Abstract
Genome-wide studies reveal that transcription by RNA polymerase II (Pol II) is dynamically regulated. To obtain a comprehensive view of a single transcription cycle, we switched on transcription of five long human genes (>100 kbp) with tumor necrosis factor-α (TNFα) and monitored (using microarrays, RNA fluorescence in situ hybridization, and chromatin immunoprecipitation) the appearance of nascent RNA, changes in binding of Pol II and two insulators (the cohesin subunit RAD21 and the CCCTC-binding factor CTCF), and modifications of histone H3. Activation triggers a wave of transcription that sweeps along the genes at ≈3.1 kbp/min; splicing occurs cotranscriptionally, a major checkpoint acts several kilobases downstream of the transcription start site to regulate polymerase transit, and Pol II tends to stall at cohesin/CTCF binding sites.
Keywords: endothelial cell, polymerase II, RNA, tumor necrosis factor alpha
Transcription by RNA polymerase II (Pol II) is at the core of gene expression and hence is the basis of all cellular activities. To generate a mature messenger RNA (mRNA), Pol II traverses a transcription cycle; this involves recruitment to an activated promoter, initiation, escape into the gene, elongation, and termination (1). Processing of the nascent transcript—which can include capping, splicing, and poly (A) addition—is coupled to polymerization, and the C-terminal domain (CTD) of the polymerase acts as a scaffold for the binding of many of the factors involved (2–4).
Genome-wide analyses using chromatin immunoprecipitation followed by microarray analysis (ChIP-chip) or deep sequencing (ChIP-seq) provide powerful means of mapping comprehensively and at high resolution where proteins bind to the DNA template. Such studies have revealed widespread pausing and abortion by engaged Pol II (5–10). Pol II dynamics have also been studied using fluorescence imaging (11–15), but it remains difficult to observe both the rapid recycling of Pol II (14) and the unstable nascent transcripts.
To overcome these problems, we focus on the temporal profile of the first cycle of transcription after switching on transcription. As Pol II transcribes at more than 3 kbp per min (12), we analyzed five genes longer than 100 kbp that could be rapidly and synchronously activated by tumor necrosis factor-α (TNFα), a potent cytokine that orchestrates the inflammatory response by sequentially activating the expression of more than 6,000 genes in cultured human umbilical vein cells (HUVECs) (16, 17). To avoid problems caused by variations in sequence-specific signal in the arrays used (18) and to increase sensitivity, we developed analytical algorithms for temporal profiling that handled each probe sequence separately. Our repeated comprehensive observations reveal a wave of transcription sweeping along the genes; a major checkpoint regulates polymerase transit ≈1–10 kbp into the genes, and polymerases tend to stall further downstream at sites where the RAD21 subunit of cohesin and CCCTC-binding factor (CTCF) bind (19, 20).
Results
A Wave of premRNA Synthesis That Sweeps Down Activated Genes.
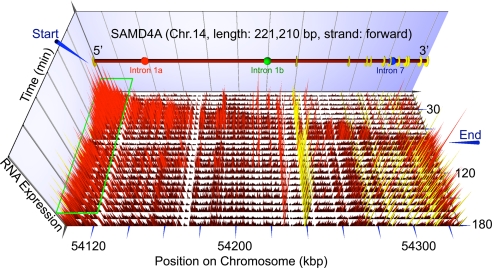
At different times after stimulation with TNFα, total nuclear RNA was purified and hybridized to a tiling microarray bearing oligonucleotides complementary to SAMD4A, a long gene of 221 kbp; signals were normalized using an algorithm (see SI Materials and Methods). In Fig. 1, the height of the red and yellow needles reflects the intensity of signal of each probe that is given by premRNA binding to intronic and exonic probes, respectively. An alternative way of presenting these results and a control with antisense probe are illustrated in Fig. S1. A wave of intronic signal (red) appears to sweep down the gene from “Start” (at 15 min) to “End” (at 75–90 min). Exonic signal (yellow) increases significantly above the basal level only after ≈75 min, when the polymerase has terminated. Similar waves are seen with other long genes, including ZFPM2 (486 kbp; Fig. S2).
