control of cell polarity in Saccharomyces cerevisiae. J Cell Biol. 1991 Aug;114(3):515–532. doi: 10.1083/jcb.114.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney F. P., Pocklington M. J., Orr E. The yeast type II myosin heavy chain: analysis of its predicted polypeptide sequence. J Muscle Res Cell Motil. 1991 Feb;12(1):61–68. doi: 10.1007/BF01781175. [DOI] [PubMed] [Google Scholar]
- Thorne K. J., Thornley M. J., Naisbitt P., Glauert A. M. The nature of the attachment of a regularly arranged surface protein to the outer membrane of an Acinetobacter sp. Biochim Biophys Acta. 1975 Apr 21;389(1):97–116. doi: 10.1016/0005-2736(75)90388-0. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursic D., Culbertson M. R. The yeast homolog to mouse Tcp-1 affects microtubule-mediated processes. Mol Cell Biol. 1991 May;11(5):2629–2640. doi: 10.1128/mcb.11.5.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinh D. B., Drubin D. G. A yeast TCP-1-like protein is required for actin function in vivo. Proc Natl Acad Sci U S A. 1994 Sep 13;91(19):9116–9120. doi: 10.1073/pnas.91.19.9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddle J. A., Karpova T. S., Waterston R. H., Cooper J. A. Movement of cortical actin patches in yeast. J Cell Biol. 1996 Mar;132(5):861–870. doi: 10.1083/jcb.132.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M. D., Holtzman D. A., Drubin D. G. The yeast actin cytoskeleton. Curr Opin Cell Biol. 1994 Feb;6(1):110–119. doi: 10.1016/0955-0674(94)90124-4. [DOI] [PubMed] [Google Scholar]
- Wertman K. F., Drubin D. G., Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992 Oct;132(2):337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Ayscough K. R., Drubin D. G. A role for the actin cytoskeleton of Saccharomyces cerevisiae in bipolar bud-site selection. J Cell Biol. 1997 Jan 13;136(1):111–123. doi: 10.1083/jcb.136.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahner J. E., Harkins H. A., Pringle J. R. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996 Apr;16(4):1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]