Abstract
An accurate algorithm is essential for effective molecular diagnosis of hereditary colorectal cancer. Here we have extended the analysis of 71 colorectal cancer cases suspected to be Lynch Syndrome cases for MSH2, MLH1, MSH6 and PMS2 gene defects. All cases were screened for mutations in MSH2, MLH1 and MSH6 and all cases where tumors were available were screened for microsatellite instability and expression of MSH2 and MLH1. Subsequently, mutation negative cases were screened for MLH1 methylation and mutations in PMS2. Of the MSI-H cases, 96% had a mismatch repair gene defect, mostly involving MSH2 or MLH1; 1 PMS2 mutation, 1 MLH1 epimutation, and no MSH6 mutations were found. Four of the 28 MSI-H cases, including 1 Amsterdam criteria case, had biallelic tumor MLH1 methylation indicating that sporadic cases can be admixed in with Lynch Syndrome cases even those meeting the strongest criteria for Lynch Syndrome. Mismatch repair gene defects were found in similar frequency in cases where tumors were and were not available. One MLH1 and 1 MSH2 deletion mutation were found in MSI-S/L cases indicating that microsatellite instability testing can exclude cases with pathogenic mutations. Our analysis support a diagnostic algorithm where cases are selected for analysis based on clinical criteria or prediction models; isolated sporadic young-onset cases can be pre-screened by tumor testing whereas familial cases may be directly subjected to molecular analysis for mutations in mismatch repair genes followed by microsatellite instability, protein expression and DNA methylation analysis to aid in the resolution of mutation negative cases.
Introduction
Lynch Syndrome, also called hereditary non-polyposis colon cancer (HNPCC), is an autosomal dominant inherited cancer predisposition syndrome characterized by predisposition to develop a number of cancers at an early age and high penetrance with mutation carriers having a significantly increased lifetime risk of developing colorectal (CRC) and other forms of cancer (1–3). Inherited defects in genes encoding components of the major post replication DNA mismatch repair (MMR) system have been found to underlie many cases of Lynch Syndrome with most of the genetic defects identified being attributable to mutations in two genes, MSH2 and MLH1 (4–6). A small proportion of cases have been shown to be caused by germline mutations in two other MMR genes, MSH6 and PMS2; however, PMS2 mutations and MSH6 mutations are often associated with weaker family histories, later ages of diagnosis and potentially a different cancer spectrum (7–13). The range of mutations identified in MSH2 and MLH1 includes missense, nonsense, frameshift, splice site mutations and deletion mutations as well as the more recently appreciated rare epimutations (4–6, 14–17). In addition, there are apparently polymorphisms in MMR genes that may cause increased risk of developing cancer (18–20). The complete loss of mismatch repair function in tumors leads to increased mutations at microsatellite sequences resulting in the microsatellite instability high (MSI-H) phenotype, although numerous studies report MSI-H Lynch Syndrome cases that lack mutations in known MMR genes; examples include (4, 5, 21).
Early detection of a germline alteration in an MMR gene definitively diagnoses Lynch Syndrome within a family, allowing for monitoring and early treatment for appropriate family members, which leads to a reduction in morbidity and mortality of mutation carriers (22, 23). Conversely, unaffected individuals can be spared unnecessary screening procedures. To achieve these benefits, it is important to have appropriate, simple criteria for identifying individuals who should receive genetic testing and efficient, accurate genetic testing methods. An essential step in the clinical diagnostic setting is to identify all cases that will prove to have a causal mutation while including as few cases as possible that lack a mutation in order to provide definitive diagnosis to as many relevant cases as possible while keeping costly uninformative genetic testing to a minimum. Therefore, efforts have been made to find the most sensitive clinical criteria, based on family history, to be used to select families for mutation detection analyses (24–27). Studies of families meeting the most restrictive criteria for Lynch Syndrome, the Amsterdam criteria, have identified germline MSH2 and MLH1 mutations with a relatively high sensitivity [~60%] and specificity [~70%] (28). In contrast, germline MSH2 and MLH1 mutations were found with a higher sensitivity [~94%] and a lower specificity [~50%] when families meeting the least restrictive criteria, the Bethesda criteria, were studied (28). Both the original and revised Bethesda criteria appear to be equally effective for selecting CRC cases with weak or no known family history of Lynch Syndrome associated cancers that are associated with MMR defects (29, 30). More recently, new patient selection algorithms have been developed that have the potential to improve the sensitivity and specificity of mutation detection (31). However, there has been little evaluation of potential criteria for identifying isolated individuals with Lynch Syndrome associated cancers other than colorectal or endometrial cancer for genetic testing. Thus, the use of restrictive criteria improves the likelihood of finding a germline mutation at the cost of excluding cases that have a germline mutation, whereas less restrictive criteria can in principal lead to the identification of most germline mutations at a cost of analyzing many cases without a germline mutation. Given this need to include many individuals in genetic studies who may not have a germline MMR defect, development of accurate and efficient genetic testing strategies is important.
There are many published studies using a diversity of genetic testing methods and strategies describing genetic defects in MMR genes in different cancers. However, there does not appear to be a generally accepted strategy for detecting genetic defects in MMR genes, possibly because a considerable amount of genetic testing is done at local research and clinical sites. Most present large-scale studies screen cases selected on the basis of family history for tumor MSI and MMR gene expression to identify cases for subsequent molecular analysis for MMR gene defects. Such strategies may not be applicable to cancer predisposition clinic based testing where it is important to find all pathogenic mutations because of reports of MSI-L/S cases with MMR gene mutations and observations that not all missense mutations result in loss of protein (4, 5, 7, 32–34); indeed a recent functional bases study of missense mutation in the MSH2 gene found that two thirds of mutations causing an MMR defect did not significantly reduce MSH2 protein levels (35). We have previously studied 71 cases of familial colorectal cancer for the presence of germline mutations in MMR genes by analyzing MSI, expression of MLH1 and MSH2 proteins and by direct sequencing of genomic DNA to detect mutations in MLH1, MSH2 and MSH6 (10, 14, 28, 33). The suspected Lynch Syndrome cases were selected as meeting at least one of several established Lynch Syndrome criteria including the Amsterdam, modified Amsterdam, Bethesda or Lynch Syndrome-like criteria (24–27). In the present study, we have extended the prior analysis by including additional methods for detecting MMR gene defects. Of the 71 total cases analyzed, an MMR gene defect was implicated in 38 (54%) of the cases, and of the 28 MSI-H cases studied, an MMR gene defect was implicated in 27 (96%) cases. Because the majority of the genetic and molecular analysis was performed in parallel rather than sequentially, the results reported can be used to guide the development of efficient screening strategies in both the near-term as well as in the future when better prescreening strategies become widely available.
