Abstract
H-, N- and K-ras4B are lipid-anchored, peripheral membrane guanine nucleotide binding proteins. Recent work has shown that Ras proteins are laterally segregated into non-overlapping, dynamic domains of the plasma membrane called nanoclusters. This lateral segregation is important to specify Ras interactions with membrane-associated proteins, effectors and scaffolding proteins and is critical for Ras signal transduction. Here we review biological, in vitro and structural data that provide insight into the molecular basis of how palmitoylated Ras proteins are anchored to the plasma membrane. We explore possible mechanisms for how the interactions of H-ras with a lipid bilayer may drive nanocluster formation.
Keywords: Ras, nanocluster, structure, lipid-raft, MD simulations
Introduction
The plasma membrane is a complex dynamic structure that exhibits heterogeneity on multiple length and time scales. The origins of this heterogeneity are diverse and include several thousand species of lipid, compartmentalization by a sub-membrane actin cytoskeleton, and the lateral assembly of sphingolipids and cholesterol into transient liquid-ordered domains. Further mixing and heterogeneity may be contributed by endocytic and exocytic processes. An important concept in modern cell biology is that this plasma membrane lateral heterogeneity imposes a non-random distribution of proteins across different nanoscale domains. The selective concentration of signalling proteins by proteolipid-based sorting to discrete areas of the cell membrane may increase the efficiency and specificity of signalling events and prevent cross-talk between different pathways.
The Ras GTPases are small lipid-anchored proteins that are associated predominantly with the inner leaflet of the plasma membrane, but exchange constantly with internal membranes, including the Golgi, ER, endosomes and mitochondria. Work over the last few years has shown that Ras proteins are organized in specific nanodomains on the plasma membrane. In fact, analysis of Ras nanodomains and those populated by GPI-anchored proteins has provided some of the clearest evidence that membrane-bound lipid-anchored proteins are non-randomly distributed on the nanoscale. In this review we will explore how new insights derived from molecular dynamics simulations of Ras proteins on model membranes when combined with recent advances in cell biology suggest an intriguing model for how lipid-anchored Ras proteins may actually drive nanodomain formation on the plasma membrane.
Ras proteins are spatially segregated on the plasma membrane
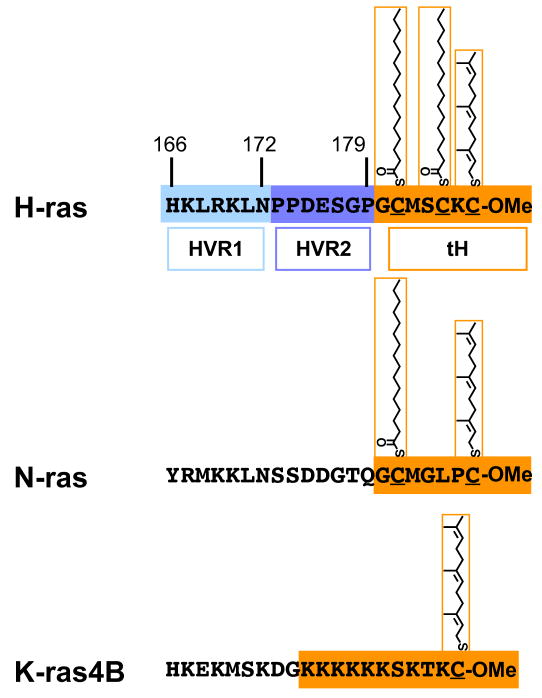
Three Ras isoforms H-, N-, and K-ras4B (hereafter referred to as K-ras) are ubiquitously expressed in mammalian cells. These proteins share a highly conserved G-domain that binds guanine nucleotides and interacts with a common set of effectors and exchange factors. The major sequence differences between isoforms are confined to the C-terminal 24-25 amino acids, called the hypervariable region. Ras proteins must be anchored to cell membranes for biological activity. The lipidic membrane anchor is attached via a series of posttranslational modifications. The first set of three modifications is directed by the C-terminal CAAX motif and is common to all Ras isoforms. Ras is first farnesylated on the cysteine residue of the CAAX motif by the cytosolic enzyme farnesyl protein transferase [1, 2]. The AAX sequence is removed by Rce1 [3] and the now C-terminal farnesyl cysteine is caboxylmethylated by Icmt [4] (Fig. 1). These two latter processing steps take place on the cytosolic surface of the ER (reviewed in Ref. [5]). Processed thus far, Ras binds with low affinity to cellular membranes and is able to autonomously exchange between them [6–8]. For plasma membrane targeting an additional second signal is necessary. In the case of N- and H-ras this is provided by mono- or dual-palmitoylation of cysteines adjacent to the prenylated cysteine, carried out by palmitoyl transferases localized to the ER and Golgi [9]. In the case of K-ras the second signal is a stretch of basic residues [10] (Fig. 1).
Figure 1.
The processed hypervariable regions of H-, N- and K-ras. In the case of H-ras, the division of the linker region of the HVR into HVR1 and 2 (blue hues) is shown. The minimal plasma membrane targeting sequences are outlined in orange and include C-terminal farnesylation and carboxylmethylation, and the palmitoyl modifications or the stretch of basic residues. The minimal plasma membrane targeting sequence of H-ras is abbreviated tH.
