Abstract
Botrytis cinerea is a pathogenic filamentous fungus which infects more than 200 plant species. The enzymes secreted by B. cinerea play an important role in the successful colonization of a host plant. Some of the secreted enzymes are involved in the degradation of pectin, a major component of the plant cell wall. A total of 126 proteins secreted by B. cinerea were identified by growing the fungus on highly or partially esterified pectin, or on sucrose in liquid culture. Sixty-seven common proteins were identified in each of the growth conditions, of which 50 proteins exhibited a Signal P motif. Thirteen B. cinerea proteins with functions related to pectin degradation were identified in both pectin growth conditions, while only four were identified in sucrose. Our results indicate it is unlikely that the activation of B. cinerea from the dormant state to active infection is solely dependent on changes in the degree of esterification of the pectin component of the plant cell wall. Further, these results suggest that future studies of the B. cinerea secretome in infections of ripe and unripe fruits will provide important information that will describe the mechanisms that the fungus employs to access nutrients and decompose tissues.
Keywords: Host-pathogen interactions, Botrytis cinerea, Matrix polysaccharides, Proteomics, Mass spectrometry
1. INTRODUCTION
B. cinerea is a pathogenic filamentous fungus which infects more than 200 plant species in a variety of organs including fruit, flowers, and leaves. The host range for B. cinerea infection includes economically important crops such as tomato, berries, chickpeas, french beans, and grapes as well as cut flowers [1]. On certain fruit hosts B. cinerea initially infects while the fruits are green and remains dormant [2–4]. This quiescent infection is superceded by a resumption of fungal growth activity once the fruit ripens [2–4]. This resumption of growth and infection leads to postharvest losses and reduction in the shelf life of perishable products. Attempts to prevent postharvest disease lead to the use of fungicides in addition to those already in use for treatment of preharvest infections. The development of new strategies against postharvest infection would be of benefit from both an economic and an environmental standpoint because of the high cost and intrusive impact of the fungicides and the loss of consumable products.
Changes during the ripening process appear to play an important role in the activation of the dormant infection. One of the major processes of ripening involves the enzymatic de-esterification and depolymerization of the cell wall components, resulting in softening of the fruit [5, 6]. All the major components of the fruit cell wall, the pectin, cellulose and hemicellulose, undergo changes during ripening. Pectin is a major component of the plant cell wall, providing mechanical stability and influencing pH and ionic properties of the wall. The pectin backbone consists mainly of α-(1–4)-linked D-galacturonic acid [7]. The galacturonic acid is highly esterified in the pectin of unripened fruit [8]. A decrease in the degree of esterification of the galacturonic acid backbone, combined with changes in the average molecular weight and neutral sugar content of cell walls, is consistent with softening and ripening [5, 8–10].
B. cinerea secretes a battery of enzymes utilized for the degradation and consumption of the host plant. Pectin degradation by B. cinerea is enabled by enzymes, including pectin methyl esterases (PMEs), exopolygalacturonases (exo PGs), endopolygalacturonases (endo PGs), pectate and pectin lyases (PLs), and rhamnogalacturonan lyase and hydrolase [11–14]. The genome of B. cinerea has multiple isoforms for most of the above enzymes, for example six isoforms of endo PG have been previously reported [15]. During plant pathogen interactions, pectin fragments called oligogalacturonides are produced which act as defense elicitors. In Fragaria vesca, a partial degree of demethylation of oligogalacturonides may be required for eliciting defense responses to B. cinera infection [16]. Furthermore, in Arabidopsis, the overexpression of the pectin methylesterase inhibitor (PMEI) resulted in an increased resistance to B. cinerea, implying the importance of the degree of esterification of pectin to plant resistance [17].
To better understand at the molecular level the complex interaction between pathogen and host, we propose to use an idealized model system that is accessible and easily manipulated, and whose results can subsequently be incorporated into a biological model. Here we demonstrate the impact of the degree of esterification of pectin on fungal secretion and report on the secretome of B. cinerea when grown in liquid culture on three different carbon nutrient sources. Two of these conditions simulate fungal interactions with expected host nutrient sources.
Specifically, B. cinerea was grown in liquid cultures with highly esterified pectin, partially esterified pectin, and sucrose as sole carbon sources. Sucrose was used to define those enzymes which can be considered constitutively expressed. Shotgun proteomics was used to study the B. cinerea secretome. We observed changes in the profile of secreted proteins that were nutrient dependant, indicating an adaptability of B. cinerea to the growth conditions.
2. MATERIALS AND METHODS
Microorganisms and culture conditions
B cinerea strain (BO5.10) was a kind gift of the laboratory of John Labavitch, University of California-Davis. Fungal stock cultures were maintained on potato dextrose agar, PDA (BD Biosciences, MD, USA) at 37°C for 14 days. Conidia were harvested with a sterile 0.01% Tween 20 (w/v) solution and spores were gently suspended with a magnetic stirrer. Erlenmeyer flasks containing 150 ml of liquid media (described below) were inoculated with the spore suspension at 106 spores ml−1, and incubated at 24°C in an orbital shaker at 200 rpm. The growth medium contained KH2PO4 (0.29 g), K2HPO4 (0.94 g), (NH4)2SO42 (1.20 g), NaCl (0.15 g), CaCl2 H2O(0.40 g), MgSO4 (0.150 g), FeSO4 7H2O (0.015 mg), ZnSO4 7H2O (0.015 g), and MnSO4 H2O (0.150 mg). The medium was supplemented with one of the three following carbon sources: Citrus pectin (0.5% w/v) (Sigma-Aldrich, MO, USA) labeled as HE pectin, 30% esterified pectin (0.5% w/v) (Hercules DL USA) labeled as PE pectin, and sucrose (0.5% w/v) (Fisher Scientific, NJ, USA). The pH was adjusted to 4 with H2SO4. The medium was sterilized at 121°C for 15 min. After seven days the supernatant cultures were collected by filtration through a No. 41 Whatman filter paper and lyophilized. After lyophilization, the cultures were resuspended in 2 mL of deionized water prior to further desalting using a HiPrep 26/10 desalting column (GE, Pistcataway, NJ). The desalted fractions containing the secreted proteins were lyophilized and stored at −20 °C until further analysis.
