Abstract
Consensus genetic linkage maps provide a genomic framework for quantitative trait loci identification, map-based cloning, assessment of genetic diversity, association mapping, and applied breeding in marker-assisted selection schemes. Among “orphan crops” with limited genomic resources such as cowpea [Vigna unguiculata (L.) Walp.] (2n = 2x = 22), the use of transcript-derived SNPs in genetic maps provides opportunities for automated genotyping and estimation of genome structure based on synteny analysis. Here, we report the development and validation of a high-throughput EST-derived SNP assay for cowpea, its application in consensus map building, and determination of synteny to reference genomes. SNP mining from 183,118 ESTs sequenced from 17 cDNA libraries yielded ≈10,000 high-confidence SNPs from which an Illumina 1,536-SNP GoldenGate genotyping array was developed and applied to 741 recombinant inbred lines from six mapping populations. Approximately 90% of the SNPs were technically successful, providing 1,375 dependable markers. Of these, 928 were incorporated into a consensus genetic map spanning 680 cM with 11 linkage groups and an average marker distance of 0.73 cM. Comparison of this cowpea genetic map to reference legumes, soybean (Glycine max) and Medicago truncatula, revealed extensive macrosynteny encompassing 85 and 82%, respectively, of the cowpea map. Regions of soybean genome duplication were evident relative to the simpler diploid cowpea. Comparison with Arabidopsis revealed extensive genomic rearrangement with some conserved microsynteny. These results support evolutionary closeness between cowpea and soybean and identify regions for synteny-based functional genomics studies in legumes.
Keywords: legume, diploid, recombinant inbred lines, segregating traits, 1536-plex genotyping
Recent progress in genome resource development for model and major crop plants has energized genetic research and fostered a surge of new initiatives in plant improvement. However, this activity has largely bypassed “orphan crops” such as cowpea [Vigna unguiculata (L.) Walp.] (2n = 2x = 22), which are crops of relevance to food security and income for subsistence farmers in developing countries (1). Despite the limited genome resources, access to most of the genes in these organisms can be gained through cDNA sequences, which represent expressed genes. Partial cDNA sequences, known as ESTs, when determined from multiple genotypes of a species, facilitate the identification of SNPs in protein-encoding genes and can be used in conjunction with mapping populations to generate genetic linkage maps that represent a gene-based framework of the genome.
Cowpea is a very important leguminous crop of the developing world. The crop is particularly important in sub-Saharan Africa, where >10 million hectares are cultivated in the semiarid Savanna and Sahelian zones of West and Central Africa (http://faostat.fao.org). Several parts of South America (particularly northeastern Brazil and Peru) and parts of south Asia (India, Myannmar), the Middle East, and the southern regions of North America are also important cowpea production regions (2). Cowpea is a particularly valuable component of low-input farming systems of resource-poor farmers because of its productivity and yield stability in the face of abiotic stress (drought, heat, low soil fertility), and the ability of the crop to enhance soil fertility for succeeding cereal or tuber crops grown in rotation (3). With its greater tolerance to heat, drought, and low soil fertility (4) and yet close evolutionary relatedness to other economically important grain legumes such as common bean (Phaseolus vulgaris) and soybean (Glycine max), cowpea can serve as a model species for crop adaptation to these stresses. Until recently only limited progress had been made in basic gene discovery and gene regulation in cowpea, with such information available for Vigna species being 2- to 5-fold less than for pea (Pisum sativum), common bean (Phaseolus vulgaris), and alfalfa (Medicago sativum) and >500-fold less than for soybean (G. max) (5).
Before this work, only cross-specific and low-density genetic linkage maps comprised mostly of anonymous markers were available for cowpea. To date, the most comprehensive genetic map of V. unguiculata consists of 11 linkage groups (LGs) spanning a total of 2,670 cM, with an average distance of ≈6 cM between markers. It includes 242 amplified fragment length polymorphism (AFLP) and 18 disease and pest resistance-related markers (6), plus 133 random amplified polymorphic DNA (RAPD), 39 restriction fragment length polymorphism (RFLP), and 25 AFLP markers from an earlier map (7), for a total of 441 markers. However, the cM distance covered by this map is three to four times greater than other published RFLP-, RAPD-, and AFLP-based cowpea linkage maps (7–9).
Here, we describe the development and implementation of high-throughput SNP genotyping in cowpea and its application to produce a high-density SNP consensus map based on genotyping 741 members of six recombinant inbred line (RIL) populations, which can be related readily to prior maps through shared markers. Synteny was investigated to G. max, a major crop of worldwide importance, and which outside of the genus Vigna and next to common bean (P. vulgaris) is the closest legume relative of cowpea (10). Synteny with M. truncatula and Arabidopsis thaliana was also surveyed.