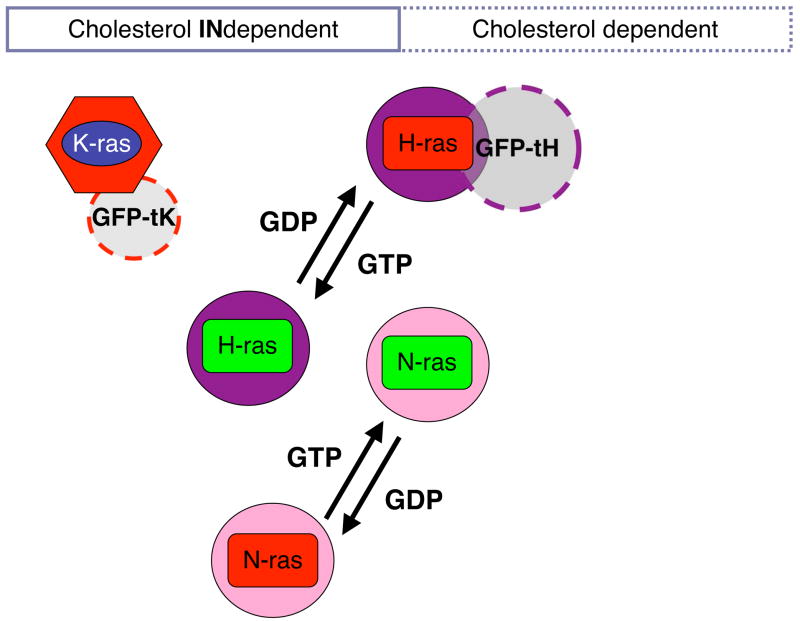
The different membrane anchors laterally segregate the Ras isoforms into spatially distinct domains on the plasma membrane. This was first shown using electron microscopy (EM) of intact apical plasma membrane sheets followed by statistical analysis of the immunogold point patterns. H-ras and K-ras cluster in nanoscale domains with a characteristic radius of 6–12 nm. Importantly, the nanodomains occupied by H-ras and K-ras do not show any significant overlap [11, 12]. Interestingly, not only are the different Ras isoforms laterally segregated, but inactive GDP-loaded H-ras occupies nanodomains that are spatially distinct from those occupied by GTP-loaded H-ras, thus H-ras exhibits GTP-dependent lateral segregation on the plasma membrane [11, 13, 14]. More recent work now suggests that this same phenomenon is also exhibited by N-ras [14], although the spatial relationship, between N- and H-ras nanodomains remains unresolved (Fig. 2). The spatial segregation of a largely conserved G-domain with an identical in vitro biochemistry to different nanodomains that differ in their protein and lipidomic content offers an ingeniously simple way for the Ras isoforms to generate their different effector activation profiles [15–17].
Figure 2.
Nanodomains of the classical Ras isoforms characterized by EM and statistical analysis of the point patterns of immunogold-labelled plasma membrane sheets. Thus, both the sensitivity of nanoclustering towards cholesterol depletion or the co-clustering of two proteins can be examined. Both H-ras and N-ras show GTP-dependent lateral segregation into nanoclusters with different sensitivities to cholesterol depletion. It is unclear whether K-ras exhibits a similar GTP-dependent lateral segregation. Note that only proteins in nanoclusters are shown; randomly distributed proteins are omitted.
Ras proteins operate in nanoclusters
An important question to consider when evaluating the clustered distribution of Ras proteins on the plasma membrane is how this non-random patterning arises. Do Ras proteins partition into pre-existing nanodomains, or do Ras proteins actively participate in the formation of nanodomains. In this context it is worth considering a recent analysis of the surface distribution of GPI-anchored proteins on the outer leaflet of the plasma membrane. Through a combination of computational simulation and homo- and hetero-FRET measurements, it was shown that GPI-anchored proteins are organized in what have been termed nanoclusters, which have a small diameter (~5 nm) and contain at most 4 proteins [18]. The formation of these nanoclusters is cholesterol-dependent in that it is abolished if plasma membrane cholesterol is depleted. Only 20–40% of GPI-anchored proteins are in nanoclusters, the remainder are randomly arrayed over the plasma membrane. Interestingly, the ratio of nanoclustered to random GPI-anchored proteins remains constant over a wide range of expression levels [18], strongly suggesting that the distribution is in someway actively generated or maintained [18, 19].
It has recently become clear that a similar nanoclustering process operates for Ras proteins on the inner leaflet of the plasma membrane. Analysis of EM-data aided by Monte-Carlo simulations of immunogold point patterns shows that approximately 30–40% of Ras proteins form nanoclusters with diameters in the range of ~6–20 nm containing 6–8 proteins [12]. Specifically, there are ~7.7 Ras proteins in a K-ras-GTP nanocluster (exemplified by K-rasG12V) compared with ~5.9 in H-ras-GTP nanoclusters (exemplified by H-rasG12V). Importantly, the lipid-modified, minimal membrane anchoring sequences of H- or K-ras alone are also sufficient to induce nanoclustering, as shown by EM [11, 12] or inferred from FRET-data [20]. The similarity in the distributions of lipidated proteins on the inner and outer leaflets of the plasma membrane suggests that common biophysical principals may operate on membrane anchors to drive nanoclustering [12, 19, 21, 22].
Further insights into Ras surface organization have been revealed using single fluorophore video tracking (SFVT) to analyze the diffusion and mobility of individual Ras proteins. In non-stimulated cells the majority of individual YFP-H-ras or -K-ras proteins diffuse as fast as phospholipids and in an apparently random, Brownian mode, while 16% of the proteins are immobile [23]. After stimulation with EGF the immobile fraction increases to approximately 50% and the diffusion rate of the fast-diffusing population drops by a factor of 3–4. Tracking of constitutively activated G12V mutants also shows periods of immobility, lasting up to 1 s, alternating with mobile periods. An independent study found similar results, with slowed diffusion upon activation of H-ras and an increase in the fraction of proteins diffusing in a confined mode [24]. Interestingly, a correlation between GTP-dependent activation and lateral mobility has also been observed with other lipid-anchored proteins. Heterotrimeric G proteins dissociate into monomeric Gα and a dimeric Gβγ subunit after GTP loading. Again, SFVT reveals that ~80% of the Gq heterotrimers are immobile compared with only ~40% of monomeric Gαq [25].
By synthesizing dynamic and nanoclustering data, we can now generate a partial picture of Ras protein dynamics and organization on the membrane: a fixed fraction of Ras protein is organized into nanoclusters, the lifetime of these nanoclusters varies, and appears longer for Ras-GTP nanoclusters (0.5–1 s) than inactive Ras-GDP clusters (<0.1 s) [26]. There is accumulating evidence from EM and SFVT experiments that the immobile Ras-GTP nanoclusters are the sites of effector recruitment and activation, and that the mobile monomeric Ras proteins do not recruit cytosolic effectors [23, 26, 27]. It is tempting to speculate that Ras-GDP nanoclusters are the immediate targets of activated growth factor receptors (GFR) that have recruited Ras exchange factors such as Sos1. The engagement of a Sos1-loaded-GFR with a Ras-GDP nanocluster would be an efficient mechanism to ensure the activation of multiple Ras molecules through a single collision event. Moreover, different Ras-GDP nanoclusters may have differential access to an activated GFR depending on the ability of that GFR to partition into, or associate with, a Ras nanocluster. Indeed, a recent FRET analysis of Ras activation is consistent with this hypothesis [28]. Taken together these data strongly suggest that Ras nanoclusters should be thought of as dynamic, transient signalling platforms [26]. The imminent signalling characteristics of such signalling nanoclusters were recently elucidated for the MAPK-pathway [27]. By combining computational modelling and experimental data it could be shown that K-ras nanoclusters operate as sensitive analogue-digital switches, which transduce graded ligand input into a fixed ERK-signalling output.