Secreted protein isolation and separation by 1D-SDS -PAGE
Lyophilized secreted proteins were resuspended in 100 μl of deionized water. 30 μl of the sample was mixed with 10 μL of 2X Laemmli buffer (Sigma-Aldrich, MO, USA) and boiled at 95°C for 5 min before loading onto the gel. Proteins were separated on a 4–12% Bis-Tris preacasted gel (Invitrogen, Carlsbad, CA) using the 1x MOPS SDS buffer (Invitrogen, Carlsbad, CA) as running buffer. SeeBlue® Plus2 Prestained Standard molecular weight standards (Invitrogen, Carlsbad, CA) were used. After electrophoresis, proteins were visualized by staining with Coomassie blue. Each gel lane was excised into three sections of equal length and destained.
In gel digestion
Excised bands were first cut into smaller pieces (1 × 1 mm), dried by vacuum centrifugation, and reduced by submerging the gel pieces in 100 mM ammonium bicarbonate solution containing 10 mM dithiothreitol for 1 h at 55°C. Excess dithiothreitol/ammonium bicarbonate was removed and the same volume of 100 mM ammonium bicarbonate containing 55 mM iodoacetamide was added and incubated for 45 min in the dark. After alkylation, the gel pieces were treated with 100 mM ammonium bicarbonate and acetonitrile sequentially and then dried by vacuum centrifuge. To the dried gel pieces, 2 μg of trypsin was added in sufficient 100 mM ammonium bicarbonate solution to submerge the gel pieces in the solution. Digestion was carried out at 37°C overnight. The gel was washed once with ammonium bicarbonate followed by acetonitrile, and twice with 5% formic acid followed by acetonitrile. Peptides were collected from the washings, dried by vacuum centrifugation, and resuspended in 0.1% formic acid solution for mass spectrometric analysis.
LC-MS/MS analysis
The peptides from each sample were analyzed in duplicate. An Agilent 1100 capillary LC (Palo Alto, CA) was attached to the mass spectrometer via a T splitter to allow infusion at ηL flow rates. Five μm diameter C18 beads (Rainin, Woburn, MA) were packed into a pulled fused silica capillary (10.5 cm × 100 μm ID) under 1000 psi pressure using nitrogen gas. Peptide samples were loaded onto the column for 45 min under the same pressure. Peptides were then eluted with a gradient using 0.1% formic acid (buffer A) and 99.9% acetonitrile/0.1% formic acid (buffer B). Following the initial wash with 95% buffer A for 10 min, peptides were eluted from the column during a 90 min linear gradient of 5–60% of buffer B at a flow rate of ~200 ηL/min directly into a LTQ linear ion trap mass spectrometer (Thermo Fisher, San Jose, CA) using a voltage of 2500V.
The instrument was set to acquire MS/MS spectra on the nine most abundant precursor ions from each MS scan with a repeat count set of 3 and repeat duration of 5 sec. Dynamic exclusion was enabled for 160 sec. Raw tandem mass spectra were converted into a peak list using ReAdW followed by mzMXL2Other algorithms [18]. The peak lists were then searched using Mascot 1.9 (Matrix Science, Boston, MA).
Database searching and protein identification
A target database was created from B. cinerea BO5.10 genes (Broad Institute, MA). A decoy database was then constructed by reversing the sequences in the normal database. Searches were performed against the target and decoy databases using the following parameters: 1) fully tryptic enzymatic cleavage with two possible missed cleavages, 2) peptide tolerance of 800 parts-per-million, 3) fragment ion tolerance of 0.8 Da, and 4) variable modifications due to carboxyamidomethylation of cysteine residues (+ 57 Da) and deamidation of asparagine residues (+1 Da). Following the database searches, statistically significant proteins were determined for each of the four samples at a 1% protein FDR using the ProValT algorithm [19].
Protein functional annotation
For proteins with no assigned functions, homology searches were performed using the BlastP program against all non-redundant protein sequences present in the NCBI database (http://www.ncbi.nlm.nih.gov/blast). Protein alignments were considered significant if they were below an e-value of 10−50. The mechanism of secretion was predicted using SignalP and SecretomeP to identify classical motifs [20].
3. RESULTS
Visualization of secreted proteins
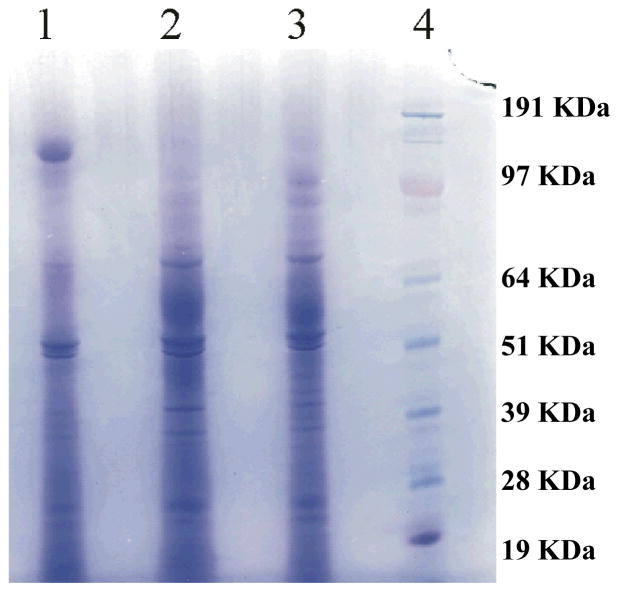
The production of B. cinerea secreted proteins was carried out in liquid culture conditions having either partially esterified (PE) pectin, highly esterified (HE) pectin, or sucrose as the carbon source. Analysis of the 1D SDS-PAGE used for the separation of the secreted proteins (Figure 1) revealed the presence of numerous proteins. The protein band patterns for B. cinerea cultured in the PE pectin and HE pectin media were very similar, while there were certain significant differences for B. cinerea grown in sucrose. To identify the B. cinerea secreted proteins in each growth condition, each gel lane was cut into three equal parts and analyzed by shotgun proteomics.
Figure 1.
Secreted proteins from B. cinerea, grown in liquid cultures with sucrose (Lane 1), HE pectin (Lane 2) or PE pectin (Lane 3) as the sole carbon source, were separated on one-dimensional SDS-PAGE and stained with Coomassie blue as described in the Materials and Methods section. Molecular weight standards are shown in Lane 4.