Results
Identification of SNPs and Development of GoldenGate Assays.
Sequencing 11 cDNA libraries generated 141,453 ESTs (Table S1). In addition, 41,505 ESTs from two libraries produced in a Generation Challenge Program project led by the International Institute of Tropical Agriculture and 160 from other libraries were included to give a total of 183,118 ESTs, which were used to create a comprehensive EST sequence assembly. HarvEST:Cowpea (http://harvest.ucr.edu) provides this information, which is also summarized in Table S1. The total number of nucleotides in the consensus sequences from contigs containing at least four members was 11.3 Mbp. Taking 10,000 as the number of high-quality SNPs, the average SNP frequency was 1 per 1.13 kbp.
The 1,536-SNP GoldenGate assay (see Materials and Methods) was applied to 759 DNA samples from mapping parents, RILs, and synthetic heterozygotes. Of these, 1,375 SNPs had satisfactory technical performance, an 89.55% success rate. Genotypes used for the 11 cDNA libraries (Table 1) contained no heterozygous loci.
Table 1.
Summary of genotypes used for SNP discovery
| Genotype | Other identifier | Origin | Type | Key traits |
|---|---|---|---|---|
| CB27 | H9–8–27 | UCR | Cultivar, MP | Heat tolerance, Fusarium wilt & root-knot nematode resistance |
| IT84S-2049 | UCR 430 | IITA | Breeding line MP | Root-knot nematodes resistance |
| 524B | UCR | Breeding line, MP | Large seed, Fusarium wilt resistance | |
| UCR 5301 | TVNu-463 | Botswana | Wild ssp. dekindtiana | Genetic diversity |
| UCR 779 | Botswana 19A | Botswana | Landrace | Cowpea aphid resistance |
| IT97K-461–4 | IITA | Breeding line | Striga gesnerioides resistance | |
| UCR 707 | Ex-Luanda | Kenya | Landrace | Genetic diversity |
| PI 418979 | China | Landrace | Asparagus bean | |
| UCR 41 | TVu-1996 | Nicaragua | Landrace | Genetic diversity |
| UCR 2563 | TVu-1522 | Iran | Landrace | Genetic diversity |
| Dan Ila | IITA | Breeding line, MP | Drought tolerance | |
| TVu 11986 | IITA | Breeding line, MP | Drought tolerance | |
| TVu 7778 | IITA | Breeding line, MP | Drought susceptible | |
| 12008D | ILRI | Landrace | Animal feed, drought tolerance | |
| 1393–2-1 | UCR | Breeding line | Chilling tolerance, large seed |
Three additional libraries contributed ESTs but were not used for SNPs because trace files were not available. MP, mapping parent for RILs used in this work or otherwise. University of California, Riverside; IITA, International Institute of Tropical Agriculture; ILRI, International Livestock Research Institute.
Polymorphism and Segregation Distortion.
A total of 986 of the 1,375 SNPs (72%) were polymorphic in at least one RIL population. In total, 410 markers exhibited segregation distortion in one or more populations; however, 360 of these had minor allele frequencies (MAFs) higher than the 0.30 threshold in at least one population and could be mapped. Of the remaining 50 with low MAF, 39 were polymorphic in only one population, and 11 were polymorphic in multiple populations but with low MAF in each case. SNPs with a low MAF (< 0.30) in each population ranged from 33 for RIL 524B × IT84S-2049 to 236 for RIL Dan Ila × TVu-7778 (Table 3). Six SNPs were discarded because they had GoldenGate Assay “no-call” frequencies >5% in the population in which they were segregating, and two SNPs were eliminated after they mapped in two different LGs.
Table 3.
Population size, SNP polymorphisms, map characteristics, and traits for RIL populations used for consensus mapping
| Population | RILs | Polymorphic SNPs | SNPs with MAF < 0.3 | Mapped SNPs | Map size, cM | Largest gap between SNPs, cM | Traits segregating |
|---|---|---|---|---|---|---|---|
| 524B × IT84S-2049 | 79 | 469 | 33 | 436 | 665 | 13.3 | AR, IGW, NR, SR, VR, SCT |
| CB27 × 24–125B-1 | 90 | 366 | 67 | 299 | 651 | 17.0 | AR, IGW, CWR, NR, M, FLTR, |
| CB46 × IT93K-503–1 | 103 | 422 | 34 | 388 | 601 | 17.1 | NR, IGW, DT, FLTR, M, MR, Y, AR, FR |
| Dan Ila × TVu-7778 | 109 | 524 | 236 | 288 | 665 | 16.3 | DT, Y, BB |
| TVu-14676 × IT84S-2246–4 | 137 | 409 | 60 | 349 | 600 | 17.9 | SR, NR |
| Yacine × 58–77 | 114 | 499 | 84 | 415 | 657 | 17.6 | FLTR, IGW, |
AR, aphid resistance; BB, bacterial blight resistance; CWR, cowpea weevil resistance; DT, drought tolerance; FR, Fusarium resistance; FLTR, flower thrips resistance; IGW, individual grain weight; M, maturity; MR, Macrophomina resistance; NR, nematode resistance; SR, Striga resistance; SCT, seedling cold tolerance; VR, virus resistance; Y, yield.