Finally, it is important to stress that the nanoclustering concept implies that Ras proteins are critically involved in actually generating the nanodomains in which they operate. In the next sections we will consider what aspects of Ras protein structure may contribute to the biological and perhaps biophysical mechanisms of nanoclustering.
The biology of Ras nanoclustering
The formation of different Ras nanoclusters is a complex interplay between multiple determinants on Ras and the plasma membrane. Cell biological experiments initially identified some of the critical determinants that drive Ras nanoclustering and GTP-dependent lateral segregation. For example, different Ras nanoclusters exhibit differential requirements for plasma membrane cholesterol. H-ras-GDP, N-ras-GTP and GFP-tH (GFP fused to the minimal membrane anchor of H-ras) nanoclusters are cholesterol-dependent, whereas H-ras-GTP, N-ras-GDP, K-ras-GTP and GFP-tK (GFP fused to the minimal membrane anchor of K-ras) nanoclusters are cholesterol-independent (Fig. 2). In addition, latrunculin treatment of cells reveals that H-ras-GDP and GFP-tH nanoclusters are also actin dependent, whereas H-ras-GTP nanoclusters are actin independent [12].
The correlation between cholesterol and actin dependence of GFP-tH and H-ras-GDP nanoclustering also extends to GPI-anchored protein nanoclustering on the outer plasma membrane. It is worth considering this association in more detail. At a fundamental level the formation of nanoclusters represents a collision, or series of collisions in 2D, between a set of lipid-anchored proteins and will therefore be influenced by the diffusive behaviour of the proteins. The plasma membrane is not a well-mixed, homogeneous medium for diffusion, and obstacles such as transmembrane proteins anchored to the cytoskeleton [29], cholesterol-dependent nanoclusters and other types of microdomains result in varying degrees of anomalous diffusion [30, 31]. For example the compartmentalized hop-diffusion of proteins visualized by high-speed single particle tracking is attributed to the effects of a submembrane actin-meshwork [32]. Interestingly, the size of these diffusion-compartments is cell-type specific [33] and recent ultrastructural analysis has confirmed that they indeed match the compartment-size imposed by the submembrane cytoskeletal meshwork [34]. It is possible that small Lo domains (which may be transient lateral assemblies of cholesterol and sphingolipid [22, 29]) require compartmentalization by a picket fence in order to promote cholesterol-dependent nanoclustering, a hypothesis that could be explored mathematically as in Nicolau et al. (2007) [31]. Alternatively there may be a direct interaction between cholesterol-dependent domains and the cytoskeleton perhaps mediated via PIP2-binding proteins. In this context, it is worth noting that cholesterol depletion leads to a redistribution and loss of plasma membrane binding of PIP2-binding proteins with concomitant reorganization of the cytoskeleton [35]. However, the requirement for an intact actin cytoskeleton to maintain nanoclustering does not always overlap with the requirement for cholesterol [12], suggesting that the link between actin and membrane cholesterol is indirect. For example, nanoclustering of K-ras-GTP is independent of cholesterol, but is still partially abrogated by disrupting the actin cytoskeleton [12].
Which components of the Ras protein are necessary for nanoclustering? In the case of H-ras the membrane interactions of the minimal membrane anchor are extensively modified by the adjacent linker region of the HVR and by the G-domain. Analysis of a large set of HVR deletion and alanine substitution mutants of H-ras in the GTP- and GDP-bound states using a combination of EM and fluorescence recovery after photobleaching (FRAP) has allowed the mapping of the critical determinants of H-ras membrane affinity and lateral segregation. The minimal membrane anchor of H-ras provides a high affinity interaction with cholesterol-dependent nanoclusters [11, 12]. The addition of the G-domain to the minimal membrane anchor results in a much lower membrane affinity, effectively equivalent to the G-domain providing a repulsive force that operates against the minimal anchor [36]. Interestingly this force is greater when the G-domain is GTP-loaded than GDP-loaded [36]. The linker region provides an additional source of membrane affinity but for cholesterol-independent nanoclusters. Analysis of this linker region in turn shows that the C-terminal half of the HVR (residues 173-179, HVR2) simply functions as a spacer, because it can be substituted by alanine residues, whereas there is an absolute requirement for some or all of the amino acid residues between 166-172 (HVR1) [36, 37]. Taken together these results led to a cell biological vector model that can account for H-ras GTP-dependent lateral segregation [36].
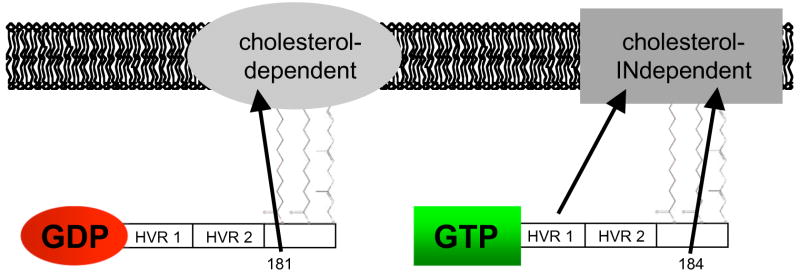
Subsequent work has shown that the palmitates in the minimal membrane anchor of H-ras also serve distinct roles. Both contribute to the overall membrane affinity of H-ras; but palmitate on Cys181 is required for efficient Golgi exit and forward trafficking to the plasma membrane whereas palmitate on 184 is essential for cholesterol-independent nanoclustering of GTP-loaded H-ras (Fig. 3) [14]. Interestingly, an H-rasG12V mutant that is mono-palmitoylated on Cys181, and thus has a minimal membrane anchor that closely mimics N-ras, exhibits cholesterol-dependent nanoclustering when GTP-loaded as does N-ras [14]. Simple vectorial models such as those in Rotblat et al. (2004) [36] and the summary model shown in Fig. 3 are useful because they explain H-ras membrane anchorage and lateral segregation by three separable forces, provided by structural elements. What such models do not provide, however, is a biophysical basis for the different vectors or forces that seemingly modulate H-ras membrane interactions. In the next sections we will consider this issue in the light of some very recent studies using model membrane systems.
Figure 3.