Shotgun proteomics
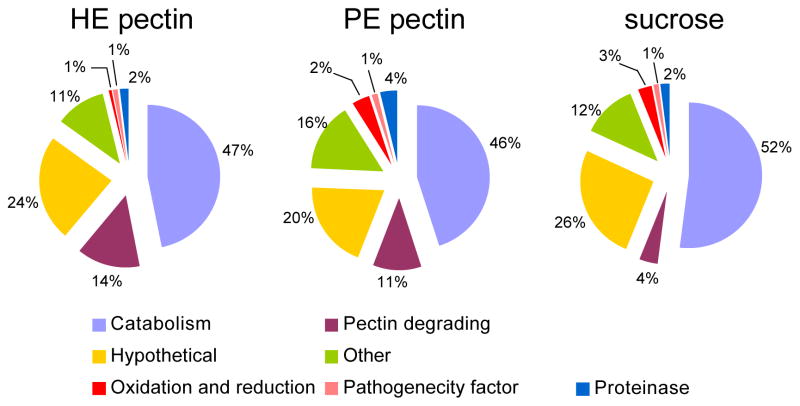
Three segments from each lane of the 1D SDS-PAGE were digested in-gel and resulting peptides were subjected to LC-MS/MS analysis. Each fraction was analyzed in duplicate. All together, a total of 126 protein groups were identified (Supplementary Information Table 1), indicating that a minimum of 126 B. cinerea proteins were present in the liquid culture after B. cinerea had grown for seven days. Each protein group had at least one discriminating peptide in that group compared to all the other identified proteins (Supplementary Information Table 2). At a 1% FDR, B. cinerea grown in PE pectin produced 105 B. cinerea proteins identified by 822 MS/MS spectra, while B. cinerea grown in HE pectin or on sucrose produced 95 and 89 proteins identified by 791 and 638 MS/MS spectra, respectively. The BlastP algorithm was used to determine the putative function of the B. cinerea proteins based on homology with other proteins, which in turn was used to classify the proteins into different categories: carbohydrate metabolism and transport (catabolism), pectin degrading enzymes (pectinases), peptidases, pathogenicity factor, hypothetical and others. More than half of the identified proteins belonged to the category of carbohydrate metabolism and transport (Figure 2). Thirty-one identified proteins did not match any annotated protein with sufficient homology to assign a putative function and were classified as hypothetical proteins.
Figure 2.
Classifications of B. cinerea proteins into different functional categories based on their putative function, when fungus was grown in liquid culture with HE pectin, PE pectin or sucrose as the sole carbon source. Identified proteins classification categories include proteins involved in carbohydrate metabolism and transport (catabolism), pectin degrading enzymes, hypothetical proteins (proteins with unknown function), oxidation and reduction, pathogenecity factor and others.
This study was designed to provide a qualitative global secretome analysis. In the present work we provide researchers with access to descriptive proteomic analysis. A limit of the shotgun approach is that it detects only protein fragments and not intact proteins and therefore cannot discern isomeric forms of proteins and posttranslational modifications.
In the current study dynamic exclusion, complexity of the sample and disparity of the protein secretions in various growth conditions impact the spectral count of the protein. Observation with small spectral counts cannot be correlated to changes in the level of protein secretion. For reasonable semi-quantification there is a requirement of the observation of many spectra for a given protein [21]. At least a three-fold difference is required for less than 4 spectral counts to determine any significant change in the protein [22]. Thus any differences in protein level that are below 4 spectral counts need to be viewed in a qualitative manner.
Proteins secreted by B. cinerea in different growth conditions
The proteins were assumed to be extracellular because they were obtained from culture media after simple filtration. The extracellular localization of the identified proteins was confirmed using the Signal P algorithm. Based on amino acid sequences, Signal P predicted that 87 proteins were secreted out of the 126 (Supplementary Information Table 1) total identified proteins. Of the 67 common proteins identified under all three growth conditions (Figure 3, Table 1), 50 were predicted to be secreted by signal P. Eighty-eight proteins were observed to be in common in both of the pectin growth conditions.
Figure 3.
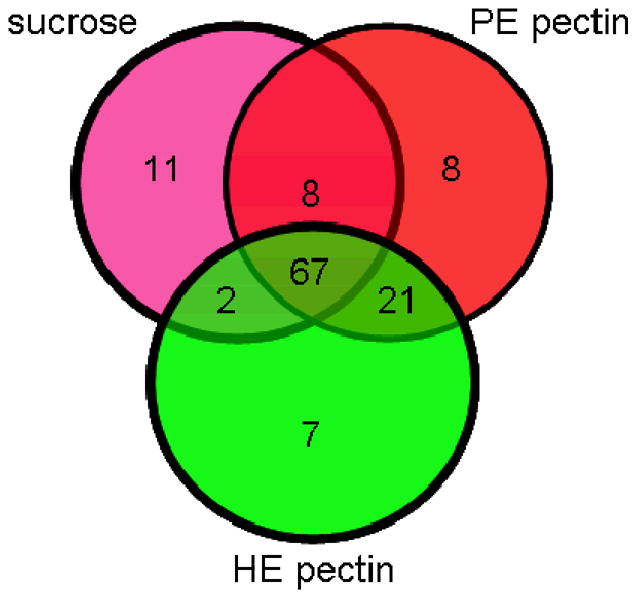
Venn diagram of the identified B. cinerea proteins using LC-MS/MS, when fungus was grown in liquid cultures using HE pectin, PE pectin or sucrose as the sole carbon source.
Table 1.
Results of B. cinerea proteins with signal P identified in all three growth conditions.