Methods
Patients
DNA samples from the Lynch Syndrome families analyzed here and Caucasian control samples have been previously described (28, 36). These Lynch Syndrome patients were collected as a clinic based series of patients meeting one of the criteria for suspect Lynch Syndrome cases including Amsterdam, Modified-Amsterdam, HNPCC-like and Bethesda criteria; the distribution of the patients among these different criteria and descriptions of the different criteria are summarized in Table 1. Previous results obtained by screening these samples by DNA sequencing for mutations in MSH2, MSH6 and MLH1, screening for germline methylation of MLH1 and tumor MSI and expression of MSH2 and MLH1 proteins have also been described (10, 14, 28, 33, 36).
Table 1.
Summary of clinical, genetic and microsatellite instability data.
| Case | Clinical Criteria | MMR Gene Defect | Other | MSI | |||
|---|---|---|---|---|---|---|---|
| AMS | M-AMS | HNPCC-like | Bethesda | Variant | |||
| Tumor Available | |||||||
| 1755 | X | X | 1,2,3,4,7 | MLH1 IVS7-2A>G Splice | MLH1 c.2147G>A V716M | H | |
| 2675 | X | X | 2,3,4,7 | MLH1 c.677G>A Splice | H | ||
| 2906 | X | X | 2,3,4 | MLH1 c.676C>T R226X | H | ||
| 357 | 4 | MLH1 IVS9-1G>T Splice | H | ||||
| 2722 | X | X | 3,4 | MLH1 c.1810A>T K604X | H | ||
| 397 | X | X | 2,3,4 | MLH1 IVS16+1G>A Splice | H | ||
| 232 | X | X | 3,4 | MLH1 c.2265G>C R755S | H | ||
| 1448 | X | X | 2,3 | MLH1 c.2104-2105ΔAG FS | MSH2 c.965G>A G322D | H | |
| 4103 | X | 2,3,4 | MSH2 c.704-705ΔAA FS | H | |||
| 1251 | 2,3,4 | MSH2 IVS5+3A>T Splice | H | ||||
| 1025 | 7 | MSH2 IVS5+3A>T Splice | H | ||||
| 1851 | X | 3 | MSH2 IVS5+3A>T Splice | H | |||
| 1754 | 2,3,4 | MSH2 c.1352-1353ΔAG FS | H | ||||
| 241 | X | X | 3 | MSH2 c.1786ΔAAT N596Δ | H | ||
| 257 | X | X | 3,4 | MLH1 Δ exons 1–13 | H | ||
| 1642 | X | X | 3,4,7 | MSH2 Δ exons 5, 6 | H | ||
| 2956 | X | X | 3,4 | MSH2 Δ exons 1-16 | H | ||
| 1103 | 4 | MLH1 germline methylation/LOH | H | ||||
| 3055 | X | X | 3 | MLH1 somatic methylation | H | ||
| 1446 | X | MSH2 no expression | H | ||||
| 1769 | X | PMS2 c.1211C>G P404R | H | ||||
| 2496 | X | MLH1 somatic methylation | H | ||||
| 245 | 4 | None | H | ||||
| 629 | X | X | 2 | MSH2 Δ exon 7 | H | ||
| 1120 | X | X | 3,4 | MLH1 Δ exon 12 | H | ||
| 1102 | 4 | MLH1 somatic methylation | H | ||||
| 2214 | X | MLH1 somatic methylation | H | ||||
| 2738 | X | X | 3,4 | MLH1 Δ exon 12 | H | ||
| 1524 | X | X | 3,4 | None | L | ||
| 261 | X | None | L | ||||
| 2911 | 2 | None | L | ||||
| 230 | 2,4 | None | L | ||||
| 1252 | X | X | None | L | |||
| 3045 | X | 2,3,4 | None | L | |||
| 2825 | X | X | None | L | |||
| 1239 | 4 | None | L | ||||
| 1648 | 3,4 | None | L | ||||
| 2228 | X | None | L | ||||
| 1657 | X | 2 | None | S | |||
| 1264 | 4 | MSH2 Δ exon 7 | S | ||||
| 1372 | 4 | None | S | ||||
| 2848 | X | None | S | ||||
| 2703 | X | None | S | ||||
| 2248 | 3,4 | None | S | ||||
| 2642 | 3,4 | None | S | ||||
| 362 | X | X | 3,4,7 | MLH1 Δ exons 16-19 | S | ||
| 2851 | X | None | L | ||||
| 487 | X | None | S | ||||
| No Tumor Available | |||||||
| 5 | X | X | 1,2,3,4 | MLH1 c.245C>T T82I | |||
| 260 | X | X | 1,2,3,4 | MLH1 c.1517T>C V506A | MLH1 c.1853AA>GC K618A | ||
| 3002 | X | 3 | MSH2 c.944G>T G315V | ||||
| 336 | X | X | 1,3,4 | MLH1 c.1852ΔAAG K618Δ | |||
| 171 | X | X | 1,3,4 | MLH1 c.2250C>G Y750X | |||
| 1846 | X | X | 1,3,4 | MLH1 c.1381A>T K461X | |||
| 2913 | X | X | 1,3,4 | MLH1 c.2198insAACA | |||
| 951 | X | 1,2,3,4 | MSH2 c.704ΔAA | ||||
| 597 | X | X | 1,2,3,4 | MLH1 Δ exon 6 | MSH2 c.815C>T A272V | ||
| 1370 | 4 | None | MSH2 c.965G>A G322D | ||||
| 170 | X | X | 1,4 | None | |||
| 2763 | X | X | 7 | None | |||
| 3012 | X | X | 1,3 | None | |||
| 419 | X | None | |||||
| 1373 | X | 3,4 | None | ||||
| 352 | X | 1,2 | None | ||||
| 2915 | X | None | |||||
| 2224 | X | 4 | None | ||||
| 1253 | X | 4 | None | ||||
| 1525 | 4 | None | |||||
| 104 | 4 | None | |||||
| 1114 | 4 | None | |||||
| 349 | 4 | None | |||||
Clinical criteria, MSH2 and MLH1 mutations detected by DNA sequencing, analysis in normal controls and MSI status have been described (Refs. 10, 14, 24–28, 32, 35). HNPCC-like; family history suggestive of Lynch Syndrome, but not fulfilling Amsterdam or Modified Amsterdam criteria. The original Bethesda Criteria were used in this study: 1 - Individuals with cancer in families that fulfill the Amsterdam criteria; 2 - Individuals with 2 Lynch Syndrome related cancers, including synchronous and metachronous CRCs or associated extracolonic cancer; 3 - Individuals with CRC and a first degree relative with CRC and/or Lynch Syndrome related extracolonic cancer and/or colorectal adenoma with one of the cancers diagnosed at < 45 years and the adenoma diagnosed at < 40 years; 4 - Individuals with CRC or endometrial cancer diagnosed at < 45 years; 5 - Individuals with right-sided CRC with an undifferentiated pattern (solid/cribform) on histopathology diagnosed at < 45 years; 6 - Individuals with signet-ring-cell type CRC diagnosed at < 45 years; 7 - Individuals with adenomas diagnosed at <40 years.
Abbreviations and definitions: AMS, Amsterdam; M-AMS, Modified Amsterdam.