Vectorial model of GTP-dependent H-ras lateral segregation as defined by EM and FRAP analysis. This model complements a previous vectorial model in Rotblat et al. (2004) [36], by adding the contributions from the individual palmitoyls and HVR1.
Ras membrane interactions from MD simulations and in vitro studies
What are the structural mechanisms that drive clustering of the diverse Ras membrane anchors? A solution to this question would flow directly from 3D structures of Ras in different activation states on an intact membrane. However, despite substantial efforts in recent years, there are only few 3D structures of membrane-bound protein domains. This is because these domains are difficult to approach by X-ray crystallography or by solution NMR [38]. Nevertheless, membrane-binding mechanisms have been elucidated by combining X-ray crystallographic or NMR structures of soluble domains of membrane-bound proteins, with biophysical and cell-biological experiments on the intact, full length membrane-bound protein [39–42]. Other methods, such as electron paramagnetic resonance (EPR), a combination of solid state NMR and related spectroscopic approaches (such as electron-nuclear double resonance) also provide useful structural insights into peptide-membrane interactions [43–47]. The emerging picture of proteins bound onto the surface of membranes is that membrane binding domains: (i) contain a pocket for the specific recognition of a unique lipid ligand (e.g. the C1, PH and FYVE domains [48, 49]), (ii) rely on the electrostatic fields generated by multiple acidic phospholipid groups (e.g. PKC-α C2 and BAR domains [50]) or (iii) use multiple covalent lipid modifications (e.g. N- and H-ras) or a combination of lipid modification and a polybasic domains (e.g. K-ras).
Membrane interactions of N-ras
Spectroscopic studies of N-ras [46, 51, 52] have significantly advanced our understanding of how Ras proteins are anchored to the plasma membrane. Analysis of a heptapeptide representing the minimal N-ras C-terminal membrane anchor (Fig. 1) incorporated into a DMPC bilayer shows that interactions between the Ras lipids and hydrophobic DMPC acyl chains provide the major force for a stable membrane anchorage [51]. A computational study carried out on the same system in turn provids some atomic details of the N-ras peptide-DMPC interactions [53]. The peptide atoms are distributed such that the amino acid backbone and a proline residue reside among the phospholipid head groups, whereas the hydrophobic side chains of Met182 and Leu184 (Fig. 1) insert into the upper acyl chain region of the hydrophobic membrane core. This observation illustrates how membrane binding of the Ras anchor can benefit from both van der Waals interactions and hydrogen bonds from the peptide backbone [53]. Furthermore, while the effect of Ras peptide insertion on the overall structure of the host lipid is minimal [51, 53], some effect on the DMPC lipids close to those of Ras is evident [53]. The acyl chains of the Ras peptide exhibit increased dynamics and reduced order, which allow them to adapt to the DMPC hydrophobic chain length [54], or POPC chain length [46]. Recently, the N-ras membrane anchor synthetically joined to the G-domain has been studied [46, 52]. The overall pattern of the anchor peptide-membrane interaction and the structure of the anchor is consistent with earlier MD predictions [53]. The anchor has a “horse-shoe” shape [52] with substantial flexibility at its termini [46]. Interestingly, AFM imaging of full length N-ras on three component lipid bilayers identifies a significant fraction of the protein at the interface of Lo and Ld regions of the membrane [55]. This observation suggests that the presence of the G-domain may, in complex model membranes, provide additional determinants for N-ras membrane interactions.
Membrane interactions of H-ras
Compared with the extensive data on N-ras, very little structural data are available on H-ras except for a solution NMR study showing that the HVR influences the structure and dynamics of the G-domain [56]. Recently, however, a MD simulation study has provided a detailed description of the structure and dynamics of full-length H–ras in a DMPC bilayer [57]. The membrane association of H-ras is mainly due to the van der Waals interactions between the lipids attached to the Ras anchor and DMPC lipids in the bilayer (Fig. 4). There are additional contributions from the Met182 side chain interacting with DMPC acyl chains, similar to the N-ras anchor peptide, and hydrogen bonding between the anchor backbone and DMPC phosphate and carbonyl-oxygen atoms. Furthermore, Ser183 and Lys185 side chains hydrogen bond with DMPC phosphate and help maintain the anchor backbone at the lipid-water interface.
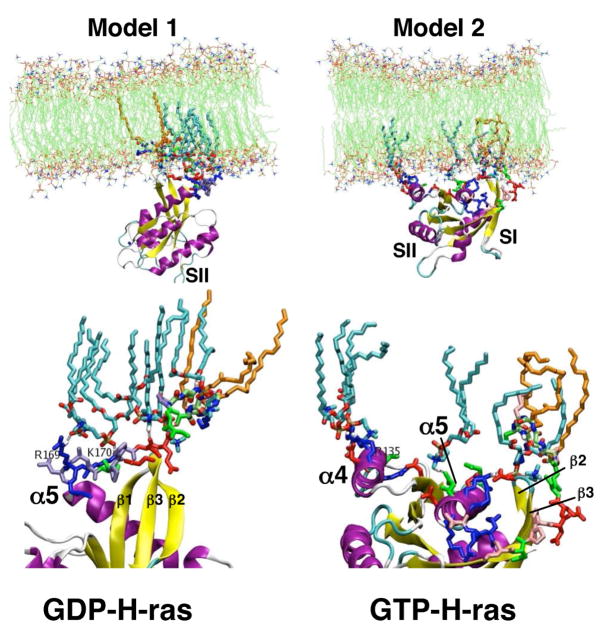
Figure 4.
The two models of membrane bound H-ras obtained from MD simulations. Model 1(left) is the predominant conformation found for H-ras-GDP, while model 2 (right) is the predominant conformation for H-ras-GTP. The blowup of the interaction sites (bottom) shows lipids (cyan) and interacting residues of H-ras in sticks. Note that model 1 is predominantly anchored via the HVR, while model 2 shows extensive contact of α-helix 4 with the membrane. Membrane lipids (cyan) with specific contacts to H-ras residues or its lipids (orange) are shown in sticks.