| Gene Ida | Putative Functionb | Signal Pc | PE pectin | HE pectin | sucrose | |||
|---|---|---|---|---|---|---|---|---|
| Scored | Spectrae | Scored | Spectrae | Scored | Spectrae | |||
| BC1G_13137.1 | Polygalacturonase | 0.986Y | 492.13 | 12 | 791.28 | 16 | 73.04 | 1 |
| BC1G_00617.1 | Pectin methylesterase | 1.000Y | 247.49 | 6 | 296.65 | 11 | 122.37 | 2 |
| BC1G_06546.1 | alpha-galactosidase precursor (Melibiase) | 0.999Y | 247.44 | 7 | 47.29 | 1 | 129.34 | 2 |
| BC1G_07622.1 | beta-glucosidase | 0.995Y | 523.25 | 13 | 396.44 | 5 | 461.52 | 11 |
| BC1G_07319.1 | 1,3-beta glucanase | 0.999Y | 792.22 | 16 | 808.75 | 17 | 532.25 | 9 |
| BC1G_10455.1 | 1,3-beta-glucanosyltransferase Gel1 | 0.998Y | 254.54 | 4 | 172.44 | 3 | 263.19 | 10 |
| BC1G_12859.1 | alpha-glucosidase precursor | 1.000Y | 443.98 | 10 | 559.85 | 10 | 552.39 | 19 |
| BC1G_04994.1 | alpha-L-arabinofuranosidase | 0.999Y | 816.83 | 15 | 460.12 | 10 | 50.56 | 2 |
| BC1G_08372.1 | alpha-L-arabinofuranosidase A | 0.995Y | 100.63 | 2 | 336.39 | 4 | 102.4 | 1 |
| BC1G_06328.1 | alpha-L-rhamnosidase | 0.992Y | 218.77 | 6 | 265.45 | 5 | 231.9 | 6 |
| BC1G_14030.1 | beta-1-3-glucanosyltransferase | 1.000Y | 409.21 | 12 | 525.03 | 10 | 420.58 | 9 |
| BC1G_10247.1 | beta-fructofuranosidase | 1.000Y | 518.21 | 11 | 331.52 | 6 | 57.64 | 2 |
| BC1G_03567.1 | beta-galactosidase | 0.998Y | 792.51 | 17 | 309.73 | 6 | 266.3 | 6 |
| BC1G_02364.1 | beta-glucosidase | 0.991Y | 608.27 | 16 | 602.61 | 9 | 273.37 | 5 |
| BC1G_10221.1 | beta-glucosidase 1 precursor | 0.816Y | 1203.24 | 29 | 651.09 | 13 | 1544.26 | 48 |
| BC1G_13690.1 | Cell wall glycosyl hydrolase YteR, putative | 0.998Y | 243.78 | 4 | 495.08 | 13 | 110.69 | 3 |
| BC1G_06509.1 | Chitin binding protein | 1.000Y | 307.19 | 5 | 308.5 | 4 | 572.64 | 11 |
| BC1G_00594.1 | Endoglucanase | 0.999Y | 586.93 | 28 | 516.98 | 26 | 290.77 | 9 |
| BC1G_13938.1 | Exo-arabinanase | 1.000Y | 396.29 | 10 | 604.94 | 15 | 92.18 | 2 |
| BC1G_01033.1 | Exo-beta-1,3-glucanase | 0.929Y | 69.41 | 1 | 137.36 | 3 | 67.03 | 1 |
| BC1G_01617.1 | Exo-polygalacturonase | 0.996Y | 1098.98 | 29 | 833.38 | 33 | 116.36 | 2 |
| BC1G_07215.1 | Family 20 chitobiase | 0.999Y | 170.5 | 3 | 99.4 | 2 | 141.94 | 3 |
| BC1G_11018.1 | Family of alpha-1,6-mannanases | 0.999Y | 480.62 | 7 | 411.05 | 9 | 631.97 | 11 |
| BC1G_00409.1 | Glcosyl transferase/cell wall glucanase | 1.000Y | 347.29 | 6 | 271.42 | 7 | 246.29 | 4 |
| BC1G_08755.1 | Glucoamylase P precursor | 0.997Y | 359.15 | 7 | 126.38 | 2 | 366.87 | 6 |
| BC1G_11898.1 | Glucosidase | 0.999Y | 507.88 | 12 | 767.12 | 17 | 748.96 | 12 |
| BC1G_12132.1 | Glycosyl hydrolase family 65 protein | 0.997Y | 152.95 | 4 | 186.69 | 4 | 286.58 | 5 |
| BC1G_09079.1 | GPI-anchored cell wall beta-1,3-endoglucanase | 0.999Y | 664.29 | 12 | 621.5 | 13 | 588.23 | 9 |
| BC1G_01204.1 | Glyoxal oxidase | 0.983Y | 347.32 | 6 | 156.57 | 2 | 366.68 | 5 |
| BC1G_00455.1 | Mannosyl-oligosaccharide alpha-1,2-mannosidase precursor | 0.998Y | 1016.64 | 29 | 950.03 | 29 | 593.02 | 12 |
| BC1G_02314.1 | Extracellular phytase | 0.995Y | 460.62 | 12 | 230.08 | 4 | 427.18 | 14 |
| BC1G_10486.1 | Glutaminase GtaA | 0.992Y | 475.9 | 11 | 465.49 | 14 | 380.16 | 11 |
| BC1G_11266.1 | Laccase | 0.999Y | 550.72 | 17 | 418.69 | 14 | 565.69 | 16 |
| BC1G_10329.1 | Laccase | 0.989Y | 266.26 | 4 | 497.02 | 8 | 186.98 | 4 |
| BC1G_11835.1 | Phytase | 0.978Y | 84.38 | 1 | 61.01 | 3 | 191.12 | 4 |
| BC1G_02965.1 | Acid phosphatase/phosphoesterase | 0.996Y | 354.4 | 5 | 310.9 | 6 | 100.75 | 2 |
| BC1G_02163.1 | Ceratoplatanin | 0.996Y | 238.17 | 8 | 238.86 | 8 | 237.05 | 9 |
| BC1G_07521.1 | Aspartate protease | 0.999Y | 413.97 | 13 | 318.55 | 15 | 417.04 | 8 |
| BC1G_03710.1 | Carboxypeptidase S1 | 0.989Y | 52.47 | 1 | 58.78 | 1 | 79.96 | 1 |
| BC1G_01393.1 | Hypothetical protein | 1.000Y | 461.2 | 14 | 486.53 | 14 | 510.93 | 9 |
| BC1G_15201.1 | Hypothetical protein | 1.000Y | 240.06 | 10 | 211.51 | 14 | 329.78 | 13 |
| BC1G_12279.1 | Hypothetical protein | 1.000Y | 41.65 | 1 | 47.4 | 1 | 66.02 | 3 |
| BC1G_12374.1 | Hypothetical protein | 0.999Y | 207.74 | 5 | 189.88 | 4 | 358.51 | 8 |
| BC1G_08393.1 | Hypothetical protein | 0.999Y | 150.34 | 3 | 258.01 | 3 | 155.89 | 3 |
| BC1G_14136.1 | Hypothetical protein | 0.999Y | 36.38 | 1 | 128.38 | 2 | 60.58 | 3 |
| BC1G_11139.1 | Hypothetical protein | 0.999Y | 61.09 | 2 | 72.08 | 3 | 72.26 | 1 |