A total of 58 SNPs, mainly from the 3 × 3 list (see Materials and Methods) or dependent on cowpea genotype UCR779 in the pairwise list, could not be mapped in any population. As a net result, 928 SNPs that had MAF at least 0.3 in one or more mapping populations were used in map construction. In pairwise comparisons, between 99 and 202 SNPs were shared by any two mapping populations. The numbers of SNPs shared between populations are summarized in Table 2.
Table 2.
Pairwise comparison of common SNP markers across cowpea RIL populations
| RIL population | 524B × IT84S-2049 | CB46 × IT93K-503–1 | TVu-14676 × IT84S-2246–4 | CB27 × 24–125B-1 | Dan Ila × TVu-7778 |
|---|---|---|---|---|---|
| Yacine × 58–77 | 168 | 162 | 161 | 119 | 123 |
| Dan Ila × TVu-7778 | 147 | 130 | 99 | 130 | |
| CB27 × 24–125B-1 | 156 | 154 | 108 | ||
| TVu14676 × IT84S-2246–4 | 137 | 134 | |||
| CB46 × IT93K-503–1 | 202 |
Individual Genetic Linkage Maps.
Elimination of RILs with nonparental alleles, excessive heterozygosity, and excessive no-calls (see Materials and Methods) resulted in the final mapping population sizes given in Table 3. Therefore, a total of 632 RILs were used in individual and consensus map building. For each of the six mapping populations, LGs were resolved without conflicting marker assignments by using JoinMap 3.0 (11) and the parameters described in Materials and Methods. The stringent mapping parameters (see Materials and Methods) adopted for individual map construction resulted in the number of LGs ranging from the expected 11 in TVu-14676 × IT84S-2246–4 to 15 in 524B × IT84S-2049. Although the additional smaller LGs could be consolidated into larger LGs using lower logarithm of odds (LOD) thresholds, this separation was tolerated before consensus mapping to minimize spurious linkage that could result in tangled consensus LGs. Maps of the six populations ranged from 600 to 665 cM and consisted of 288–436 SNP markers. The largest gap between mapped SNPs ranged from 13.3 to 17.9 cM for individual maps (Table 3). Average marker distances ranged from 2.31 cM in Dan Ila × TVu-7778 to 1.52 cM in 524B × IT84S-2049.
Consensus Genetic Linkage Map.
Because of the high prevalence of segregation distortion in the Dan Ila × TVu-7778 population, a framework consensus map was generated by using five populations. The Dan Ila × TVu-7778 population was added while making sure that no LG or marker order conflicts were introduced.
Within-LG marker assignment was attempted first by using JoinMap; however, major reshuffling of marker order was observed compared with individual maps. Previous studies have encountered the same limitation when JoinMap was used to construct high-density consensus maps (12, 13). Therefore, in our mapping protocol, homologous LGs were used to generate consensus LGs one at a time with MergeMap (12) to establish marker order (see Materials and Methods). Then map distances were added by using the cM values generated by JoinMap for each LG. The resulting consensus map contained 928 SNP markers on 619 unique map positions distributed over 11 LGs, covering a total genetic distance of 680 cM. This is an average marker distance of 0.73 cM, or one SNP per 668 kbp considering the cowpea genome to be 620 Mbp. Coincidentally, the map contains almost exactly an average of 1.0 map positions per Mbp (619 map positions/620 Mbp). The average distance between unique map positions was 1.09 cM (680 cM/619 map positions).
Marker density was generally consistent throughout the map with only 19 instances where distances between adjacent markers were >4 cM and only one region on VuLG1 where the distance was >10 cM. VuLG1 also had the least marker density with an average distance of 1.23 cM, whereas VuLG3 had the highest marker density with an average distance of 0.49 cM between markers. The remaining LGs had average marker distances <1.0 ranging from 0.60 to 0.98 cM. LG sizes ranged from 44.75 cM for the smallest to 85.24 cM for the largest. Number of markers, average marker distance, and corresponding LGs from previous publications are summarized in Table 4 and marker distribution across LGs is shown in Fig. S1.
Table 4.