The rest of the H-ras protein variously interacts with the surface of the membrane, but most interestingly the MD simulations suggest two modes of Ras binding to the DMPC bilayer. The two modes, referred to as model 1 and model 2 (Fig. 4) differ in the orientation of the G-domain relative to the membrane plane and in the interactions of positively charged residues with membrane phosphates. In model 1, which is the preferred conformation of GDP-bound H-ras, basic amino acids in the HVR including Arg169 and Lys170 interact with DMPC phosphates. In model 2, which is the preferred conformation of GTP-bound H-ras, basic amino acids in α-helix 4 of the G-domain (Arg128 and Arg135) interact with DMPC phosphates. Differences between model 1 and 2 also involve the membrane core penetration of the Ras lipids. The Ras acyl chains populate the entire hydrophobic core in model 1, in an extended conformation, but only the lower leaflet in model 2 (Fig. 4) [57]. FRET experiments (D. Abankwa and J. F. Hancock, unpublished data) and biological assays of mutant H-ras proteins with alanine substitutions in α-helix 4 and HVR1 [57] validate the importance of these regions in Ras-membrane interactions. It is possible that these observations may also be relevant to N-ras, which appears to undergo substantial, but as yet poorly defined, structural changes when tethered to a lipid monolayer [58].
It is interesting to note that the G-domain switch 1 region is more accessible to cytosolic effectors in model 2 than model 1. This is also true for other residues implicated in specific effector interactions, such as Tyr64 with phosphoinositide 3-kinase γ. It has been noticed previously that Ras-effector complexes exhibit different angles of interaction on the plasma membrane [59]. The observed change in orientation of the G-domain relative to the plane of the membrane on GTP-binding may therefore be important for optimal effector recruitment by membrane-bound H-ras. Another difference between model 1 and 2 is that the anchor peptide is more deeply buried in the DMPC bilayer in model 1 than model 2. This observation predicts that membrane-bound Ras thioesterases would have easier access to the thioester linkages of palmitate 181 and 184 in model 2 than model 1. This different accessibility may account for the faster turnover rates of palmitate on GTP-loaded H-ras compared to GDP-loaded H-ras that is observed in vivo [7, 8, 60].
A model for H-ras nanodomain formation
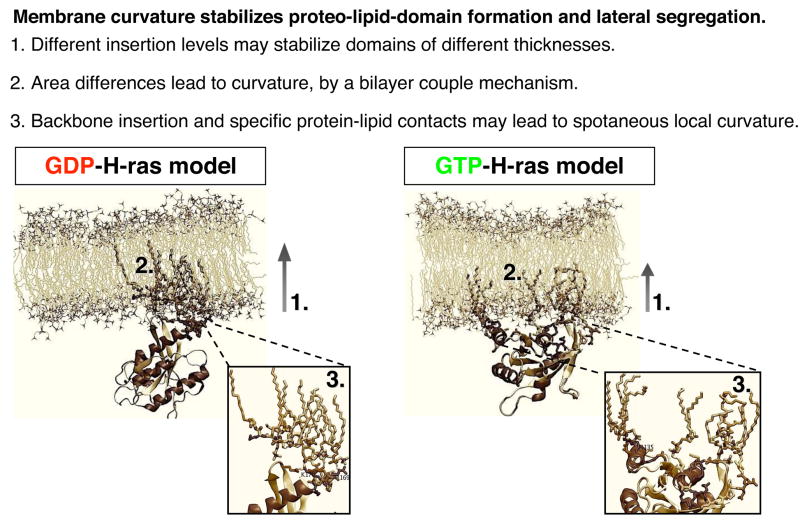
The limited structural and thermodynamic data currently available do not yet allow for a complete deduction of how H-ras proteins drive the formation of signalling nanodomains. However, some insights can be gained by considering recent structural data in conjunction with physics-based theoretical studies of membrane mechanical properties. In this context the induction of membrane curvature is especially relevant since curvature is associated with lipid sorting and the attendant probability of forming lipid domains [61]. In brief, surface-bound membrane proteins, depending on the depth of membrane penetration, can induce membrane-curvature by three related mechanisms (for an excellent review see [62]). Scaffold mechanisms rely on the intrinsic curvature of a protein, or network of proteins, with a rigid tertiary structure and a curved surface to mechanically bend the lipid bilayer, e.g. the BAR domain proteins and clathrin-adaptor protein complexes. Local spontaneous curvature mechanisms involve monolayer deformation caused by proteins with amphipathic helices that become embedded in the lipid bilayer, e.g. epsin and the GTPase Sar1. Bilayer couple mechanisms operate when protein domains penetrate only one leaflet of a bilayer causing an area difference between the inner and outer leaflets and a subsequent compensatory curvature. The latter two mechanisms often operate together. The direct involvement of these mechanisms for protein nanocluster formation is suggested by a recent coarse grain simulation, which showed that curvature imposed by proteins results in a net attractive force, which results in the aggregation of these proteins [63].
In both model 1 and model 2 the insertion of Ras into the inner leaflet will produce an area difference between the two leaflets. According to the bilayer-couple mechanism the membrane will develop a curvature to compensate. In addition, the interfacial localization of the anchor backbone may perturb the bilayer via a local spontaneous curvature mechanism. This is consistent with the local effect of H-ras on the DMPC lipid structure observed during the MD simulations [57]. Moreover, the area of the inner leaflet occupied by Ras appears to be greater in model 2 (GTP) than model 1 (GDP) suggesting that the two conformations may have different propensities to induce curvature. These effects whilst small for single Ras proteins will be enhanced as Ras nanoclusters form. The depth of insertion and the ordering of the acyl chains also differ between the two H-ras conformations. The extended, ordered acyl chains of GDP-bound model 1 are well placed to interact with and perhaps capture and stabilize transient Lo domains in the plasma membrane. In accordance with current views of the ‘raft hypothesis’ [22] H-Ras-GDP/Lo complexes would be the basic building block for a cholesterol-dependent nanocluster.
The orientation of the G-domain, which is regulated by GTP to GDP exchange [57] controls the insertion depth of the anchor and acyl chains and determines whether GTP- or GDP clustering is favoured. Consideration of the energetics of Ras-membrane interaction illustrates how this switching may occur. The insertion level of the anchor into model membranes depends, at least in part, on whether the backbone amides preferentially interact with the phosphate- or the lipid glycerol-oxygen atoms. Hydrogen bonding between an amide and a glycerol carbonyl will be stronger given the more apolar nature of its micro-environment. Thus, deeper insertion of the lipid tails is possible because the anchor has the potential to gain in free energy through enhanced hydrogen bonding from the backbone, as well as increased van der Waals interactions from complete insertion of the lipid tails and hydrophobic side chains. This scenario implies that GDP-H-ras will always insert deeper, and consequently segregate to or capture Lo domains, aided by the extended acyl-chains.