Other Variant: Known polymorphisms or variants found in normal controls (MSH2 G322D, MLH1 K618A), variants found in normal controls and in conjunction with a more definitive pathogenic mutation (MLH1 V716M) or variants not found in normal controls but found in conjunction with a more definitive pathogenic mutation (MSH2 A272V).
PMS2 c.1211C>G P404R was not found in 184 normal control DNAs.
Screening of an additional 39 suspected Lynch Syndrome cases revealed three additional deletions: case 3919 with deletion of MLH1 exon 19, case 3769 with deletion of MSH2 exons 1 – 7 and case 3173 with deletion of MSH2 exons 3 – 6.
MLPA
Deletion analysis was performed with both the MLPA P003 MLH1/MSH2 and MLPA P008 Lynch Syndrome-2 Exon Deletion Test Kit’s (MRC-Holland) according to the manufacturer’s protocol. A 5µl aliquot of 100 ng genomic DNA was heated to 98˚C for 5 min and cooled to room temperature. A 3 µl aliquot of SALSA probe and MLPA buffer was added to the DNA and heated to 95˚C for 1 min and then incubated at 60˚C for 16 hrs to hybridize the probes and DNA. Ligase-65 and Ligase-65 buffers were then added yielding a final volume of 40 µl which was then incubated at 54˚C for 15 min. The ligated products were then heated to 98˚C for 5 min and kept at 4˚C until the initiation of PCR using the manufacture’s PCR Protocol 2. The PCR product was mixed with formamide and ROX-500 standard. Subsequent electrophoresis was performed on an ABI 3730 sequencer. Genescan and Genotyper software (Applied Biosystems) was used to collect and export the peak areas to an Excel spreadsheet. Individual peak areas were divided by the sum of that samples total peak areas for a relative peak area. A ratio of each probe’s relative peak area and the average relative peak areas of the controls corresponded to the number of copies for that individual probe. A ratio of 1.0 corresponded to two copies of that particular probe and a ratio of 0.5 corresponded to a single copy.
Primer Design
Regions of Alu repeats were found with RepeatMasker (http://repeatmasker.org/cgi-bin/WEBRepeatMasker) and excluded as areas for primer design. Primers for breakpoint and promoter PCR and DNA sequencing were chosen from genomic DNA sequence using the Primer3 web interface (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The PMS2 exon PCR and sequencing primers were previously published as follows: Exons 1, 2, 6–9 (37); Exons 3–5, 10, 12–15 (38); and exon 11a, 11b (two separate PCR reactions) (39). The primers used for sequencing PMS2 and the MSH2 and MLH1 promoters are listed in Table 2 and Table 3.
Table 2.
PMS2 sequencing primers
| Gene | Exon | Forward | Reverse | Size(bp) | Source |
|---|---|---|---|---|---|
| PMS2 | 1 | 5′-ggtcacgacggagaccg-3′ | 5′ ccatgttccccccatttcc-3′ | 400 | 36 |
| PMS2 | 2 | 5′-tgtttcttgtaactgatttctc-3′ | 5′ cttaactacaacaacattcacag-3′ | 233 | 36 |
| PMS2 | 3 | 5′-actgatagcatgggtccg-3′ | 5′ caaaattctgagacatgtga-3′ | 201 | 37 |
| PMS2 | 4 | 5′-acactgtcttgggaaatg-3′ | 5′ attaattttcagagaggtttc-3′ | 278 | 37 |
| PMS2 | 5 | 5′-ctcaaccatttagatcttga-3′ | 5′ aataaagcatttctcaataat-3′ | 451 | 37 |
| PMS2 | 6 | 5′-acttgagctgtgtaattcc-3′ | 5′ cccgctataatcactagagc-3′ | 289 | 36 |
| PMS2 | 7 | 5′-gtccactgtgtctttattag-3′ | 5′ agctctcaggataaaatgttc-3′ | 204 | 36 |
| PMS2 | 8 | 5′-taatccctttcactctgg-3′ | 5′ ccataaactgcctattatcag-3′ | 237 | 36 |
| PMS2 | 9 | 5′-ggggctgggaacatttgtc-3′ | 5′ atagcagagctgtagaatttc-3′ | 215 | 36 |
| PMS2 | 10 | 5′-attaagcccttccgtattt-3′ | 5′ ggaaacacattagctaaaagc-3′ | 742 | 37 |
| PMS2 | 11a | 5′-gtcctctcaccatttcagg-3′ | 5′ agtttggctgaggcaaaactc-3′ | 621 | 38 |
| PMS2 | 11b | 5′-actgcagcagcgaatatgc-3′ | 5′ aaaaaagaaaattttagataaaaagag-3′ | 519 | 38 |
| PMS2 | 12 | 5′-gtgtacaggtctgaaaacttg-3′ | 5′ cgcctggccaactagata-3′ | 1000 | 37 |
| PMS2 | 13 | 5′-cacttagctgagtagtgttgttattt-3′ | 5′ tgaacacctgaaagagaggaaac-3′ | 704 | 37 |
| PMS2 | 14 | 5′-tccaaaaagcattttgtgagtt-3′ | 5′ gagttcaaggtcacagagaacg-3′ | 780 | 37 |
| PMS2 | 15 | 5′-aactactaaaacgttgaacc-3′ | 5′ tttttgagacacagtcttgt-3′ | 726 | 37 |
Table 3.
MLH1 and MSH2 promotor primers
| MLH1 Promoter Primer | Coordinate† | |
| F1 | 5′-AACCCTTTCACCATGCTCTG-3′ | −1469 |
| F2 | 5′-TACATGCTCGGGCAGTACCT-3′ | −881 |
| R2 | 5′-TGAAGAGAGAGCTGCTCGTG-3′ | −749 |
| R1 | 5′-GCTCACGTTCTTCCTTCAGC-3′ | −77 |
| MSH2 Promoter Primer | Coordinate | |
| F1 | 5′-CACCTCCCAGGTTCAAGAGA-3′ | −1491 |
| F2 | 5′-GCCTCAGCCCTGCTAATATC-3′ | −930 |
| R2 | 5′-CGGTAGCTCACGCCTGTAAT-3′ | −795 |
| R1 | 5′-CCCACACCCACTAAGCTGTT-3′ | −42 |
The coordinate given is the nucleotide position of the 5′ nucleotide of the primer.
Breakpoint PCR and Sequencing
Intronic primers distal to the deleted exons were used to amplify the breakpoints of those samples with deletions limited to exons not including the most 5’ and 3’ most exons of the gene of interest. For the SKOV3 cell line, which is hemizygous for an MLH1 deletion (40), primer pairs comprising 200–500 bp amplicons were designed and tested every 5–10 kb downstream of the 3′ end of the gene until a PCR product was observed. Once the breakpoint was localized at 5 to 10 kb resolution, this mapping procedure was repeated with more closely spaced amplicons until it was possible to locate the breakpoint precisely enough to then amplify across it. The Expand Long Template PCR System (Roche) was used to PCR amplify across the breakpoints. PCR reactions of 25 µl were performed using final concentrations of 1x Buffer 2, 500 µM dNTPs, 300 nM primers (each), 2U polymerase, and 50–100 ng genomic DNA. Cycling conditions consisted of: 93˚C for 2 min; followed by 10 cycles of 93˚C for 10 sec, 55˚C for 30 sec, 68˚C for 45 sec up to 8 min depending on the experiment; followed by 20 cycles of 93˚C for 10 sec, 55˚C for 30sec, 68˚C for 45 sec to 8 min +20 sec per cycle; one cycle of 68˚C for 7 min; and finally hold at 4˚C. The PCR products were analyzed on 1% agarose gels run in TBE. Pre-sequence clean-up of the PCR template was performed in 20 µl reactions with 2U SAP and 10U EXO1 (US Biochemicals) and then sequencing was performed on an ABI 3730 using procedures provided by the manufacturer.