| BC1G_00198.1 | Hypothetical protein | 0.999Y | 1004.38 | 21 | 736.07 | 15 | 1005.62 | 29 |
| BC1G_08801.1 | Hypothetical protein | 0.982Y | 199.37 | 5 | 86.34 | 1 | 51.09 | 1 |
| BC1G_10095.1 | Hypothetical protein | 0.951Y | 156.41 | 2 | 48.69 | 1 | 76.79 | 1 |
| BC1G_13215.1 | Hypothetical protein | 0.820Y | 144.78 | 2 | 113.98 | 1 | 122.66 | 2 |
Protein identification number provided by the broad institute B. cinerea BO5.10 database
Name/function was assigned based on sequence similarity when blasted using NCBI non redundant database
Signal P prediction value using algorithm SignalP3.0 server (http://www.cbs.dtu.dk/services/SignalP/)
Protein score calculated by adding individual non redundant peptide Mascot score over a threshold score at 1% False Discovery Rate as calculated by ProValT
Total number of spectra matched to proteins in all replicates in a growth condition of B. cinerea
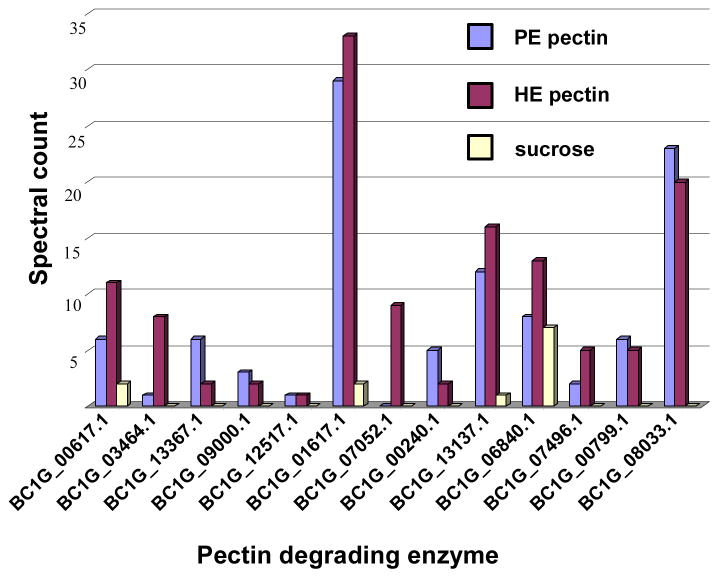
Pectin degrading enzymes
Thirteen pectin degrading enzymes were identified in the culture filtrates following B. cinerea growth on the three carbon sources (Table 2 and Figure 4). The pectinases included three PMEs, three PLs, two endo PGs, two exo PGs, and two PGs of unknown mode of action. These 12 proteins were found in culture filtrates following growth on either source of pectin. Only one pectate lyase was uniquely observed in the highly esterified pectin growth medium. The two B. cinerea endo PGs, PG2 and PG6, were only observed when B. cinerea was grown in either pectin source but not when grown in sucrose. After growth in sucrose, we were able to identify only four enzymes, BC1G_00617.1, BC1G_013137.1, BC1G_01617.1 and BC1G_06840.1, out of 13 pectin degrading enzymes identified from pectin growth conditions, and these had spectral counts at least three-fold less than those observed when B. cinerea was grown with either source of pectin. Spectral counts are the number of MS/MS spectra identified as belonging to specific peptides and provide a semi-quantitative estimate of the relative protein abundance in the analyzed sample [22]. Only one B. cinerea PME was identified as secreted when B. cinerea was grown in sucrose and it had a similar number of spectral counts compared to the cultures grown in pectins. Three proteins, BC1G_06840.1, BC1G_07946.1 and BC1G_00799.1, were identified with pectin degrading functions yet lacked a classical signal peptide for secretion.
Table 2.
Results of the pectinases identified in the current study.
| Gene Ida | Putative Functionb | Signal Pc | PE pectin | PE pectin | HE pectin | HE pectin | sucrose | sucrose |
|---|---|---|---|---|---|---|---|---|
| Scored | Spectrae | Scored | Spectrae | Scored | Spectrae | |||
| BC1G_00617.1 | Pectin methylesterase | 1.000Y | 247.49 | 6 | 296.65 | 11 | 122.37 | 2 |
| BC1G_03464.1 | Rhamnogalacturonase | 1.000Y | 59.08 | 1 | 142.64 | 8 | - | - |
| BC1G_13367.1 | Endopolygalacturonase 2 | 1.000Y | 179.84 | 6 | 53.75 | 2 | - | - |
| BC1G_09000.1 | Pectate lyase | 1.000Y | 141.08 | 3 | 160.45 | 2 | - | - |
| BC1G_12517.1 | Pectate lyase, putative | 1.000Y | 44.42 | 1 | 55.61 | 1 | - | - |
| BC1G_01617.1 | Exo-polygalacturonase | 0.996Y | 1098.98 | 29 | 833.38 | 33 | 116.36 | 2 |
| BC1G_07052.1 | Pectate lyase | 0.997Y | - | - | 384.47 | 9 | - | - |
| BC1G_00240.1 | Similar to exopolygalacturonase | 0.990Y | 389.71 | 5 | 143.36 | 2 | - | - |
| BC1G_13137.1 | Polygalacturonase | 0.986Y | 492.13 | 12 | 791.28 | 16 | 73.04 | 1 |
| BC1G_06840.1 | Pectin methylesterase | 0.232N | 332.97 | 8 | 436.87 | 13 | 363.34 | 7 |
| BC1G_07496.1 | Polygalacturonase | 0.000N | 106.26 | 2 | 230.48 | 5 | - | - |
| BC1G_00799.1 | Pectin methylesterase | 0.002N | 204.22 | 6 | 225.48 | 5 | - | - |
| BC1G_08033.1 | Polygalacturonase 6 | 0.999Y | 712.92 | 23 | 564.83 | 20 | - | - |
Protein identification number provided by the broad institute B. cinerea BO5.10 database
Name/function was assigned based on sequence similarity when blasted using NCBI non redundant database
Signal P prediction value using algorithm SignalP3.0 server (http://www.cbs.dtu.dk/services/SignalP/)
Protein score calculated by adding individual non redundant peptide Mascot score over a threshold score at 1% False Discovery Rate as calculated by ProValT
Total number of spectra matched to proteins in all replicates in a growth condition of B. cinerea
Figure 4.