Consensus genetic map, correspondence of LGs to previous maps, and synteny with soybean (GmChr) and Medicago (MtChr) highlighted by number of cowpea orthologs in parentheses
| Consensus VuLG | Markers | Length | Ouédraogo et al. (6) LG | Muchero et al. (8) LG | GmChr (Cowpea orthologs) | MtChr (Cowpea orthologs) |
|---|---|---|---|---|---|---|
| 1 | 69 | 85.2 | 4 | 6 | 18(27), 9(10), 13(9), 8(8), 15(5) | 7(25), 2(12), 3(7), 4(7) |
| 2 | 116 | 84.0 | 4, 11 | 5, 9 | 10(43), 20(39), 2(13) | 1(76), 7(15), 4(12), 5(11), 2(5) |
| 3 | 168 | 81.8 | 3 | 5, 10 | 5(43), 8(39), 17(33), 7(15), 13(14), 16(5) | 4(41), 8(36), 5(23), 2(20), 3(12), 6(6) |
| 4 | 68 | 66.4 | 9 | 7, 8 | 19(19), 3(13), 11(9), 18(9), 16(5) | 3(22), 7(22), 5(5) |
| 5 | 75 | 62.6 | 5 | 11 | 14(37), 2(15), 17(9) | 5(38), 1(14), 3(9) |
| 6 | 93 | 59.1 | 8 | 3 | 15(32), 8(15), 9(10), 13(8), 18(9), 19(6) | 2(37), 3(11), 5(11), 7(7), 1(6), 4(6), 8(6) |
| 7 | 72 | 52.9 | 5 | 1 | 11(21), 1(20), 2(9), 9(9) | 5(42), 1(6), 8(6), 7(5) |
| 8 | 65 | 49.0 | 7, 10 | 2 | 6(27), 4(26), 15(5) | 3(29), 8(9), 4(7) |
| 9 | 66 | 48.4 | 1 | 1 | 12(30), 11(11), 15(9), 6(7) | 4(17), 2(15), 8(8), 3(7) |
| 10 | 77 | 46.0 | 6 | 2, 3 | 7(23), 3(20), 1(10), 16(10), 8(6) | 8(21), 4(14), 7(12), 5(8), 2(7) |
| 11 | 59 | 44.8 | 2 | 2, 3 | 16(15), 13(10), 19(9), 9(8), 2(5) | 6(12), 4(11), 5(6), 1(5), 7(5) |
Of the 928 SNP-harboring cowpea unigenes, 921were annotated based on sequence homology with soybean at e-scores of 1.00e-10 or better. The annotation information for the mapped SNPs is available from HarvEST:Cowpea (http://harvest.ucr.edu and www.harvest-web.org). The HarvEST BLAST server (http://138.23.191.145/blast/index.html) provides the mapped SNP unigenes as a searchable database.
Synteny with Soybean.
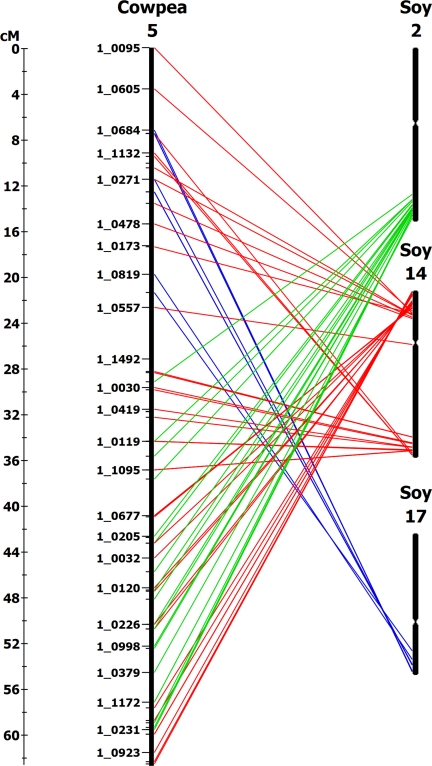
Extensive macrosynteny and microsynteny were observed between cowpea and soybean. Only 7 of the 928 genes placed on the cowpea consensus map did not have a soybean hit of e-10 or better. Of the remaining 921 genes, 789 were in regions highly syntenic and collinear with soybean chromosomal (GmChr) regions, which represents ≈85% of the cowpea genome covered by the current map. The ranked order of syntenic regions of soybean is included in Table 4. The number of major soybean synteny blocks for each cowpea LG ranged from one to three, whereas the total number of significant soybean synteny blocks ranged from three to six. Seven of 11 LGs had major synteny with two soybean chromosomes, among which four (VuLG2, VuLG3, VuLG5, and VuLG7) had synteny along the entire LG. For example, VuLG5 was completely syntenic with soybean 14 as shown in Fig. 1. An additional example is shown in Fig. S2.
Fig. 1.
Macrosynteny and collinearity between cowpea LG VuLG5 and soybean chromosomes GmChr2, GmChr14, and GmChr17.