Sequencing-based mutation screening of MLH1/MSH2 promoters and PMS2 exons
The MLH1 promoter was amplified with the primers MLH1 promoter F1 and MLH1 promoter R1 yielding a 1393 bp PCR product that was then sequenced with the same primers as well as with MLH1 promoter F2 and MLH1 promoter R2. The MSH2 promoter was amplified with the primers MSH2 promoter F1 and MSH2 promoter R1 yielding a 1450 bp PCR product and sequenced with the same primers as well as with MSH2 promoter F2 and MSH2 promoter R2 (see Table 3). The promoter regions of MLH1 and MSH2 were amplified in 25 µl volumes consisting of 1x PCII buffer, 150 µM dNTP’s, 200 nM primers (each), 5U Klentaq, and 25–50 ng genomic DNA. The cycling conditions were as follows: 94˚C for 3 min; 8 cycles of 94˚C for 30 sec, 63˚C (-1˚C/cycle) for 30sec, 72˚C for 1 min; then 32 cycles of 94˚C for 30 sec, 55˚C for 30 sec, 72˚C for 1 min; and finally hold at 4˚C. The PMS2 exons were amplified with the same reaction and cycling conditions as above except for exons 1 and 12. For these latter exons, the reaction conditions consisted of 1x PCR buffer II (Roche), 1.5 mM MgCl2, 100 µM dNTP’s, 200 nM primers (each), 1U Amplitaq (Roche), and 25–50 ng genomic DNA. Exon 12 PCR used the same cycling conditions as above. Exon 1 PCR used the following cycling conditions: 94˚C for 3 min; 35 cycles of 94˚C for 30 sec, 52˚C for 30 sec, 72˚C for 1 min; and finally hold at 4˚C. Pre-sequence clean-up and sequencing were performed as previously described (10).
MLH1 methylation
Methylation of the MLH1 promoter in tumor and blood DNA samples was analyzed by both methylation specific PCR and bisulfite DNA sequencing as previously described (14). Samples scored as somatic methylation did not have methylated species present in DNA from blood and appeared to have only methylated DNA species present in tumor DNA, and hence likely showed biallelic methylation (41–43).
Results
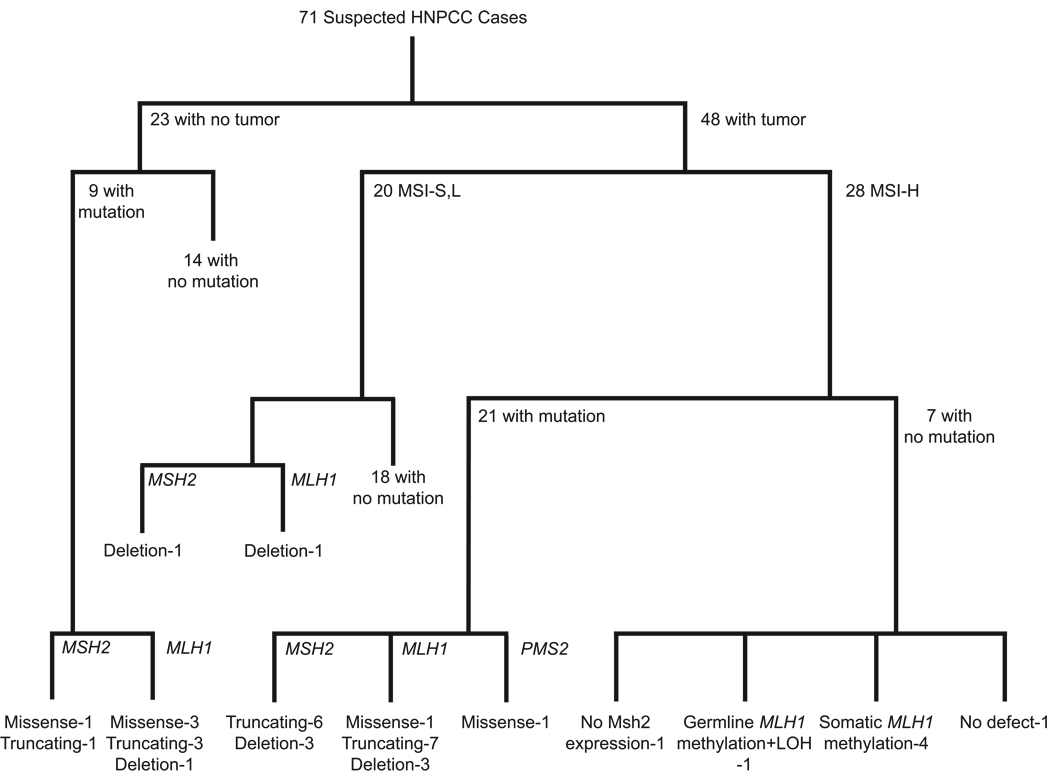
We have been exhaustively analyzing a series of 71 colorectal cancer cases suspected of being Lynch Syndrome cases by different clinical criteria (Table 1) for germline defects in MMR genes. Through several previously published studies and the work described here, these cases have been analyzed by the following strategy (10, 14, 28, 33, 36). First, the coding region and intron-exon junctions of the MSH2, MSH6 (including the MSH6 promoter) and MLH1 genes were sequenced in DNA from blood of all cases. Tumors, when available, were screened for MSI using 5 and 10 microsatellite marker panels recommended by the NCI consensus groups (44, 45) and were screened for expression of Msh2 and Mlh1 protein by immunohistochemistry. All tests were performed on all samples independent of any results obtained. Second, for all cases where mutations were not initially found, DNA from blood was screened for deletion mutations in MSH2 and MLH1 and all mutation negative MSI-H cases were also screened for germline and tumor MLH1 methylation. Third, all cases in which an alteration in MSH2, MSH6 or MLH1 or MLH1 methylation was not found were then screened for deletions in PMS2, MSH6, MUTYH, MLH3 and TACSTD1 including the region between MSH2 and TACSTD1. Finally, the coding region, intron-exon junctions and promoter regions of the PMS2 gene and the promoters of MSH2 and MLH1 were sequenced in DNA from blood of all MSI-H cases where mutations had not been found. The results from this analysis, some of which has been published and discussed previously (10, 14, 28, 33, 36, 46), are summarized in Table 1 and Figure 1.