Spectral count of thirteen PDEs secreted by B. cinerea grown in three different liquid cultures (sucrose, HE pectin or PE pectin as the sole carbon source). The spectral count for each PDE is the sum of the spectra identified to that PDE obtained from two replicate LC-MS/MS analyses at 1% FDR.
4. DISCUSSION
Proteomics
Proteomics was used to study the impact of different degrees of pectin esterification on the proteins secreted by B. cinerea when pectin was the sole carbon source for growth in culture. Previously, proteomic studies in other filamentous fungi have shown substrate dependent secretion. Aspergillus flavus, a filamentous fungus, has shown unique rutin degrading enzymes when grown in the presence of rutin in liquid culture, and absence of those enzymes when grown in potato dextrose in liquid culture [23]. Although fruit, which serves as a natural host for B. cinerea, is a much more complex environment than a single carbon source liquid culture medium, changes in the extent of pectin esterification occur as fruits ripen. Therefore, differences observed in protein secretion following infection with B. cinerea could be a consequence of the differences in the degree of esterification. Modeling the changes in fruit pectins required a simple system.
Studying the changes in a mixture of proteins between two or more similar systems generally requires the use of equal concentrations of sample. In the present study, changes occur both in terms of number and in overall concentration of secreted proteins depending on the growth conditions. Fungi are known to exhibit substrate dependant secretion; therefore, a comparison based on equal protein concentration might result in an inaccurate representation of the relative amounts of the secreted proteins. To accommodate this concern we chose to use equal volume as the basis for our comparison. The proteins are collected in a standard volume of solution, and analyses were conducted on a volume-to-volume basis. If the proteins secreted by the fungus in different systems results in too large a change in concentration, then quantitative comparison of individual proteins using this method may not be possible. However a comparison between profiles of proteins is possible and the results from an equal volume analysis represent the most accurate reflection of the secretion profile.
The gel analysis suggested that the total concentrations of proteins secreted in the different media were similar, validating our comparison between profiles. Overall we were able to identify 126 B. cinerea proteins. Sixty-seven proteins were observed in all three systems, indicating that these proteins may be constitutively secreted at a minimal detectable level in all growth conditions and thus their presence is not dependent on the carbon source. Among these proteins, 50 had an N-terminal signal peptide motif, confirming their secretion. On the basis of putative function, 35 of these were classified as carbohydrate metabolism and transport proteins, 11 as hypothetical proteins, one as a peptidase, one as a pathogenicity factor and two others. Thirty-five proteins were identified with only one peptide identification. Generally proteins secreted by the fungi are small and have some post translation modification. Hence it is difficult to get high percentage coverage of the amino acid sequence of the protein using mass spectrometry.
Pectin degrading enzymes
Pectinases play an important role in cell wall degradation and successful invasion. Endo PGs are one of the most widely studied classes of pectin degrading enzymes. Endo PGs hydrolyze the internal (1–4) linkage between D-galacturonic acid units of pectin [24]. Previously, six endo PGs have been identified from B. cinerea [25]. However, it has been suggested that B. cinerea can secrete up to 13 endo PG isoenzymes when post translational modifications are taken into account [26]. The secretion of different isoforms can be explained by differences in substrate, pH, and environmental conditions [11]. The deletion mutants of both BcPG1 and BcPG2 showed reduced virulence on multiple hosts [25]. In this study we identified endoPG 2 (BC1G_13667.1) and endoPG 6 (BC1G_08033.1) when the fungus was grown in either HE or PE pectin as a carbon source in liquid culture. The absence of all endo PG isoforms following growth in sucrose indicates the secretion of endo PGs is carbon source dependant. This is consistent with previous work demonstrating that the expression of the endo PG gene family has a differential pattern that depends on host tissue, stage of infection and temperature [27]. There was no significant change in the secretion of endo PGs as a result of the differences in the degree of pectin esterification. Previously, it had been suggested that a basic level of gene expression exists for two PGs (BcPG1 and BcPG2) in liquid culture growth conditions [11]. In the current study, we were unable to detect any BcPG1 in the culture media. There is a possibility that the BcPG1 protein was present, but not in sufficient quantity for detection via shotgun proteomics, or it is possible that it was not detected as the result of extensive post translation modifications. Post translational modifications have been reported on other endo PG isoforms, notably BcPG6 [28], which were, in fact, detected in our analysis. The controlled secretion of endo PGs and the role of the PG inhibiting proteins in the plant wall as a defense mechanism against B. cinerea infections emphasize the importance of BcPG [29, 30].
PMEs de-esterify pectin, releasing methanol and polygalacturonic acid. This de-esterification facilitates the subsequent action of PGs and PLs [24]. On certain hosts, BCPME1 has been shown to be an essential determinant of B. cinerea virulence [24, 25]. We were able to identify three different PMEs, BC1G_00617.1, BC1G_6840.1 and BC1G_00799.1, which were secreted by B. cinerea. Two PMEs, BC1G_00617.1 and BC1G_6840.1, were identified as secreted in all three growth conditions, implying that these PMEs may be a constituent of the secretion profile which is independent of the carbon substrate.
PL cleaves polygalacturonic acid into oligogalacturonides via beta-elimination [24], but the role of pectate lyases in B. cinerea infections has not been investigated previously. Three pectate lyases, BC1G_07052.1 BC1G_09000.1 and BC1G_12517.1, were identified as secreted only under the pectin growth conditions. Of interest is a pectate lyase, BC1G_07052.1, which was only observed in HE pectin.
Other pectinases identified were two exo PGs, BC1G_01617.1 and BC1G_00240.1, identified in both pectin growth conditions. BC1G_01617.1 was identified in the sucrose growth medium, but with a spectral count of 2 as compared to 29 and 33 spectral counts for material from the cultures grown in both PE and HE pectin. Two other PGs, BC1G_13137.1 and BC1G_07496.1 were detected in both pectin growth mediums. BC1G_13137.1 was also identified in the sucrose growth medium, but with a spectral count of 1 compared to spectral counts of 12 and 16 in PE pectin and HE pectin medium. BC1G_03464.1 is a putative rhamnogalacturonase that hydrolyzes the internal (1–2) linkage between units of pectin in rhamnogalacturonan I, and was identified in both of the pectin growth conditions and not in sucrose [14].