Synteny with M. truncatula.
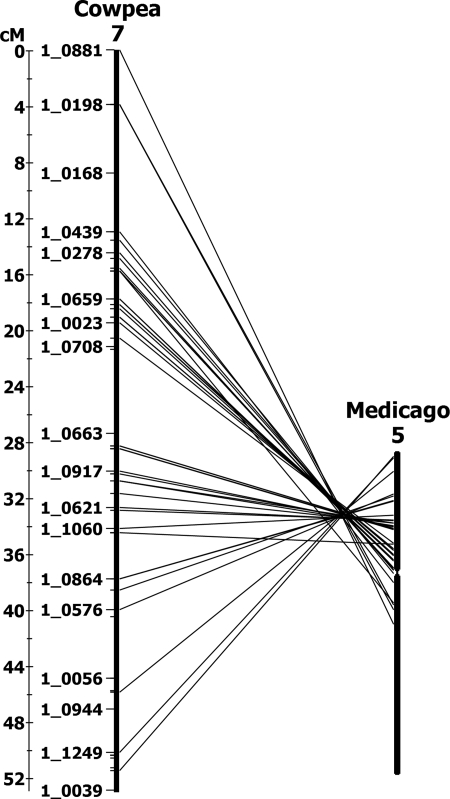
As expected, synteny was reduced in M. truncatula compared with soybean. However, regions with extensive macrosynteny, microsynteny, and collinearity were observed. Of the 928 EST-derived SNPs mapped in the cowpea consensus map, 809 had significant M. truncatula hits, with 759 (82%) in regions defined by synteny. Extensive chromosomal rearrangement was evident, with cowpea LGs typically chimeric to three or more M. truncatula chromosomal segments (Table 4). Regardless, blocks of extensive synteny and collinearity were evident, because 10 of the 11 cowpea LGs had at least 17 corresponding loci on a Medicago chromosome (Table 4). Specifically, the entire VuLG7 exhibited extensive synteny and collinearity with a section of MtChr5 (Fig. 2). Similarly, VuLG2 had extensive macrosynteny with MtChr1 where 76 of 116 cowpea loci had corresponding orthologs on MtChr1 (Table 4). Also, one block of VuLG3 was syntenic and collinear with the lower section of MtChr4.
Fig. 2.
Macrosynteny and collinearity between cowpea linkage VuLG7 and M. truncatula chromosomes MtChr6.
Synteny with Arabidopsis.
Major chromosomal rearrangement was observed between cowpea and Arabidopsis such that no macrosynteny was evident. Significant microsynteny was observed in some regions, but collinearity was markedly reduced relative to legume reference genomes. The strongest instance of cowpea–Arabidopsis microsynteny and collinearity was an ≈14-cM section of VuLG1 and AtChr1 where gene order exhibited only minor differences.
Discussion
In this study, the high-throughput Illumina GoldenGate SNP assay was tested and validated for cowpea by using six RIL mapping populations. The effectiveness and utility of this approach in cowpea was demonstrated by the successful genotyping of 1,375 of 1,536 SNP loci (89.55%) among 741 RILs derived from six crosses, synthetic heterozygotes, and parental genotypes. Further, the quality of the data was highlighted by the incorporation of 928 (67%) of these 1,375 markers into a consensus genetic map. Given that this oligonucleotide pool assay was developed for cowpea and none of the SNPs were validated in practice before this work to our knowledge, the 90% success rate supports the efficacy of strategies adopted in developing EST-derived SNPs from cowpea.
The gene-based consensus map described here will enable integration of information from populations developed from diverse crosses, enable whole genome molecular marker-assisted selection, and facilitate linkage disequilibrium studies, association mapping, and synteny-based genomics in cowpea. All of these possibilities should benefit from the dense marker placement (0.73 cM/marker) in the current map. Significantly, almost all syntenic blocks were in subtelomeric regions of soybean chromosomes, suggesting that these are gene-rich and that the heterochromatic pericentric regions are not densely populated with markers in the cowpea map. Although LGs <50 cM for VuLG8, VuLG9, VuLG10, and VuLG11 might suggest incomplete maps, this observation could result from high within-LG marker ordering LODs. All four LGs had lengths >50 cM using LODs between 1 and 3. To minimize spurious linkages, we used LODs 7 or higher. The map size reported here was consistent with previously published map sizes of 972 cM (7), 643 cM (8), and 669.8 cM (9). However, the Ouédraogo et al. (6) map was ≈3–4 times longer with a total distance of 2,670 cM. The same mapping population (524B × IT84S-2049) used to construct that map was used in the current study, and its individual SNP-based map size was 665 cM, well within the expected range, based on the average observed across the individual maps.