Figure 1.
Flow diagram summarizing the results of mutation detection analyses. The mutation analysis strategy and order of mutation testing is described under "Results".
Using MLPA, 4 of the 71 families were found to have a deletion covering all or part of the MSH2 gene and 5 of the 71 families were found to have a deletion covering part of the MLH1 gene (Table 1). To identify additional deletions, we subsequently screened an additional 39 families obtained from another study and identified two additional MSH2 deletions and one additional MLH1 deletion. In addition, analysis by MLPA confirmed the MSH2 and MLH1 deletions we previously reported in the LoVo and SKOV3 tumor cell lines (40). By using a large selection of primer pairs designed to potentially amplify across deletion breakpoint junctions, we were ultimately able to amplify and sequence 4 MLH1 and 3 MSH2 deletion breakpoint junctions and then confirm each breakpoint junction by amplification with an optimized pair of PCR primers followed by sequencing (Table 4). This analysis showed that 3 of the 7 deletion events involved Alu elements flanking both sides of the deleted region and 6 of the deletion events involved flanking microhomologies ranging from 1 to 21 base pairs in length. One of the deletions (Cases 1120 and 2738, Exon 12) had the same breakpoint as that of a previously published deletion (47). None of the remaining deletions appeared to correspond to previously described deletions identified by either sequencing deletion breakpoint junctions or through identification of primer pairs for amplifying breakpoint junctions even though in several cases the same combinations of exons were found to be deleted (17, 48–50).
Table 4.
MSH2 and MLH1 Deletion Breakpoints
| Deleted Region | Breakpoint | Sequence† |
|---|---|---|
| MLH1 exon6# | 5′ w.t. (c.453+769_799) | GACCAGCCTGACCAACATGGaGAAACcCCAT |
| Case 597 | GACCAGCCTGACCAACATGGCGAAACGCCAT | |
| 3′ w.t. (c.546-483_513) | tACCAGCCTGgCCAACATGGCGAAACGCCAT | |
| MLH1 exon12 | 5′ w.t. (c.1039–2315_2345) | TCCAATTTAATTCCAAcaCtGtctacttgga |
| Cases 1120 & 2738 | TCCAATTTAATTCCAAAGCAGGAATAATAAT | |
| 3′ w.t. (c.1409+812_842) | aaagctggagaaaaggAGCAGGAATAATAAT | |
| MSH2 exon5–6# | 5′ w.t. (c.793-397_427) | TCTAACCTCACAAGGTtgaAagggcctAatT |
| Case 1642 | TCTAACCTCACAAGGTCTGACCTTGAGATCT | |
| 3′ w.t. (c.1077-491_521) | ggcgggtctcaAActTCTGACCTTGAGATCT | |
| MSH2 exon3–6# | 5′ w.t. (c.367-267_297) | CCCCATCTCTACTAAAAATACAAAAAttaGc |
| Case 3173 | CCCCATCTCTACTAAAAATACAAAAAAATGA | |
| 3′ w.t. (c.1077–2095_2125) | CCCCgTCTCTACTAAAAATACAAAAAAATGA | |
| MSH2 exon3–8# | 5′ w.t. (c.367–446_476) | CCCAAAGTGTTGGGATTACAggcgTGAGCCA |
| Cell line LoVo | CCCAAAGTGTTGGGATTACAAGTATGAGCCA | |
| 3′ w.t. (c.1387-692_722) | CCCAAAGTGcTGGGATTACAAGTATGAGCCA | |
| MLH1 exon4–19 | 5′ w.t. (c.360_c.380+10) | TGATGGAAAGTGTGCATacAggtataGTGct |
| Cell line SKOV3 | TGATGGAAAGTGTGCATCGACTCCAGGTGGC | |
| 3′ w.t. (*93,487_93,517) | gcccctcAtagcTaCATCGACTCCAGGTGGC |
The sequence following the case number is the breakpoint junction and flanking sequence, with the junction sequence underlined. The nucleotide coordinates in () are the coordinates of the upstream and downstream target sequences.
Breakpoints occur in Alu repeat region
Of the initial 28 MSI-H cases analyzed, 8 did not have a mutation in MSH2, MSH6 or MLH1. Subsequent analysis found that one of these cases had a germline missense variant in PMS2 that was not found in 184 normal control DNAs. The resulting PMS2 amino acid substitution did not affect a conserved amino acid and is in a region that is homologous to a region of bacterial MutL for which no structural information is available and which can tolerate different deletions without effecting MMR (51). One of the 8 cases showed apparent germline methylation of MLH1 with associated LOH of the non-methylated allele in the tumor (14). Four of the cases showed apparent bi-allelic methylation of the MLH1 promoter in the tumor consistent with the idea that these cases might represent sporadic cancers (14, 32, 42, 43, 52). One of the cases had no detectable alteration in any gene analyzed but showed loss of MSH2 expression in the tumor; this case is a candidate to have a mutation in a region that was not sequenced such as an intron or is a candidate for germline or somatic methylation of MSH2 (16), although this sample did not appear to have a deletion eliminating the TACSTD1 termination codon associated with germline MSH2 methylation (16). In total, only 1 out of 28 MSI-H cases could not be linked to loss of function or an alteration in a known MMR gene. One MLH1 and 1 MSH2 deletion, and no other mutations were found among the 20 MSI-L/S cases (10% mutation frequency). The yield and distribution of mutations in cases without available tumors was essentially the same as that in cases where a tumor was available.
Discussion
In the present study, we have extended our prior analysis of 71 suspected Lynch Syndrome cases as defined by a diversity of clinical criteria for MMR defects using methods that had not been previously applied to these cases. This extensive analysis has yielded a number of key results. First, among the 28 clearly MMR defective cases as evidenced by their MSI-H signature, it was possible to link 27 of the cases (96%) to a defect in a known MMR gene and in most of these cases it was possible to identify the underlying mutation at the DNA level. This is arguably the highest or among the highest frequencies reported (4, 5, 21). Second, consistent with previous experience, the vast majority of mutations detected (97 %) were in the MSH2 or MLH1 genes with only 1 mutation in PMS2 and no mutations in MSH6 detected (4–6, 21). Third, we detected mutations in cases where tumor samples were not available at the same frequency as in cases where tumor samples were available. Fourth, our ability to detect mutations and provide insight into the genetics of a high proportion of cases did not require the use of complex techniques like "conversion of diploidy to haploidy" (5). Fifth, of the 7 MSI-H cases where no mutation was found at the DNA sequence level, 4 had somatic silencing of MLH1 and were likely sporadic cases (14, 32, 42, 43, 52), 1 had an MLH1 epimutaton (14, 15), 1 lacked expression of MSH2 protein and in 1 case we were not able to develop any insight into the underlying MMR defect. And finally 1 MSH2 mutation and 1 MLH1 mutation were detected among 20 MSI-S, L cases indicating that recommended criteria for MSI testing may misclassify some MMR defective tumors. In sum, these results suggest that it is possible to detect the genetic basis for MMR defects in virtually every suspected Lynch Syndrome case that actually is associated with a MMR defect and, as discussed below, provide a guide to efficient mutation detection strategies.