When grown in the sucrose media, B. cinerea produced 12 proteins which were not observed in the other media. Of these 12 proteins, five (BC1G_00882.1, BC1G_14570.1, BC1G_05168.1, BC1G_10120.1 and BC1G_15832.1) were classified as hypothetical proteins. Of the remaining proteins, two were alpha amylases (BC1G_02623.1 and BC1G_02333.1), one was an alpha glucosidase (BC1G_11115.1), one was a member of the glycosyl hydrolase family 95 (BC1G_08975.1), one was a putative 3-isopropylmalate dehydrogenase Leu2A (BC1G_14880.1), one was acatalase (BC1G_12856.1), and one was a Cu/Zn superoxide dismutase (BC1G_00558.1). Thus, while the secretome of B. cinerea in sucrose growth medium has similar characteristics compared to secretomes in pectin media, the total number of pectinases and the total spectral count of pectinases secreted were lower. Hence, the secretion of pectinases, except for PMEs, depends on the carbon source of growth.
Secreted proteins
Out of the 126 proteins identified in culture filtrates following fungal growth, only 87 proteins had a signal peptide motif according to the signalP algorithm [20]. This N-terminal motif indicating that the “protein is likely to be secreted” is frequently used to confirm the extracellular nature of proteins identified. The absence of such a predicted N-terminal motif in 39 proteins indicates that these might be intracellular proteins resulting from cell lysis, as the identification of internal proteins in secretome studies due to cell lysis is an inherent feature of secretome proteomics. However, the handling of the material here did not involve any treatments likely to result in cell lysis. In addition, there were three pectinases predicted not to contain the signal peptide and hence predicted by the algorithm to not be secreted. The pectinases are known to be secreted and thus we postulate that a nonconventional N-terminal motif exists in certain B. cinerea proteins for secretion. A non-classical method of secretion is known to exist in yeast and the Cu/Zn superoxide dismutase of B. cinerea, which plays an important role in french bean virulence, had previously been confirmed to be secreted by a non-classical pathway in such organisms as Aspergillus fumigates and Claviceps purpurea [31, 32].
Identification of function
Most of the proteins identified from the B. cinerea B05.10 database are hypothetical proteins with unknown function; therefore the BlastP algorithm was used to assign putative functions to proteins by comparing the identified hypothetical proteins to the proteins in the NCBI nr database. Almost three-quarters of the identified proteins were assigned putative functions using an e value threshold of e−50; however, one-quarter of the identified proteins remained hypothetical. Most of these hypothetical proteins were similar to other fungal hypothetical proteins below the required threshold score of e−50; however, there were a few proteins with no significant alignments with any other proteins in the NCBI nr database. These hypothetical proteins, if unique, might be good targets for future biological studies.
Conclusion
Shotgun proteomics was successfully used to identify the secretome of B. cinerea grown in three culture conditions which differed by the carbon nutrients provided. We were able to identify a total of 126 B. cinerea proteins, 67 of which were observed in all three growth conditions. Thirteen pectinases were identified as secreted by B. cinerea when grown in all culture conditions. The secretion of most of the pectinases depended on the carbon substrate used by the fungus for growth. However, secretion of two pectin methyl esterases is independent of the carbon substrate. There were no major differences in protein secretion when B. cinerea was grown in liquid culture with 30% vs 80% esterified pectin. Because both the growth of B. cinerea and the secretion of proteins were similar in cultures containing differently esterified pectins, it is likely that the activation of B. cinerea from the dormant state to active infection is not solely dependant on changes in the degree of esterification of the pectin component of the plant cell wall. However, these results suggest that future studies of the B. cinerea secretome in infections of ripe and unripe fruits will provide important information that will describe the mechanisms that the fungus employs to access nutrients and decompose tissues.
Supplementary Material
Supplementary Table 1: Summary of Botrytis cinerea proteins identified in various growth conditions
Supplementary Table 2: Summary of Botrytis cinerea proteins and peptides identified
Acknowledgments
This work was supported in part by NSF “Systems Biology Approach to Tomato Fruit Susceptibility to a Necrotrophic Fungus” (IOB-0544504), DOE “Structures and Functions of Oligosaccharins” (DE-FG02-96ER20221), the DOE Center for Plant and Microbial Complex Carbohydrates (DE-FG05-93ER20097), and the NIH/NCRR Integrated Technology Resource for Biomedical Glycomics (P41 RR018502).