Fig. 1.
Transcription waves visualized using microarrays. HUVECs were stimulated with TNFα, samples collected every 7.5 min for 3 h, and total nuclear RNA purified and hybridized to a tiling microarray bearing 25-mers complementary to SAMD4A. The vertical axis gives intensity of signal detected by intronic and exonic probes (marked by red and yellow needles, respectively). Gene length and genomic location are shown at the front, probe positions within the gene from left to right; and time after stimulation from top to bottom. Blue arrowheads indicate the “Start” and “End” of the first wave of transcription that sweeps down the gene; green rectangle marks the position of probes continuously yielding signal between 7.5–180 min. Intronic targets for RNA FISH probes (1a, red; 1b, green; 7, blue) are indicated on the gene map.
Abortive Transcription.
Polymerases make many abortive transcripts of a few tens of nucleotides before forming stable elongation complexes (6, 8–10). However, probes covering the first thousands of nucleotides from the transcription start site (TSS) yield signal between 15 and 180 min (Fig. 1, green rectangle), and polymerases seem to escape downstream only for a limited interval (i.e., 15–30 min) to initiate the first wave. A similar pattern is seen with all long genes studied (Fig. S2). This points to a checkpoint that regulates escape, but here the checkpoint seems to act on a second polymerase once it has sensed that there is already the first on the gene (even though it might be >100 kbp downstream). This is a major checkpoint, as signal given by probes in the first three 1,000-nucleotide windows in intron 1 of ZFPM2 is more than two times higher than that in downstream windows (Fig. 2A).
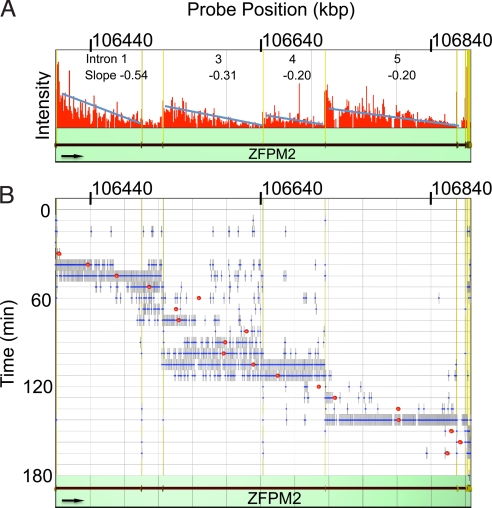
Fig. 2.
Speed of the transcription wave on ZFPM2. (A) Decay of RNA. Sum total signals given by all probes in 1,000 base windows between 15 and 180 min was calculated and shown by red bars (height reflects intensity). Blue lines were obtained by linear regression (slope indicated in each intron), showing that signal declines to zero from beginning to end of each intron. (B) Speed of wave front. For each probe on the tiling array, a blue dot indicates where expression first reaches 50% of the maximum (using 40–70% of the maximum yields similar velocities; Fig. S3), and we use this to define the wave front. A red dot marks the average position of all blue dots at one time point. The velocity of the wave front was calculated by linear regression using the red dots (red line). Vertical axis shows time after stimulation. Genomic location is shown on top of each column. Direction of the gene is shown by the arrow.
Cotranscriptional Splicing and Intron Degradation.
Figure 1 shows the wave reaches the middle of the second intron 60–75 min after stimulation; then, there is little signal in intron 1 (except close to the promoter, within the green rectangle). This is consistent with co-transcriptional splicing and degradation of RNA in the first intron while the polymerase is still transcribing the second. Quantitative analysis confirms this; summing signals given by all probes in 1,000-nucleotide windows between 15 and 180 min gives a “saw-tooth” pattern (Fig. 2A) with little signal at 3′ ends of introns. Suppose that a constant number of Pol II molecules elongate at constant speed (without aborting) and that co-transcriptional splicing occurs at each intron-exon boundary; then the slopes of the blue regression lines in Fig. 2A should be constant, as they are (for further discussion, see Fig. S3). The initial part of the first exon is deliberately excluded from this analysis, as the changing levels at the checkpoint distort the picture (Fig. S3). These calculations support the idea that once a polymerase passes through the checkpoint, it usually then reaches the terminus (12). Similar patterns are seen with the other genes (Fig. S3).