EST-derived SNP markers provide an additional level of utility in genomic studies because they tag actual genes that, when incorporated into genetic maps, may be used for synteny-based cross-species genome comparison. Soybean genome duplication was evident because a single cowpea haplotype was typically syntenic with regions on at least two different soybean chromosomes. Further, chromosomal rearrangement was apparent between the two species, because each cowpea LG was syntenic to regions on at least three and up to six soybean chromosomes. Although genome duplication in soybean will present a challenge in translational genomics with cowpea, extensive macrosynteny and collinearity in duplicated regions compared with cowpea should facilitate such efforts. The observation that 789 of the 928 EST-derived SNP markers mapped in regions exhibiting synteny and conserved collinearity suggests that ≈85% of the cowpea genome covered by this map has strong correspondence to at least one soybean genomic region. Therefore, studies in cowpea should benefit from the extensive soybean genome sequence resource in identifying genetic determinants of traits of economic importance. Similarly, because cowpea is one of the most productive crops in hot, low rainfall, and poor soil environments (2, 4), studies in soybean and common bean should benefit from validation of trait determinants in the relatively simple and nonduplicated genome of cowpea. With exception of the complexity resulting from genome duplication, similar conclusions can be drawn about synteny with M. truncatula that, despite significant chromosomal rearrangement between the two species, was extensive enough to result in 82% of the mapped cowpea genes lying in regions defined by synteny. The extensive synteny and collinearity observed between the soybean genome and the current cowpea map provides additional support for the mapping accuracy of gene-derived SNP markers within the consensus map.
The utility of the nonlegume Arabidopsis genome sequences in cowpea studies seems likely to be bottlenecked by extensive chromosomal rearrangement that results in only short segments of the two genomes having synteny. This limitation notwithstanding, the substantial residual microsynteny between the cowpea genome and this well-studied genome should provide an additional resource to complement the extensive synteny with soybean and M. truncatula.
Materials and Methods
Plant Material for cDNA Libraries.
A diverse set of 10 cowpea genotypes chosen to represent cowpea diversity worldwide was used to isolate RNA for cDNA library construction and EST sequencing (Table S1). Genotype designation, country of origin, type (e.g., landrace or improved cultivar), and important traits are summarized in Table 1. Two rounds of inbreeding by single-seed descent in greenhouses were carried out immediately before library construction to reduce or eliminate any residual heterozygosity that may have been present. Details of tissue types, growth conditions, and the RNA extraction protocol are given in SI Text.
cDNA Libraries and Sequencing.
Nine cDNA libraries were constructed from 9 of the genotypes listed in Table S1 by using the SuperScript Plasmid System with Gateway Technology for cDNA Synthesis and Cloning (Invitrogen) and the pCMV·SPORT6 vector for libraries UCRVU04 to UCRVU12. Libraries UCRVU02 and UCRVU03 were constructed by using the λZAP cDNA Synthesis Kit (Stratagene) and a Uni-ZAP XR vector. All libraries were sequenced by using the Sanger dideoxy chain termination method. Details of cDNA construction and sequencing are included in SI Text.
Description of Assembly P12.
ESTs from the above 11 and 6 existing cowpea cDNA libraries, a total of 17 libraries, were included in an assembly that we refer to as assembly P12, produced by using CAP3 (14) with parameter settings p = 75, d = 200, f = 250, h = 90, −t = 1,200. These additional six libraries are described in refs. 15–17 and http://lifesciencedb.jp/ddbj/ff_view.cgi?accession=FF557182. The ESTs included in assembly P12 covered a diverse range of tissues (seed, root, primary root, root hair, nodule, hypocotyl, epicotyl, young trifoliate leaf, leaf, auxiliary bud, and shoot meristem) and growth conditions (unstressed, drought-stressed).
SNP Identification.
SNPs were identified by using two methods. One method began with 45 pairwise genotype comparisons of ESTs from eight cultivated cowpea genotypes (524B, IT84S-2049, IT97K-461–4, PI 418979, UCR 41, UCR 707, UCR 779, UCR 2563), one wild cowpea genotype (UCR 5301), and mixed-genotype ESTs treated as a single genotype. For pairwise genotype comparisons, a SNP was accepted only if there were at least two sequences from each genotype and agreement between opposite strands from a single clone. For all SNP-finding methods, a base call was used only if its Phred quality value was at least 25 and the position was at least 25 bases from the end of an EST sequence and not inside a window of five bases containing three or more Phred values <25. This method yielded 5,101 SNPs. The second method of SNP detection ignored the cowpea genotype and equally considered each EST from all 14 libraries from which trace files were available. Two of these 14 libraries each contained a mixture of ESTs from four cowpea genotypes (Dan Ila, TVu-11986, TVu-7778, 12008D) (http://lifesciencedb.jp/ddbj/ff_view.cgi?accession=FF557182). Using this latter method, two lists of SNPs were generated, one more restrictive requiring three examples of each allele (3 × 3 list) and the other requiring only two examples of each allele (2 × 2 list). The 3 × 3 list contained 3,175 SNPs, of which 2,392 were also in the pairwise list. The union of the pairwise and 3 × 3 lists contained 5,884 SNPs. The 2 × 2 list contained 12,733 SNPs, of which 7,037 were unique to this list. The total number of SNPs in all three lists was therefore 12,921, but because the 2 × 2 list is a less reliable source of SNPs we refer to the total number of high confidence SNPs as ≈10,000. HarvEST:Cowpea contains a database running in FoxPro, so the SNP finding algorithm was encoded as a program compatible with the FoxPro environment (http://harvest.ucr.edu).