After analysis for mutations in MSH2 and MLH1, there were 8 MSI-H cases that did not have mutations in these two genes; 1 met Amsterdam criteria, 1 met modified Amsterdam criteria, 3 met the even weaker Lynch Syndrome-like criteria and 3 were isolated early onset cases without a significant associated family history. Extensive analysis of these cases revealed insights into the nature of their underlying MMR defects in all but one case and several of these cases deserve further comment. We were not able to find any molecular basis for a MMR defect in an individual diagnosed with CRC before 45 years of age and having no family history of CRC; we did not have immunohistochemistry data for this case that might have provided further insight into the nature of the MMR defect. One case meeting modified Amsterdam criteria did not have a mutation in MSH2 or in the TACSTD1 transcription termination region but the tumor from this case did not express MSH2 protein; potentially this case has an MSH2 epimutation (14–16), although we lacked sufficient tumor DNA for analysis. One case meeting modified Amsterdam criteria had a PMS2 mutation. An MLH1 epimutation was found in an individual diagnosed with CRC before 45 years of age and having no family history of CRC. And, finally, in 4 cases we found somatic methylation of MLH1 indicating they were sporadic cases (14, 32, 42, 43, 52). Three of these cases had weak family histories of CRC or were isolated CRC cases diagnosed before 45 years so it is not surprising that they might be sporadic CRC cases. However, 1 of these latter cases met Amsterdam criteria suggesting it was a sporadic case within a potential Lynch Syndrome family. These results indicate both that exhaustive analysis of the ~25% of MSI-H cases initially lacking mutations in MSH2 or MLH1 can ultimately implicate known MMR genes in virtually all of the cases and that one should not ignore the possibility that some of the cases are sporadic, particularly when analyzing cases meeting some of the weaker Bethesda criteria for Lynch Syndrome. A further implication of these results is that it is possible that some suspected Lynch Syndrome cases that are MSI-S, L or for which no tumor is available but lack a mutation in MSH2 or MLH1 may be sporadic cases and that in families with a strong family history of appropriate cancers a second individual should be analyzed.
We observed 1 MSH2 and 1 MLH1 deletion mutation among 20 MSI-S, L cases, which would be predicted to not be MMR defective (40, 45, 53) and hence not associated with defects in either MSH2 or MLH1. Other studies have also observed mutations in MSH2 and MLH1 in MSI-S/L Lynch Syndrome cases, although most of the mutations observed could not be definitively classified as pathogenic mutations (4). It is possible that MSI testing misclassifies some fraction of true MSI-H cases as MSI-S, L. It is known that no individual microsatellite is unstable in 100% of MSI-H tumors (45). We have calculated for MSI-H being defined as 4 out of 10 microsatellites being unstable, 17, 6 and 1% of true MSI-H cases would be misclassified as MSI-L if on average each microsatellite was unstable in 50, 60 and 70% of MMR defective tumors, respectively; use of larger numbers of microsatellites will reduce misclassification and using fewer will increase misclassification. As a consequence, MSI status should probably not be used to eliminate suspected Lynch Syndrome cases from testing for mutations in MSH2 or MLH1 by direct DNA sequencing and deletion screening, particularly in cases where family history makes a strong prediction of Lynch Syndrome.
The suspected Lynch Syndrome cases described here were predominantly analyzed by different methods in parallel without using any one set of results to stratify samples for subsequent analysis. Because we were able to link 96% of the obviously MMR defective MSI-H cases to a specific gene, our results can be used to guide efficient screening of suspected Lynch Syndrome cases for MMR defects. Based on our results, we recommend that in cases where clinical criteria suggest genetic risk for Lynch Syndrome, initial screening for mutations in MMR genes should involve analyzing genomic DNA for mutations in MSH2 and MLH1 by direct DNA sequencing and MLPA. This will detect essentially all mutations in MSH2 and MLH1 and the vast majority of all mutations in MMR genes. Mutation negative cases should then be analyzed for MSI status and for protein expression by IHC, which will identify those cases for which additional, more detailed analysis is warranted. The most useful subsequent analysis would be screening for mutations in PMS2, methylation (silencing) of MLH1, methylation (silencing) of MSH2 and potentially "Conversion Analysis" (5) to resolve difficult cases. Prescreening our suspected Lynch Syndrome cases for MSI status and for protein expression by IHC would have reduced the yield of mutations by misclassifying MSI-H cases as MSI-L cases and because not all mutations in MSH2 and MLH1 alter protein expression levels (4, 5, 7, 33, 34); of the 28 cases where a tumor sample was available where we identified an MMR defect, 2 would have been omitted by MSI prescreening and 6 would have been omitted by prescreening for expression of MSH2 and MLH1 (see data of Ref. 34 for protein expression data). In contrast, screening sporadic CRC cases for MSI status and for protein expression by IHC would be highly effective because silencing of MLH1 that underlies these cases results in significant MMR defects and substantive loss of MLH1 protein expression; such data could possibly be used in making treatment choices for sporadic CRCs as MMR status is thought to predict the sensitivity to some commonly used therapeutic agents (54, 55). Because mutation screening in cases where tumors are not available, and particularly mutation screening in MSI-S, L cases, will increase the number of cases screened where a mutation will not be found relative to just screening MSI-H cases, the use of newer patient selection algorithms might be considered to provide greater sensitivity and specificity in mutation detection compared to using the Bethesda criteria for patient selection (31).
In considering the choice of testing methods, its should be noted that high-quality DNA sequencing for mutation testing is readily available and has become relatively inexpensive, MLPA analysis for genome rearrangements is also inexpensive and can easily be performed by any facility offering capillary based sequencing using a commercially available kit, and both MSI and IHC testing are readily available in many clinical testing laboratories. Commercially available testing at a number of clinical labs now includes both complete gene sequencing and testing for genome rearrangements. In contrast, breakpoint sequencing is a research problem not a routine test, and does not provide information that is generally useful for diagnostic purposes. Testing for gene specific DNA methylation does not appear to be widely available at clinical testing sites; however, there are many different platforms for methylation analysis available to the research community.
We did not find MSH6 mutations in our suspected Lynch Syndrome cases but have found MSH6 mutations in familial cases not meeting any Lynch Syndrome criteria (10). Others have found MSH6 mutations at low frequency in Lynch Syndrome cases including those with atypical family histories (8, 11–13, 56). MSH6 mutations have also been associated with inherited predisposition to endometrial cancer and have been found in Lynch Syndrome families with endometrial cancer (8, 11, 56). These results suggest that a critical evaluation of family history might be used to identify suspected Lynch Syndrome cases for analysis for MSH6 mutations, and certainly only cases lacking defects in MSH2 or MLH1 should be considered for analysis of MSH6.