Velocity of the Wave.
We calculated the average speed of the wave as follows. In Fig. 2B, a blue dot for each probe indicates the first time when its expression reaches 50% of the maximum (and so marks the wave front); a red dot marks the average position of all blue dots at one time. The slope of the linear regression line drawn through the red dots and then reflects the velocity of the front. We estimate that the waves travel at 3.3, 3.2, 2.9, and 3.2 kb/min down ZFPM2, EXT1, SAMD4A, and ALCAM, respectively (Fig. S4B), giving an average of 3.1 kb/min. This is faster than 1.7–2.5 kb/min on human DMD (24), but slower than the 4.3 kb/min seen on an artificial array of ≈3-kbp human genes (12). As many regulatory genes activated by TNFα have long and conserved introns (e.g., ZFPM2, SAMD4A, NFKB1), the time spent transcribing these introns must profoundly affect temporal control by the TNFα/NFkB network (16, 17); long introns, which are conserved among many vertebrates, allow polymerases to convert space into time.
Transcription Wave Detected by RNA Fluorescence in Situ Hybridization.
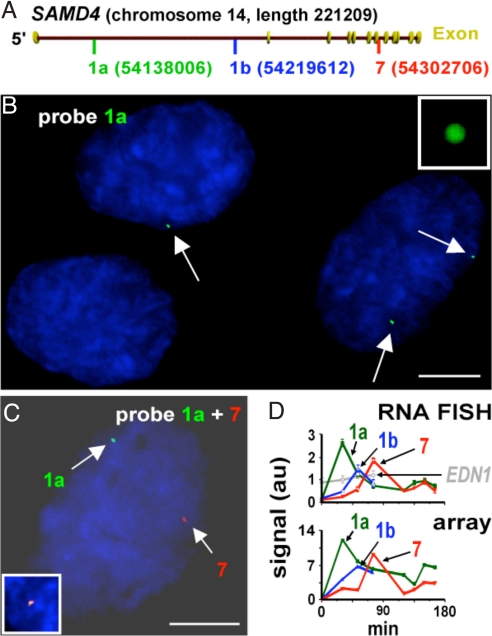
Because tiling arrays provide information on average RNA levels in ≈106 cells, cell-to-cell variation was monitored by fluorescence in situ hybridization (RNA FISH) (21, 22), using oligonucleotide probes able to detect a single transcript (23); intronic regions giving the strongest signals in tiling arrays were selected for analysis (SI Materials and Methods, Table S1). Probe 1a contains five 50-mers and carries ≈22 fluors; it is complementary to ≈600 bp in intron 1 of SAMD4A (Fig. 3A). Immediately after stimulation, it yields essentially no signal (Table S2). By 30 min, half of the cells possessed one or two, but never more than two, discrete nuclear foci (Fig. 3B, Table S2), which we assume mark a nascent transcript at one (or both) alleles. Later, the total number of foci in the population declined, but the intensity and size remained constant (Fig. 3D; Table S2). Probe 1b directed against a more 3′ region of the same intron revealed an analogous wave that peaked later (at 60 min), and one against intron 7 later still (Fig. 3D, Table S2). Total signal in the population mimics the changes seen in arrays (Fig. 3D). In contrast, antisense probes (against regions 1a, 1b, 7) yield essentially no signal, and a probe targeting an intron in EDN1 gave an unchanging signal (microarrays showed EDN1 expression was unaffected by stimulation). These results show that at least half the cells in the population respond.
Fig. 3.