SNP Selection for the Illumina GoldenGate Assay.
SNPs were selected for an Illumina GoldenGate assay only from the 5,884 SNPs in the union of the pairwise and 3 × 3 lists described above. A SNP was eliminated from further consideration if it was within 30 bases of an intron (deduced by alignment with the soybean genome sequence version 0.1B) or within 49 bases of the end of the consensus sequence. Other filters eliminated SNPs with more than two alternative nucleotides or an Illumina SNP score <0.4. After applying these filters, there were 4,118 remaining SNPs contained in 2,102 unique assembly P12 unigenes, which matched 1,948 unique soybean gene models. With the limitation to only one SNP related to each soybean gene model, prioritization of SNPs for inclusion in the 1,536-SNP cowpea GoldenGate assay from the 1,948 is described in detail in SI Text and Dataset 1.
SNP Annotations.
HarvEST:Cowpea contains annotations of the unigenes in assembly P12. Annotations include the best BLAST hits to soybean Glyma 1 (ftp://ftp.jgi-psf.org/pub/JGI_data/Glycine_max/Glyma1/), Medicago truncatula MT 2 (www.medicago.org/genome/downloads/Mt2), Arabidopsis TAIR 8 (ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR8_genome_release/), UniProt UniRef-90 Aug 16, 2008 (ftp://ftp.uniprot.org/pub/databases/uniprot/uniref/uniref90/uniref90.fasta.gz), and the position on the soybean, Medicago, and Arabidopsis chromosomes.
DNA Sources.
Parental genotypes and RILs from six mapping populations (Table 2) were genotyped for each of the 1,536 SNPs by using the GoldenGate assay. DNA isolation and preparation for genotyping are described in SI Text.
Data Processing.
Raw data from the GoldenGate assay were transformed to genotype calls initially by using Illumina BeadStudio 3.2 software with the genotyping module. Data from all samples were viewed to manually set 1,536 archetypal clustering patterns. The cluster positioning was guided by “synthetic heterozygotes” of each RIL population made by mixing parental DNAs in a 1:1 mass ratio. These samples assisted with the identification of true heterozygotes, which are expected to occur in F9–F10 RILs at a frequency of ≈0.1–0.2%. Customized workspaces were produced for each mapping population to further optimize genotype calls by using minor adjustments of the cluster positions. Fig. S3 illustrates typical workspace scenarios and the respective decisions taken. The no-call threshold was set to 0.15, which in some cases necessitated a manual override of the genotyping call exported from the BeadStudio software; these cases were limited to those that were plainly evident by eye and not in conflict in the final genetic map. Genotype calls were exported as spreadsheets from the BeadStudio 3.2 software.
Data processing before mapping excluded SNPs that had poor technical performance in the GoldenGate assay and SNPs with a MAF <0.30 (see SI Text). In addition, RILs with excessive heterozygosity, nonparental alleles, and no-calls were excluded from further analysis. An acceptable standard for each of these parameters for each mapping population was determined empirically by visual inspection of the distribution of the above quality metrics and looking for obvious break points. In addition, individual SNP markers with no-calls >5% were discarded from individual mapping datasets, even if they passed the initial SNP technical quality assessment.
Individual Maps and Consensus Map Construction.
Six F2-derived RIL populations developed by inbreeding and single-seed descent were used in this study. These were selected based on population robustness, parental diversity, and segregation of economically important agronomic traits (Table 2). Individual maps were constructed by using JoinMap 3.0 (11) at LOD grouping thresholds between 4 and 8. Within-LG marker ordering was done by using mapping LODs of 7 or higher because at lower LODs marker order shifted significantly and there was a marked decline in LG cM lengths that stabilized at higher LODs. The Kosambi mapping function (19) was used to convert recombination frequencies to cM, and the consensus map was constructed with MergeMap (12) with individual map inputs from JoinMap. MapChart 2.2 (19) was used for graphical representation of the consensus map. Comparison with previous maps (6, 8) was facilitated by constructing individual maps incorporating current SNPs with AFLP, RFLP, and RAPD markers from the previous maps. This comparison allowed SNP markers to be used as anchors in indentifying homologous LGs.