How is it then that the HVR and GTP-loaded H-ras direct localization to cholesterol-independent nanoclusters? Because the linker carries several charges and three prolines, it is to be expected that it prefers to remain solvated. When considering the G-domain, it appears that it plays two contrasting roles. On the one hand, because the G-domain rigidifies the linker [57], it reduces the entropic advantage of a solvated linker. This implies that the G-domain enhances the preference for deeper membrane penetration. On the other hand, the linker may need to be solvent-accessible to functionally interact with other molecules, possibly galectin-1 as will be discussed in the next section, which requires interfacial localization. This can be achieved by the ability of the G-domain to force the linker to adopt a solvent-accessible conformation. As the two calculated structures (Fig. 4) suggest, these two roles can be balanced by shifting the orientation of the G-domain relative to the membrane plane. Therefore, the linker and the G-domain regulate the insertion level of the anchor, which may support, from an energetic perspective, alternative mechanisms of nanoclustering.
Role of scaffold proteins in intact cells
In intact cells additional proteins influence Ras nanodomain organization. One such protein is galectin-1, a prenyl-binding protein that is recruited to, or stabilized on, the plasma membrane by interacting with GTP-loaded H-ras. The determinants on H-ras required for galectin-1 interaction overlap with those required for GTP-dependent lateral segregation. In order to bind to galectin-1 in intact cells H-ras-GTP must be prenylated and have an HVR of the correct length with an intact HVR1. However, neither of the two anchor palmitates is required for H-ras-GTP to bind galectin-1 (Y. Kloog and J. F. Hancock, unpublished data). These and other results suggest that galectin-1 may sequester the prenyl group and leave H-ras-GTP anchored to the membrane by palmitates alone. Interpreting these data in the context of the MD modelling, it seems probable that galectin-1 preferentially interacts with and stabilizes the model 2 conformation of H-ras. In this conformation the anchor peptide is buried less deeply in the bilayer (Fig. 4) such that the prenyl group may be more available for sequestration by galectin-1. In model 2 the HVR1 is also more solvent accessible. The consequence of this mode of galectin-1-H-ras interaction, which leaves all other determinants of Ras membrane binding unaffected, would be to facilitate the cholesterol-independent nanodomain formation driven by H-ras-GTP (Fig. 5).
Figure 5.
Summary of mechanisms by which membrane insertion of Ras could drive nanoclustering. Differences in membrane insertion of GDP- and GTP-loaded H-ras provide some intriguing insights in the biophysical principals that may drive activation-dependent lateral segregation.
In support of these concepts, ectopic expression of galectin-1 increases the extent of H-ras-GTP nanoclustering and also the radius of the nanoclusters [11, 26], whereas knockdown of galectin-1 expression significantly abrogates H-ras-GTP nanoclustering [11]. Modulation of H-ras nanoclustering by galectin-1 levels also correlates closely with changes in H-ras signal output [64–66]. Interestingly, another galectin family member, galectin-3, predominantly interacts with activated K-ras and increases Raf-1 and phosphoinositide 3-kinase activation [67]. It will be interesting to see whether these signalling effects are also due to modified nanoclustering of K-ras, and in turn whether MD simulations of K-ras yield new insight into domain formation by this Ras isoform.
Conclusion
The MD simulated structure of membrane bound H-ras, when combined with experimental results from molecular cell biological, EM and SFVT studies, leads to an intriguing model for how the H-ras protein may drive the formation of nanoclusters on the plasma membrane. However, much more work needs to be done in order to gain a thorough understanding of the molecular and dynamic aspects of nanoclustering. This will involve using diverse approaches to explore the mechanisms and consequences of nanoclustering of lipid-anchored proteins on the inner and outer plasma membrane. Computational simulation of nanocluster formation may be possible, using a combination of atomistic and more coarse grained modelling. Such simulations could shed some light on the cooperativity of the nanoclustering process. The dynamics of formation and disassembly of nanoclusters remain poorly characterized, as does the mechanism whereby the cluster-to-monomer ratio is maintained at relatively constant levels as protein expression levels vary. Current biophysical data suggest that nanoclusters are short-lived and highly dynamic. In the near future, high-end imaging, fluorescence correlation spectroscopy (FCS), simultaneous FRAP and anisotropy imaging and further exploitation of FRET techniques should provide more detailed quantitative kinetic data. Important questions to answer include: what is the exchange rate of nanocluster constituents, does nanocluster assembly and disassembly occur stochastically, and can it be regulated by the cell? More generally, it will be interesting to explore what emergent signalling properties flow from organizing Ras proteins into dynamic, transient signalling platforms [27]. These and other questions will continue to focus attention and interest on the membrane interactions of Ras and other lipid-anchored proteins as we strive to understand the complexities of the plasma membrane and the full extent to which it acts as a master regulator for signalling networks that are assembled thereon.
Acknowledgments
DA is a fellow of the Swiss National Science Foundation (FNS). AAG thanks the Commission for the Promotion of Young Academics of the University of Zurich, the Center for Theoretical Biological Physics of the University of California San Diego and the San Diego Super Computer Center. JFH is supported by grants from the National Institutes of Health (USA), National Health and Medical Research Council (Australia) and Australian Research Council (ARC). The IMB is a Special Research Center of the ARC.