A transcription wave visualized by RNA FISH. HUVECs were fixed at 0, 30, 52.5 and 75 min after stimulation with TNFα, and nascent SAMD4A or EDN1 RNA were detected by FISH using intron probes 1a, 1b, or 7, or an intron probe against EDN1 (labeled with Alexa 488), cells counterstained with DAPI, and images were collected. SAMD4 intron 1a (green), 1b (red), and 7 (blue) peaked as a wave of transcription passed through each region with time, whereas EDN1 signal (gray) remains constant (D, upper graph). Similar variations were given by relevant probes in microarrays (D, lower graph). (A) SAMD4A locus showing probe positions. (B) A typical field 30 min after stimulation obtained using probe 1a. Cells have 0, 1, or 2 green foci/cell (arrows) marking nascent RNA at one or other allele. Intensities are normalized relative to fluorescent beads (inset) to permit comparison between different experiments. Bar, 5 μm. (C) A nucleus 150 min after stimulation using probes 1a and 7; it contains one red and one green focus marking nascent RNA from each intron; yellow foci are never seen. (Inset) Positive control showing yellow focus given by probes 1a (green) and 1a-1 (red) 30 min after induction; these probes target intronic RNA sequences lying 1,000 nucleotides apart. Bar, 5 μm. (D) RNA FISH and arrays give similar results. (Top) Signals (i.e., size in pixels × intensity × number of foci; in arbitrary units [au]) were obtained by single (open symbols; as in B) or double labeling (closed symbols, as in C). (Bottom) Similar variations are given by relevant probes in arrays.
We confirmed that introns were removed co-transcriptionally by double labeling using probes 1a (green) and 7 (red). Although some cells possess foci of one or another (or both) colors (Fig. 3C), no foci contain both red and green signal and so appear yellow (Table S3). This suggests intron 1 must be removed before intron 7 is made. These results confirmed that in individual cells, a wave of transcription runs along activated genes, and that splicing occurs co-transcriptionally.
Stalling/Slowing of Pol II at Cohesin-Binding Sites.
The binding of Pol II was examined by ChIP-chip using antibodies against phospho-serine 5 in the heptad repeats in the CTD of the largest catalytic subunit (Rpb1); phospho-serine 5 is associated with transcriptional initiation and elongation (4, 24). Little Pol II was bound to SAMD4A (Fig. 4A, Fig. S5A) or EXT1 (Fig. S5B) before stimulation (at 0 min), although some (presumably “paused”) Pol II was detected near the TSS (indicated by the single asterisk in Fig. 4A). After 30 min, Pol II was bound to the 5′ half of the gene, and after 60 min it was bound more to 3′ (although there was still some binding near the TSS; double asterisks in Fig. 4A). These results confirm those obtained using arrays and FISH.
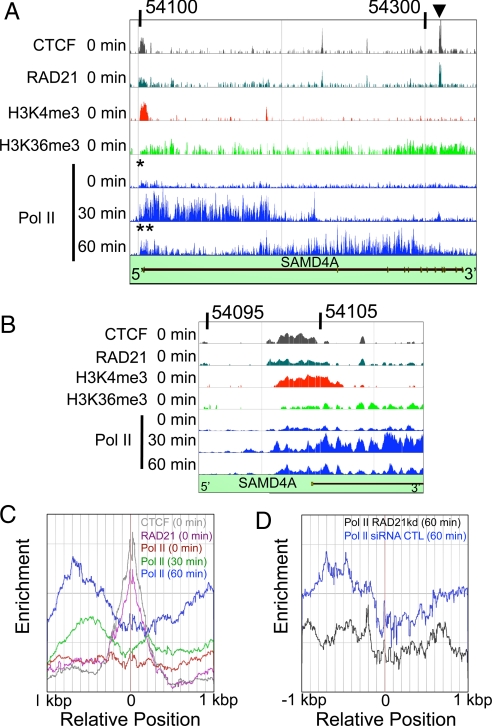
Fig. 4.