Acknowledgements
The authors would like to thank Rick Fishel at the Ohio State University Medical School for helpful discussions and comments on the manuscript and Christopher Putnam and Jason Chan at the Ludwig Institute for help with statistical calculations concerning microsatellite instability measurements. This work was supported by NIH grants CA85759 to S. Syngal and GM50006 to R. Kolodner
Footnotes
Conflicts of Interest
R. Kolodner is an inventor on patents owned by the Dana-Farber Cancer Institute covering sequences of the MSH2, MLH1 and PMS2 genes, and reagents related to these genes including antibodies for detecting the proteins encoded by these genes. S. Syngal has in the past served in an advisory role to Myriad Genetics, Inc.
References
- 1.Vasen HF, Wijnen JT, Menko FH, et al. Cancer risk in families with hereditary nonpolyposis colorectal cancer diagnosed by mutation analysis. Gastroenterology. 1996;110:1020–1027. doi: 10.1053/gast.1996.v110.pm8612988. [DOI] [PubMed] [Google Scholar]
- 2.Marra G, Boland CR. Hereditary nonpolyposis colorectal cancer:the syndrome, the genes, and historical perspectives. J Natl Cancer Inst. 1995;87:1114–1125. doi: 10.1093/jnci/87.15.1114. [DOI] [PubMed] [Google Scholar]
- 3.de la Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer. 2004;4:769–780. doi: 10.1038/nrc1453. [DOI] [PubMed] [Google Scholar]
- 4.Lagerstedt Robinson K, Liu T, Vandrovcova J, et al. Lynch syndrome (hereditary nonpolyposis colorectal cancer) diagnostics. J Natl Cancer Inst. 2007;99:291–299. doi: 10.1093/jnci/djk051. [DOI] [PubMed] [Google Scholar]
- 5.Casey G, Lindor NM, Papadopoulos N, et al. Conversion analysis for mutation detection in MLH1 and MSH2 in patients with colorectal cancer. Jama. 2005;293:799–809. doi: 10.1001/jama.293.7.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peltomaki P, Vasen H. Mutations associated with HNPCC predisposition -- Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Senter L, Clendenning M, Sotamaa K, et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology. 2008;135:419–428. doi: 10.1053/j.gastro.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wijnen J, de Leeuw W, Vasen H, et al. Familial endometrial cancer in female carriers of MSH6 germline mutations. Nat Genet. 1999;23:142–144. doi: 10.1038/13773. [DOI] [PubMed] [Google Scholar]
- 9.Hendriks YM, Wagner A, Morreau H, et al. Cancer risk in hereditary nonpolyposis colorectal cancer due to MSH6 mutations:impact on counseling and surveillance. Gastroenterology. 2004;127:17–25. doi: 10.1053/j.gastro.2004.03.068. [DOI] [PubMed] [Google Scholar]
- 10.Kolodner RD, Tytell JD, Schmeits JL, et al. Germ-line msh6 mutations in colorectal cancer families. Cancer Res. 1999;59:5068–5074. [PubMed] [Google Scholar]
- 11.Hampel H, Frankel W, Panescu J, et al. Screening for Lynch syndrome (hereditary nonpolyposis colorectal cancer) among endometrial cancer patients. Cancer Res. 2006;66:7810–7817. doi: 10.1158/0008-5472.CAN-06-1114. [DOI] [PubMed] [Google Scholar]
- 12.Akiyama Y, Sato H, Yamada T, et al. Germ-line mutation of the hMSH6/GTBP gene in an atypical hereditary nonpolyposis colorectal cancer kindred. Cancer Res. 1997;57:3920–3923. [PubMed] [Google Scholar]
- 13.Miyaki M, Konishi M, Tanaka K, et al. Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet. 1997;17:271–272. doi: 10.1038/ng1197-271. [DOI] [PubMed] [Google Scholar]
- 14.Gazzoli I, Loda M, Garber J, et al. A hereditary nonpolyposis colorectal carcinoma case associated with hypermethylation of the MLH1 gene in normal tissue and loss of heterozygosity of the unmethylated allele in the resulting microsatellite instability-high tumor. Cancer Res. 2002;62:3925–3928. [PubMed] [Google Scholar]
- 15.Hitchins MP, Wong JJ, Suthers G, et al. Inheritance of a cancer-associated MLH1 germ-line epimutation. N Engl J Med. 2007;356:697–705. doi: 10.1056/NEJMoa064522. [DOI] [PubMed] [Google Scholar]
- 16.Ligtenberg MJ, Kuiper RP, Chan TL, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat Genet. 2008 doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 17.van der Klift H, Wijnen J, Wagner A, et al. Molecular characterization of the spectrum of genomic deletions in the mismatch repair genes MSH2, MLH1, MSH6, and PMS2 responsible for hereditary nonpolyposis colorectal cancer (HNPCC) Genes Chromosomes Cancer. 2005;44:123–138. doi: 10.1002/gcc.20219. [DOI] [PubMed] [Google Scholar]
- 18.Lipkin SM, Rozek LS, Rennert G, et al. The MLH1 D132H variant is associated with susceptibility to sporadic colorectal cancer. Nat Genet. 2004;36:694–699. doi: 10.1038/ng1374. [DOI] [PubMed] [Google Scholar]
- 19.Raptis S, Mrkonjic M, Green RC, et al. MLH1 −93G>A promoter polymorphism and the risk of microsatellite-unstable colorectal cancer. J Natl Cancer Inst. 2007;99:463–474. doi: 10.1093/jnci/djk095. [DOI] [PubMed] [Google Scholar]
- 20.Mrkonjic M, Raptis S, Green RC, et al. MSH2 118T>C and MSH6 159C>T promoter polymorphisms and the risk of colorectal cancer. Carcinogenesis. 2007;28:2575–2580. doi: 10.1093/carcin/bgm229. [DOI] [PubMed] [Google Scholar]
- 21.Liu B, Parsons R, Papadopoulos N, et al. Analysis of mismatch repair genes in hereditary non-polyposis colorectal cancer patients. Nat Med. 1996;2:169–174. doi: 10.1038/nm0296-169. [DOI] [PubMed] [Google Scholar]
- 22.Jarvinen HJ, Aarnio M, Mustonen H, et al. Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology. 2000;118:829–834. doi: 10.1016/s0016-5085(00)70168-5. [DOI] [PubMed] [Google Scholar]
- 23.Wagner A, van Kessel I, Kriege MG, et al. Long term follow-up of HNPCC gene mutation carriers:compliance with screening and satisfaction with counseling and screening procedures. Fam Cancer. 2005;4:295–300. doi: 10.1007/s10689-005-0658-9. [DOI] [PubMed] [Google Scholar]
- 24.Vasen HF, Mecklin JP, Khan PM, et al. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC) Dis Colon Rectum. 1991;34:424–425. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- 25.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez-Bigas MA, Boland CR, Hamilton SR, et al. National Cancer Institute Workshop on Hereditary Nonpolyposis Colorectal Cancer Syndrome:meeting highlights and Bethesda guidelines. J Natl Cancer Inst. 1997;89:1758–1762. doi: 10.1093/jnci/89.23.1758. [DOI] [PubMed] [Google Scholar]