Abbreviations
- DMPC
1,2-dimyristoyl-sn-Glycero-3-phosphocholine
- EM
electron microscopy
- FCS
fluorescence correlation spectroscopy
- FRAP
fluorescence recovery after photobleaching
- FRET
fluorescence resonance energy transfer
- MβCD
Methyl-β-cyclodextrin
- MD
molecular dynamics
- POPC
1-Palmitoyl-2-Oleoyl-sn-Glycero-3-Phosphocholine
- SFVT
single fluorophore video tracking
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hancock JF, Magee AI, Childs JE, Marshall CJ. Cell. 1989;57:1167–77. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- 2.Casey PJ, Solski PA, Der CJ, Buss JE. Proc Natl Acad Sci U S A. 1989;86:8323–7. doi: 10.1073/pnas.86.21.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergo MO, Lieu HD, Gavino BJ, Ambroziak P, Otto JC, Casey PJ, Walker QM, Young SG. J Biol Chem. 2004;279:4729–36. doi: 10.1074/jbc.M310081200. [DOI] [PubMed] [Google Scholar]
- 4.Pillinger MH, Volker C, Stock JB, Weissmann G, Philips MR. J Biol Chem. 1994;269:1486–92. [PubMed] [Google Scholar]
- 5.Hancock JF. Nat Rev Mol Cell Biol. 2003;4:373–84. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 6.Silvius JR, Bhagatji P, Leventis R, Terrone D. Mol Biol Cell. 2006;17:192–202. doi: 10.1091/mbc.E05-05-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI. Science. 2005;307:1746–52. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 8.Goodwin JS, Drake KR, Rogers C, Wright L, Lippincott-Schwartz J, Philips MR, Kenworthy AK. J Cell Biol. 2005;170:261–72. doi: 10.1083/jcb.200502063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linder ME, Deschenes RJ. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 10.Hancock JF, Paterson H, Marshall CJ. Cell. 1990;63:133–9. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 11.Prior IA, Muncke C, Parton RG, Hancock JF. J Cell Biol. 2003;160:165–70. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plowman SJ, Muncke C, Parton RG, Hancock JF. Proc Natl Acad Sci U S A. 2005;102:15500–5. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prior IA, Harding A, Yan J, Sluimer J, Parton RG, Hancock JF. Nat Cell Biol. 2001;3:368–75. doi: 10.1038/35070050. [DOI] [PubMed] [Google Scholar]
- 14.Roy S, Plowman S, Rotblat B, Prior IA, Muncke C, Grainger S, Parton RG, Henis YI, Kloog Y, Hancock JF. Mol Cell Biol. 2005;25:6722–33. doi: 10.1128/MCB.25.15.6722-6733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehrhardt A, David MD, Ehrhardt GR, Schrader JW. Mol Cell Biol. 2004;24:6311–23. doi: 10.1128/MCB.24.14.6311-6323.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voice JK, Klemke RL, Le A, Jackson JH. J Biol Chem. 1999;274:17164–70. doi: 10.1074/jbc.274.24.17164. [DOI] [PubMed] [Google Scholar]
- 17.Yan J, Roy S, Apolloni A, Lane A, Hancock JF. J Biol Chem. 1998;273:24052–6. doi: 10.1074/jbc.273.37.24052. [DOI] [PubMed] [Google Scholar]
- 18.Sharma P, Varma R, Sarasij RC, Ira, Gousset K, Krishnamoorthy G, Rao M, Mayor S. Cell. 2004;116:577–89. doi: 10.1016/s0092-8674(04)00167-9. [DOI] [PubMed] [Google Scholar]
- 19.Mayor S, Rao M. Traffic. 2004;5:231–40. doi: 10.1111/j.1600-0854.2004.00172.x. [DOI] [PubMed] [Google Scholar]
- 20.Abankwa D, Vogel H. J Cell Sci. 2007;120:2953–2962. doi: 10.1242/jcs.001404. [DOI] [PubMed] [Google Scholar]
- 21.Sharma DK, Brown JC, Choudhury A, Peterson TE, Holicky E, Marks DL, Simari R, Parton RG, Pagano RE. Mol Biol Cell. 2004;15:3114–22. doi: 10.1091/mbc.E04-03-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hancock JF. Nat Rev Mol Cell Biol. 2006;7:456–62. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murakoshi H, Iino R, Kobayashi T, Fujiwara T, Ohshima C, Yoshimura A, Kusumi A. Proc Natl Acad Sci U S A. 2004;101:7317–22. doi: 10.1073/pnas.0401354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lommerse PH, Snaar-Jagalska BE, Spaink HP, Schmidt T. J Cell Sci. 2005;118:1799–809. doi: 10.1242/jcs.02300. [DOI] [PubMed] [Google Scholar]
- 25.Perez JB, Segura JM, Abankwa D, Piguet J, Martinez KL, Vogel H. J Mol Biol. 2006;363:918–30. doi: 10.1016/j.jmb.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 26.Hancock JF, Parton RG. Biochem J. 2005;389:1–11. doi: 10.1042/BJ20050231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian T, Harding A, Inder K, Plowman S, Parton RG, Hancock JF. Nat Cell Biol. 2007;9:905–14. doi: 10.1038/ncb1615. [DOI] [PubMed] [Google Scholar]
- 28.Fukano T, Sawano A, Ohba Y, Matsuda M, Miyawaki A. Cell Struct Funct. 2007;32:9–15. doi: 10.1247/csf.06019. [DOI] [PubMed] [Google Scholar]
- 29.Kusumi A, Koyama-Honda I, Suzuki K. Traffic. 2004;5:213–30. doi: 10.1111/j.1600-0854.2004.0178.x. [DOI] [PubMed] [Google Scholar]
- 30.Nicolau DV, Jr, Burrage K, Parton RG, Hancock JF. Mol Cell Biol. 2006;26:313–23. doi: 10.1128/MCB.26.1.313-323.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicolau DV, Jr, Hancock JF, Burrage K. Biophys J. 2007;92:1975–87. doi: 10.1529/biophysj.105.076869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujiwara T, Ritchie K, Murakoshi H, Jacobson K, Kusumi A. J Cell Biol. 2002;157:1071–81. doi: 10.1083/jcb.200202050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murase K, Fujiwara T, Umemura Y, Suzuki K, Iino R, Yamashita H, Saito M, Murakoshi H, Ritchie K, Kusumi A. Biophys J. 2004;86:4075–93. doi: 10.1529/biophysj.103.035717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morone N, Fujiwara T, Murase K, Kasai RS, Ike H, Yuasa S, Usukura J, Kusumi A. J Cell Biol. 2006;174:851–62. doi: 10.1083/jcb.200606007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwik J, Boyle S, Fooksman D, Margolis L, Sheetz MP, Edidin M. Proc Natl Acad Sci U S A. 2003;100:13964–9. doi: 10.1073/pnas.2336102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rotblat B, Prior IA, Muncke C, Parton RG, Kloog Y, Henis YI, Hancock JF. Mol Cell Biol. 2004;24:6799–810. doi: 10.1128/MCB.24.15.6799-6810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaumot M, Yan J, Clyde-Smith J, Sluimer J, Hancock JF. J Biol Chem. 2002;277:272–8. doi: 10.1074/jbc.M108423200. [DOI] [PubMed] [Google Scholar]