Stalling of Pol II analyzed using chromatin immunoprecipitation. HUVECs were stimulated with TNFα and harvested after 0, 30, and 60 min; then binding of CTCF, RAD21, modified histones (H3K4me3, H3K36me3), and elongating Pol II (phospho-Ser-5 modification) to SAMD4A was analyzed by ChIP-chip (CTCF, RAD21, H3K4me3, H3K36me3, and Pol II). Numbers on top of A and B show the location of the genomic region of Chr14 (SAMD4A) analyzed. (Vertical axes) Enrichment of binding. (A) Binding to SAMD4A. CTCF and RAD21 are often found together, consistent with the binding of a functional insulator complex. Asterisk shows where (engaged) Pol II binds at 0 min near the TSS, suggesting that it might be paused or poised. Double asterisk shows where Pol II binds near the TSS at 60 min. Arrowhead shows representative colocalization site of RAD21 and CTCF ≈210 kilonucleotides downstream of the TSS. (B) Binding at the TSS of SAMD4A. H3K4me3 and CTCF/RAD21 bind in/around the TSS. At 0 min, engaged Pol II also binds to this region. At 30 min, Pol II binding spreads into the gene, and after 60 min it becomes more concentrated around the TSS again. (C) Enrichments (number densities) of engaged Pol II near 35 sites distant from the TSS that were marked by bound RAD21 (pink) and CTCF (gray). At 0 min (red line), Pol II binds symmetrically around the RAD21/CTCF. At 30 min (green line), the amount of Pol II binding increases significantly. At 60 min (blue line), Pol II binding increases further and becomes concentrated upstream of the RAD21/CTCF; this is consistent with polymerase stalling. (D) An 80% reduction in RAD21 levels (achieved using siRNA) at 60 min (black) destroys the accumulation 5′ of the RAD21/CTCF site that is seen with a control siRNA (blue).
As some Pol II was bound near the TSS, we investigated the distribution of various other markers, namely, two histone modifications (H3K4me3 and H3K36me3), and two insulator proteins (the cohesin subunit RAD21 and CTCF) (Fig. 4A, Fig. S5 A and B). The distribution of bound Pol II overlaps that of H3K4me3, a marker for active chromatin (Fig. 4A, Fig. S5A), as previously described (8, 9). RAD21 and CTCF were often, but not invariably, bound to the same sites at the same time (Fig. 4A), again as seen previously (19, 20). Closer inspection (Fig. 4B, Fig. S5 C–G) shows that RAD21 and CTCF often bind near the boundaries of regions rich in Pol II and H3K4me3. It was previously reported that histone H3K4 and H3K36 tri-methylation are associated with the elongating polymerase (25), but we could not see the correlation (Fig. 4A, Fig. S5 A and B). At 60 min (i.e., after the wave had passed), the amount of paused Pol II increased near the TSS, the boundaries of which were again marked by RAD21/CTCF binding (Fig. S5 A and B).
We next examined the averaged distribution of Pol II around 35 RAD21/CTCF binding sites distant from the TSS (Fig. 4C); sites were selected as described in Fig. S6. At 0 time, little Pol II (red line) was found at the sites (gray and purple lines). However, after 30 and 60 min, Pol II (green and blue lines) accumulated upstream, while being excluded from 200–400 bp around the sites. By definition, all 35 sites bound both CTCF and RAD21, and Pol II accumulated on the 5′ side of 33 of the 35 sites. This suggests bound RAD21/CTCF stalls/slows the polymerase (25). To confirm this, we performed ChIP-chip after “knocking-down” RAD21 levels by 80% using siRNA; less Pol II (Fig. 4D, black line) was now bound near RAD21/CTCF sites, compared to the control (Fig. 4D, blue line). We conclude that Pol II tends to stall at bound RAD21/CTCF.