List of abbreviations
- B
Botrytis
- PMEs
pectin methyl esterases
- exo PGs
exopolygalacturonases
- endo PGs
endopolygalacturonases
- PLs
pectate and pectin lyases
- PMEI
pectin methylesterase inhibitor
- PE
partially esterified pectin
- HE
highly esterified pectin
References
- 1.Williamson B, Tudzynski B, Tudzynski P, KANV, Jan AL. Botrytis cinerea: the cause of grey mould disease. Mol Plant Pathol. 2007;8:561–580. doi: 10.1111/j.1364-3703.2007.00417.x. [DOI] [PubMed] [Google Scholar]
- 2.Adaskaveg JE, Foerster H, Thompson DF. Identification and etiology of visible quiescent infections of Monilinia fructicola and Botrytis cinerea in Sweet Cherry fruit. Plant Dis. 2000;84:328–333. doi: 10.1094/PDIS.2000.84.3.328. [DOI] [PubMed] [Google Scholar]
- 3.Prins TW, Tudzynski B, Tudzynski P, Von A, et al. Infection strategies of Botrytis cinerea and related necrotrophic pathogens. Fungal Pathol. 2000:33–64. [Google Scholar]
- 4.Prusky D. Pathogens quisescence in postharvest diseases. Ann Rev Phytopathol. 1996;34:413–434. doi: 10.1146/annurev.phyto.34.1.413. [DOI] [PubMed] [Google Scholar]
- 5.Prasanna V, Prabha TN, Tharanathan RN. Fruit ripening phenomena - An overview. Crit Rev Food Sci Nutr. 2007;47:1–19. doi: 10.1080/10408390600976841. [DOI] [PubMed] [Google Scholar]
- 6.Brownleader M, Jackson P, Mobasheri A, Pantelides A, et al. Molecular aspects of cell wall modifications during fruit ripening. Crit Rev Food Sci Nutr. 1999;39:149–164. doi: 10.1080/10408399908500494. [DOI] [PubMed] [Google Scholar]
- 7.Willats WGT, McCartney L, Mackie W, Knox JP. Pectin: cell biology and prospects for functional analysis. Plant Mol Biol. 2001;47:9–27. [PubMed] [Google Scholar]
- 8.Steele NM, McCann MC, Roberts K. Pectin modification in cell walls of ripening tomatoes occurs in distinct domains. Plant Physiol. 1997;114:373–381. doi: 10.1104/pp.114.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wakabayashi K, Hoson T, Huber DJ. Methyl de-esterfication as a major factor regulating the extent of pectin depolymerization during fruit ripening: a comparison of the action of avocado (Persea americana) and tomato (Lycopersicon esculentum) polygalacturonases. J Plant Physiol. 2003;160:667–673. doi: 10.1078/0176-1617-00951. [DOI] [PubMed] [Google Scholar]
- 10.Reeve RM. Histological and histochemical changes in developing and ripening peaches. II. The cell walls and pectins. Am J Bot. 1959;46:241–248. [Google Scholar]
- 11.Wubben JP, Ten Have A, Van Kan J, Visser J. Regulation of endopolygalacturonase gene expression in Botrytis cinerea by galacutonic acid, ambient pH and carbon catabolite, repression. Curr Gen. 2000;37:152–157. doi: 10.1007/s002940050022. [DOI] [PubMed] [Google Scholar]
- 12.Schols HA, Geraeds C, Searle-Van Leeuwen MF, Kormelink FJM, Voragen AGJ. Rhamnogalacturonase: a novel enzyme that degrades the hairy regions of pectins. Carbohydr Res. 1990;206:105–115. [Google Scholar]
- 13.Kapat A, Zimand G, Elad Y. Biosynthesis of pathogenixity hydrolytic enzymes by Botrytis cinerea during infection of bean leaves and in vitro. Mycol Res. 1998;102:1017–1024. [Google Scholar]
- 14.Chen HJ, Smith DL, Starrett DA, Zhou D, et al. Cloning and characterization of a rhamnogalacturonan hydrolase gene from Botrytis cinerea. IUBMB Life. 1997;43:823–838. doi: 10.1080/15216549700204641. [DOI] [PubMed] [Google Scholar]
- 15.Wubben JP, Mulder W, ten Have A, Van Kan J, Visser J. Cloning and partial characterization of endopolygalacturonase genes from Botrytis cinerea. Appl Environ Microbiol. 1999;65:1596–1602. doi: 10.1128/aem.65.4.1596-1602.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osorio S, Castillejo C, Quesada MA, Medina-Escobar N, et al. Partial demethylation of oligogalacturonides by pectin methyl esterase 1 is required for eliciting defence responses in wild strawberry (Fragaria vesca) Plant J. 2008;54:43–55. doi: 10.1111/j.1365-313X.2007.03398.x. [DOI] [PubMed] [Google Scholar]
- 17.Lionetti V, Railoa A, Camardella L, Giovane A, et al. Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol. 2007;143:1871–1880. doi: 10.1104/pp.106.090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedriolo PGA, Eng JK, Hubley R, Vogelzang M, et al. A common open representation of mass spectrometry data and its application to proteomics research. Nature Biotechnol. 2004;22:1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 19.Weatherly DB, Atwood JA, Minning TA, Cavola C, et al. A heuristic method for assigning a false-discovery rate for protein identifications from Mascot database search results. Mol Cell Proteom. 2005;4:762–772. doi: 10.1074/mcp.M400215-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 21.Bantscheffet M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: A critical review. Anal Bioanal Chem. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 22.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, et al. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Medina ML, Haynes PA, Breci L, Francisco WA. Analysis of secreted proteins from Aspergillus flavus. Proteomics. 2005;5:3153–3161. doi: 10.1002/pmic.200401136. [DOI] [PubMed] [Google Scholar]
- 24.Valette-Collet O, Cimerman A, Reignault P, Levis C, Boccara M. Disruption of Botrytis cinerea pectin methylesterase gene Bcpme 1 reduces virulence on several host plants. Mol Plant Microbe Interact. 2003;16:360–367. doi: 10.1094/MPMI.2003.16.4.360. [DOI] [PubMed] [Google Scholar]
- 25.Van Kan J. Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 2006;11:247–253. doi: 10.1016/j.tplants.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 26.ten Have A, Breuil WO, Wubben JP, Visser J, Van Kan J. Botrytis cinerea endopolygalacturonase genes are differentially expressed in various plant tissues. Fungal Genet Biol. 2001;33:97–105. doi: 10.1006/fgbi.2001.1269. [DOI] [PubMed] [Google Scholar]
- 27.ten Have A, Mulder W, Visser J, Van Kan J. The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol Plant Microbe Interact. 1998;11:1009–1016. doi: 10.1094/MPMI.1998.11.10.1009. [DOI] [PubMed] [Google Scholar]
- 28.Xie M, Krooshof G, Benen J, Atwood JA, et al. Post-translational modifications of recombinant B. cinerea EPG 6. Rapid Commun Mass Spectrom. 2005;19:3389–3397. doi: 10.1002/rcm.2194. [DOI] [PubMed] [Google Scholar]
- 29.Johnston DJ, Williamson D, McMillan GP. The interaction in planta of polygalacturonases from Botrytis cinerea with a cell wall-bound polygalacturonase-inhibiting protein (PGIP) in raspberry fruits. Ann Rev Phytopathol. 1994;45:1837–1843. [Google Scholar]
- 30.De Lorenzo G, D’Ovidio R, Cervone F. The role of polygalacturonase-1 inhibiting proteins (PGIPs) in defense against pathogenic fungi. Ann Rev Phytopathol. 2001;39:313–335. doi: 10.1146/annurev.phyto.39.1.313. [DOI] [PubMed] [Google Scholar]
- 31.Nombela C, Gil C, Chaffin LJ. Non-conventional protein secretion in yeast. Trends Microbiol. 2006;14:15–21. doi: 10.1016/j.tim.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 32.Bendtsen JD, Kiemer L, Fausboll A, Brunak S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005;5:58. doi: 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Summary of Botrytis cinerea proteins identified in various growth conditions
Supplementary Table 2: Summary of Botrytis cinerea proteins and peptides identified