- 38.Hurley JH. Biochim Biophys Acta. 2006;1761:805–11. doi: 10.1016/j.bbalip.2006.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stahelin RV, Karathanassis D, Bruzik KS, Waterfield MD, Bravo J, Williams RL, Cho WW. Journal of Biological Chemistry. 2006;281:39396–39406. doi: 10.1074/jbc.M607079200. [DOI] [PubMed] [Google Scholar]
- 40.Cho W, Stahelin RV. Biochimica Et Biophysica Acta-Molecular and Cell Biology of Lipids. 2006;1761:838–849. doi: 10.1016/j.bbalip.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 41.Stahelin RV, Hwang JH, Kim JH, Park ZY, Johnson KR, Obeid LM, Cho WH. Journal of Biological Chemistry. 2005;280:43030–43038. doi: 10.1074/jbc.M507574200. [DOI] [PubMed] [Google Scholar]
- 42.Cho WH, Stahelin RV. Annual Review of Biophysics and Biomolecular Structure. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 43.Rauch ME, Ferguson CG, Prestwich GD, Cafiso DS. J Biol Chem. 2002;277:14068–76. doi: 10.1074/jbc.M109572200. [DOI] [PubMed] [Google Scholar]
- 44.Fanucci GE, Cadieux N, Kadner RJ, Cafiso DS. Proc Natl Acad Sci U S A. 2003;100:11382–7. doi: 10.1073/pnas.1932486100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rufener E, Frazier AA, Wieser CM, Hinderliter A, Cafiso DS. Biochemistry. 2005;44:18–28. doi: 10.1021/bi048370d. [DOI] [PubMed] [Google Scholar]
- 46.Reuther G, Tan KT, Vogel A, Nowak C, Arnold K, Kuhlmann J, Waldmann H, Huster D. J Am Chem Soc. 2006;128:13840–6. doi: 10.1021/ja063635s. [DOI] [PubMed] [Google Scholar]
- 47.Bringezu F, Majerowicz M, Wen S, Reuther G, Tan KT, Kuhlmann J, Waldmann H, Huster D. Eur Biophys J. 2006 doi: 10.1007/s00249-006-0119-x. [DOI] [PubMed] [Google Scholar]
- 48.Hurley JH, Anderson DE, Beach B, Canagarajah B, Ho YS, Jones E, Miller G, Misra S, Pearson M, Saidi L, Suer S, Trievel R, Tsujishita Y. Trends Biochem Sci. 2002;27:48–53. doi: 10.1016/s0968-0004(01)02022-9. [DOI] [PubMed] [Google Scholar]
- 49.Misra S, Miller GJ, Hurley JH. Cell. 2001;107:559–62. doi: 10.1016/s0092-8674(01)00594-3. [DOI] [PubMed] [Google Scholar]
- 50.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. Science. 2004;303:495–9. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 51.Huster D, Vogel A, Katzka C, Scheidt HA, Binder H, Dante S, Gutberlet T, Zschornig O, Waldmann H, Arnold K. J Am Chem Soc. 2003;125:4070–9. doi: 10.1021/ja0289245. [DOI] [PubMed] [Google Scholar]
- 52.Reuther G, Tan KT, Kohler J, Nowak C, Pampel A, Arnold K, Kuhlmann J, Waldmann H, Huster D. Angew Chem Int Ed Engl. 2006;45:5387–90. doi: 10.1002/anie.200504266. [DOI] [PubMed] [Google Scholar]
- 53.Gorfe AA, Pellarin R, Caflisch A. J Am Chem Soc. 2004;126:15277–86. doi: 10.1021/ja046607n. [DOI] [PubMed] [Google Scholar]
- 54.Vogel A, Katzka CP, Waldmann H, Arnold K, Brown MF, Huster D. J Am Chem Soc. 2005;127:12263–72. doi: 10.1021/ja051856c. [DOI] [PubMed] [Google Scholar]
- 55.Nicolini C, Baranski J, Schlummer S, Palomo J, Lumbierres-Burgues M, Kahms M, Kuhlmann J, Sanchez S, Gratton E, Waldmann H, Winter R. J Am Chem Soc. 2006;128:192–201. doi: 10.1021/ja055779x. [DOI] [PubMed] [Google Scholar]
- 56.Thapar R, Williams JG, Campbell SL. J Mol Biol. 2004;343:1391–408. doi: 10.1016/j.jmb.2004.08.106. [DOI] [PubMed] [Google Scholar]
- 57.Gorfe AA, Bayer M-H, Abankwa D, Hancock JF, McCammon JA. J Med Chem. 2007;50:674–84. doi: 10.1021/jm061053f. [DOI] [PubMed] [Google Scholar]
- 58.Meister A, Nicolini C, Waldmann H, Kuhlmann J, Kerth A, Winter R, Blume A. Biophys J. 2006;91:1388–401. doi: 10.1529/biophysj.106.084624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herrmann C. Curr Opin Struct Biol. 2003;13:122–9. doi: 10.1016/s0959-440x(02)00007-6. [DOI] [PubMed] [Google Scholar]
- 60.Baker TL, Zheng H, Walker J, Coloff JL, Buss JE. J Biol Chem. 2003;278:19292–300. doi: 10.1074/jbc.M206956200. [DOI] [PubMed] [Google Scholar]
- 61.Roux A, Cuvelier D, Nassoy P, Prost J, Bassereau P, Goud B. Embo J. 2005;24:1537–45. doi: 10.1038/sj.emboj.7600631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zimmerberg J, Kozlov MM. Nat Rev Mol Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 63.Reynwar BJ, Illya G, Harmandaris VA, Muller MM, Kremer K, Deserno M. Nature. 2007;447:461–4. doi: 10.1038/nature05840. [DOI] [PubMed] [Google Scholar]
- 64.Paz A, Haklai R, Elad-Sfadia G, Ballan E, Kloog Y. Oncogene. 2001;20:7486–93. doi: 10.1038/sj.onc.1204950. [DOI] [PubMed] [Google Scholar]
- 65.Elad-Sfadia G, Haklai R, Ballan E, Gabius HJ, Kloog Y. J Biol Chem. 2002;277:37169–75. doi: 10.1074/jbc.M205698200. [DOI] [PubMed] [Google Scholar]
- 66.Rotblat B, Niv H, Andre S, Kaltner H, Gabius HJ, Kloog Y. Cancer Res. 2004;64:3112–8. doi: 10.1158/0008-5472.can-04-0026. [DOI] [PubMed] [Google Scholar]
- 67.Elad-Sfadia G, Haklai R, Balan E, Kloog Y. J Biol Chem. 2004;279:34922–30. doi: 10.1074/jbc.M312697200. [DOI] [PubMed] [Google Scholar]