Discussion
Here we use TNFα to switch on transcription of five human genes rapidly and synchronously. As the genes are all longer than 100 kbp, and as samples are collected every 7.5 min for 3 h, there is ample time to monitor one complete transcription cycle. We use tiling microarrays and RNA FISH to follow the appearance of nascent RNA, and ChIP-chip to monitor changes in binding of Pol II and two insulator proteins (the cohesin subunit, RAD21, and CTCF), as well as two histone modifications (H3K4me3, H3K36me3). High-resolution data collection followed by statistical analysis in living human cells revealed precise mode that TNFα induces a wave of nascent RNA and Pol II to sweep along the genes from promoter to terminus (Figs. 1, 2, and 4A); these results are consistent with the polymerase initiating soon after stimulation, and transcribing ≈3.1 kbp/min (Fig. S4). Once RNA in a 3′ intron is seen, RNA in more 5′ introns has disappeared; for example, RNA FISH revealed that RNA from intron 7 is never found with its counterpart from intron 1 (Fig. 3C). Moreover, once RNA at the 3′ end of a long intron appears, more 5′ sequences in the same intron have disappeared (Figs. 1 and 2A). Here, splicing and degradation of intronic RNA occur co-transcriptionally, with degradation of intronic RNA beginning even before the polymerase reaches the next exon, and well before it terminates. These results contrast with a genome-wide analysis that indicates that premRNA splicing in yeast is predominantly posttranscriptional (26); but the results are consistent with observations pointing to a functional coupling between transcription and splicing (27, 28).
It is easy to imagine how a brief pulse of initiation soon after stimulation would trigger the wave. However, polymerases continue to initiate throughout the period analyzed, with intronic signal falling significantly 1–10 kbp into the gene (e.g., after the green rectangle in Fig. 1). This could be because polymerases speed up, or, more likely, abort. Because polymerases continue to initiate throughout the period analyzed, the checkpoint must operate to allow only the first ones to pass through while forcing later ones to abort, as only then can the widening trough between the peak of newly-initiated transcripts at the TSS and the advancing wave be generated. This checkpoint is much further into the gene than another major one acting only a few tens of nucleotides from the TSS (5, 6, 8–10). It seems able to sense whether another (pioneering) polymerase is already transcribing the gene, and we can only speculate on how it might do so.
Although the wave apparently progresses steadily down the gene, detailed analysis shows the polymerase tends to stall or slow at sites where RAD21, a subunit of cohesin, binds (Fig. 4A, Fig. S5 A and B). RAD21 often bound to the same sites as CTCF (Fig. 4A, arrowhead); and, as CTCF physically links cohesin to chromatin (29), these complexes could act as steric barriers to the elongating polymerase. This notion was supported by statistical analysis of 35 sites; Pol II tended to accumulate just upstream of these sites (Fig. 4C), and “knocking down” RAD21 abolishes this accumulation (Fig. 4D). Although the effects at these sites are clearly more subtle than those occurring at the major checkpoint within 1 to 10 kbp of the TSS, this finding provides evidence that elongation of Pol II can be regulated by an epigenetic modification.
In conclusion, we obtained a comprehensive view of a complete transcription cycle on several long human genes. One challenge now is to see whether the rich levels of regulation that we observe in these long genes are also found in genes of more average length.
Materials and Methods
Cell Culture.
Before stimulation, HUVECs were serum starved in EBM-2 (Clonetics) containing 0.5% fetal bovine serum (FBS) for 18 h, treated ± 10 ng/ml TNFα, and harvested at various times thereafter (17).
Tiling Microarrays.
We designed tiling microarrays (NimbleGen) covering ZFPM2, EXT1, SAMD4A, ALCAM, NFKB1, and EDN1. Probes (25 nucleotides) were selected so that the median of probes locate with 12 base interval, and designed to minimize cross hybridization with duplicated and repeated sequences in the human genome (SI Materials and Methods) (5, 30). Total RNA was purified using ISOGEN (NipponGene, Japan) and hybridized to microarrays (SI Materials and Methods).
RNA FISH.