- 27.Vasen HF, Watson P, Mecklin JP, et al. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 28.Syngal S, Fox EA, Eng C, et al. Sensitivity and specificity of clinical criteria for hereditary non-polyposis colorectal cancer associated mutations in MSH2 and MLH1. J Med Genet. 2000;37:641–645. doi: 10.1136/jmg.37.9.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez-Moranta F, Castells A, Andreu M, et al. Clinical performance of original and revised Bethesda guidelines for the identification of MSH2/MLH1 gene carriers in patients with newly diagnosed colorectal cancer:proposal of a new and simpler set of recommendations. Am J Gastroenterol. 2006;101:1104–1111. doi: 10.1111/j.1572-0241.2006.00522.x. [DOI] [PubMed] [Google Scholar]
- 30.Pinol V, Castells A, Andreu M, et al. Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. Jama. 2005;293:1986–1994. doi: 10.1001/jama.293.16.1986. [DOI] [PubMed] [Google Scholar]
- 31.Green RC, Parfrey PS, Woods MO, et al. Prediction of Lynch syndrome in consecutive patients with colorectal cancer. J Natl Cancer Inst. 2009;101:331–340. doi: 10.1093/jnci/djn499. [DOI] [PubMed] [Google Scholar]
- 32.Poynter JN, Siegmund KD, Weisenberger DJ, et al. Molecular characterization of MSI-H colorectal cancer by MLHI promoter methylation, immunohistochemistry, and mismatch repair germline mutation screening. Cancer Epidemiol Biomarkers Prev. 2008;17:3208–3215. doi: 10.1158/1055-9965.EPI-08-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wahlberg SS, Schmeits J, Thomas G, et al. Evaluation of microsatellite instability and immunohistochemistry for the prediction of germ-line MSH2 and MLH1 mutations in hereditary nonpolyposis colon cancer families. Cancer Res. 2002;62:3485–3492. [PubMed] [Google Scholar]
- 34.Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 35.Gammie AE, Erdeniz N, Beaver J, et al. Functional characterization of pathogenic human MSH2 missense mutations in Saccharomyces cerevisiae. Genetics. 2007;177:707–721. doi: 10.1534/genetics.107.071084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Syngal S, Fox EA, Li C, et al. Interpretation of genetic test results for hereditary nonpolyposis colorectal cancer:implications for clinical predisposition testing. Jama. 1999;282:247–253. doi: 10.1001/jama.282.3.247. [DOI] [PubMed] [Google Scholar]
- 37.Nicolaides NC, Carter KC, Shell BK, et al. Genomic organization of the human PMS2 gene family. Genomics. 1995;30:195–206. doi: 10.1006/geno.1995.9885. [DOI] [PubMed] [Google Scholar]
- 38.De Vos M, Hayward BE, Picton S, et al. Novel PMS2 pseudogenes can conceal recessive mutations causing a distinctive childhood cancer syndrome. Am J Hum Genet. 2004;74:954–964. doi: 10.1086/420796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakagawa H, Lockman JC, Frankel WL, et al. Mismatch repair gene PMS2:disease-causing germline mutations are frequent in patients whose tumors stain negative for PMS2 protein, but paralogous genes obscure mutation detection and interpretation. Cancer Res. 2004;64:4721–4727. doi: 10.1158/0008-5472.CAN-03-2879. [DOI] [PubMed] [Google Scholar]
- 40.Boyer JC, Umar A, Risinger JI, et al. Microsatellite instability, mismatch repair deficiency, and genetic defects in human cancer cell lines. Cancer Res. 1995;55:6063–6070. [PubMed] [Google Scholar]
- 41.Kane MF, Loda M, Gaida GM, et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 42.Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veigl ML, Kasturi L, Olechnowicz J, et al. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc Natl Acad Sci U S A. 1998;95:8698–8702. doi: 10.1073/pnas.95.15.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition:development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 45.Dietmaier W, Wallinger S, Bocker T, et al. Diagnostic microsatellite instability:definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749–4756. [PubMed] [Google Scholar]
- 46.Bandipalliam P, Garber J, Kolodner RD, et al. Clinical presentation correlates with the type of mismatch repair gene involved in hereditary nonpolyposis colon cancer. Gastroenterology. 2004;126:936–937. doi: 10.1053/j.gastro.2004.01.038. [DOI] [PubMed] [Google Scholar]
- 47.Li L, McVety S, Younan R, et al. Distinct patterns of germ-line deletions in MLH1 and MSH2:the implication of Alu repetitive element in the genetic etiology of Lynch syndrome (HNPCC) Hum Mutat. 2006;27:388. doi: 10.1002/humu.9417. [DOI] [PubMed] [Google Scholar]
- 48.Nakagawa H, Hampel H, de la Chapelle A. Identification and characterization of genomic rearrangements of MSH2 and MLH1 in Lynch syndrome (HNPCC) by novel techniques. Hum Mutat. 2003;22:258. doi: 10.1002/humu.9171. [DOI] [PubMed] [Google Scholar]
- 49.Viel A, Petronzelli F, Della Puppa L, et al. Different molecular mechanisms underlie genomic deletions in the MLH1 Gene. Hum Mutat. 2002;20:368–374. doi: 10.1002/humu.10138. [DOI] [PubMed] [Google Scholar]
- 50.De Lellis L, Curia MC, Catalano T, et al. Combined use of MLPA and nonfluorescent multiplex PCR analysis by high performance liquid chromatography for the detection of genomic rearrangements. Hum Mutat. 2006;27:1047–1056. doi: 10.1002/humu.20386. [DOI] [PubMed] [Google Scholar]
- 51.Guarne A, Ramon-Maiques S, Wolff EM, et al. Structure of the MutL C-terminal domain:a model of intact MutL and its roles in mismatch repair. Embo J. 2004;23:4134–4145. doi: 10.1038/sj.emboj.7600412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuismanen SA, Holmberg MT, Salovaara R, et al. Genetic and epigenetic modification of MLH1 accounts for a major share of microsatellite-unstable colorectal cancers. Am J Pathol. 2000;156:1773–1779. doi: 10.1016/S0002-9440(10)65048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strand M, Prolla TA, Liskay RM, et al. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 54.Casorelli I, Russo MT, Bignami M. Role of mismatch repair and MGMT in response to anticancer therapies. Anticancer Agents Med Chem. 2008;8:368–380. doi: 10.2174/187152008784220276. [DOI] [PubMed] [Google Scholar]
- 55.Meyers M, Wagner MW, Mazurek A, et al. DNA mismatch repair-dependent response to fluoropyrimidine-generated damage. J Biol Chem. 2005;280:5516–5526. doi: 10.1074/jbc.M412105200. [DOI] [PubMed] [Google Scholar]
- 56.Wagner A, Hendriks Y, Meijers-Heijboer EJ, et al. Atypical HNPCC owing to MSH6 germline mutations: analysis of a large Dutch pedigree. J Med Genet. 2001;38:318–322. doi: 10.1136/jmg.38.5.318. [DOI] [PMC free article] [PubMed] [Google Scholar]