Transcripts were detected using RNA FISH (SI Materials and Methods). Probe 1a consists of five 50-mers with ≈22 fluors (Table S1) and is complementary to ≈600 nucleotides in the middle of intron 1 of SAMD4A (Fig. 3A); a probe of this type is able to detect a single transcript (21). Sense signals were detected by probes the position and sequence of which are given in Fig. 1A, Fig. 3A, and Table S1. The antisense probes (against regions 1a, 1b, 7) yielded essentially no signal, and a probe targeting an intron in EDN1 gave an unchanging signal (microarrays showed that EDN1 expression was unaffected by stimulation).
ChIP-Chip.
Detailed experimental procedures for ChIP followed by analysis using microarrays, and for the analysis of Pol II stalling at cohesion/CTCF sites are described in SI Materials and Methods. The stalling profiles of Pol II at RAD21 binding sites are given in Fig. 4 and Fig. S6.
Supplementary Material
Acknowledgments.
This work was supported by the Special Coordination Fund for Promoting Science and Technology and by grants from the New Energy and Industrial Technology Development Organization, the National Institute of Biomedical Innovation of Japan, the Strategic Information and Communications R&D Promotion Program of the Ministry of Internal Affairs and Communications, the Genome Network Project, Japan Grants-in-Aid for Scientific Research (s) 16101006, 19800009, and 19310129 from the Ministry of Education, Culture, Sports, Science and Technology, and the Japan Society for the Promotion of Science. A.P. is supported by The Wellcome Trust. M.X. thanks Oxford University (Clarendon Award), the U.K. Government (Overseas Research Student Award), and the E.P. Abraham Research Fund for support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.A.Y. is a guest editor invited by the Editorial Board.
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GPL5828 and GSE9036, GSE9055, and GSE15157).
This article contains supporting information online at www.pnas.org/cgi/content/full/0902573106/DCSupplemental.
References
- 1.Kornberg RD. The molecular basis of eukaryotic transcription. Proc Natl Acad Sci USA. 2007;104:12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 3.Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 4.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 5.Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 6.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura H, Sugaya K, Cook PR. The transcription cycle of RNA polymerase II in living cells. J Cell Biol. 2002;159:777–782. doi: 10.1083/jcb.200206019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, et al. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 14.Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT. Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol Cell. 2007;28:978–990. doi: 10.1016/j.molcel.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 15.Rafalska-Metcalf IU, Janicki SM. Show and tell: Visualizing gene expression in living cells. J Cell Sci. 2007;120:2301–2307. doi: 10.1242/jcs.008664. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann A, Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunol Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 17.Inoue K, Kobayashi M, Yano K, Miura M, Izumi A, et al. Histone deacetylase inhibitor reduces monocyte adhesion to endothelium through the suppression of vascular cell adhesion molecule-1 expression. Arterioscler Thromb Vasc Biol. 2006;26:2652–2659. doi: 10.1161/01.ATV.0000247247.89787.e7. [DOI] [PubMed] [Google Scholar]
- 18.Royce TE, Rozowsky JS, Bertone P, Samanta M, Stolc V, et al. Issues in the analysis of oligonucleotide tiling microarrays for transcript mapping. Trends Genet. 2005;21:466–475. doi: 10.1016/j.tig.2005.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 20.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 22.Xu M, Cook PR. The role of specialized transcription factories in chromosome pairing. Biochim Biophys Acta. 2008;1783:2155–2160. doi: 10.1016/j.bbamcr.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Tennyson CN, Klamut HJ, Worton RG. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat Genet. 1995;9:184–190. doi: 10.1038/ng0295-184. [DOI] [PubMed] [Google Scholar]
- 24.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tardiff DF, Lacadie SA, Rosbash M. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol Cell. 2006;24:917–929. doi: 10.1016/j.molcel.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das R, Dufu K, Romney B, Feldt M, Elenko M, et al. Functional coupling of RNAP II transcription to spliceosome assembly. Genes Dev. 2006;20:1100–1109. doi: 10.1101/gad.1397406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, et al. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, et al. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci USA. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.