Synteny.
Synteny between cowpea and G. max, M. truncatula, or Arabidopsis was determined based on sequence comparison between cowpea unigenes harboring a mapped SNP and reference genome sequences (see SNP Annotations) using BLASTX (cut-off value of e-10) against translated gene models. Map coordinates of the cowpea unigenes were compared with the chromosomal positions of the corresponding genes with the highest homology in the reference genome. Synteny was visualized by using HarvEST:Cowpea (http://harvest.ucr.edu or www.harvest-web.org).
Supplementary Material
Acknowledgments.
We thank E. Lindquist, C. Pennacchio, S. Lucas, M. Wang, and J. Bristow of the U.S. Department of Energy Joint Genome Institute (Walnut Creek, CA) for EST sequencing of libraries UCRVU04 through UCRVU12; M. Timko (University of Virginia, Charlottesville) for providing seeds of the Dan Ila × TVu-7778 and IT84S-2246 × TVu-14676 RIL populations; C. Sudhakar (Sri Krishnadevaraya University, Anantapur, India) for production of one of the cDNA libraries; and A. Farmer (National Center for Genome Resources, Santa Fe, NM) for a list of soybean gene models related to a National Science Foundation project led by D. Cook (University of California, Davis). This work was supported by the Generation Challenge Program through a grant from the Bill and Melinda Gates Foundation and U.S. Agency for International Development Collaborative Research Support Program Grants GDG-G-00-02-00012-00 and EDH-A-00-07-00005.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905886106/DCSupplemental.
References
- 1.Delmer DP. Agriculture in the developing world: Connecting innovations in plant research to downstream applications. Proc Natl Acad Sci USA. 2005;102:15739–15746. doi: 10.1073/pnas.0505895102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehlers JD, Hall AE. Cowpea (Vigna unguiculata L. Walp) Field Crops Res. 1997;53:187–204. [Google Scholar]
- 3.Sanginga N, et al. Sustainable resource management coupled to resilient germplasm to provide new intensive cereal–grain–legume–livestock systems in the dry savanna. Agric Ecosys Environ. 2003;100:305–314. [Google Scholar]
- 4.Hall AE. Breeding for adaptation to drought and heat in cowpea. Eur J Agron. 2004;21:447–454. [Google Scholar]
- 5.Timko M, et al. Sequencing and analysis of the gene-rich space of cowpea. BMC Genomics. 2008;9:103. doi: 10.1186/1471-2164-9-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ouédraogo JT, et al. An improved genetic linkage map for cowpea (Vigna unguiculata L.) Combining AFLP, RFLP, RAPD, biochemical markers, and biological resistance traits. Genome. 2002;45:175–188. doi: 10.1139/g01-102. [DOI] [PubMed] [Google Scholar]
- 7.Menéndez CM, Hall AE, Gepts P. A genetic linkage map of cowpea (Vigna unguiculata) developed from a cross between two inbred, domesticated lines. Theor Appl Genet. 1997;95:1210–1217. [Google Scholar]
- 8.Muchero W, Ehlers J, Close T, Roberts P. Mapping QTL for drought stress-induced premature senescence and maturity in cowpea [Vigna unguiculata (L.) Walp.] Theor Appl Genet. 2009;118:849–863. doi: 10.1007/s00122-008-0944-7. [DOI] [PubMed] [Google Scholar]
- 9.Ubi BE, Mignouna H, Thottappilly G. Construction of a genetic linkage map and QTL analysis using a recombinant inbred population derived from an intersubspecic cross of cowpea [Vigna unguiculata (L.) Walp.] Breed Sci. 2000;50:161–172. [Google Scholar]
- 10.Young ND, Mudge J, Ellis TN. Legume genomes: More than peas in a pod. Curr Opin Plant Biol. 2003;6:199–204. doi: 10.1016/s1369-5266(03)00006-2. [DOI] [PubMed] [Google Scholar]
- 11.Van Ooijen JW, Voorrips RE. JoinMap 3.0: Software for the Calculation of Genetic Linkage Maps. Wageningen, The Netherlands: Plant Research International; 2001. [Google Scholar]
- 12.Wu Y, Close TJ, Lonardi S. In: Computational Systems Bioinformatics: CSB 2008 Conference. Markstein P, Xu Y, editors. Stanford, CA: Imperial College Press; 2008. pp. 285–296. [Google Scholar]
- 13.Yap IV, et al. A graph-theoretical approach to comparing and integrating genetic, physical, and sequence-based maps. Genetics. 2003;165:2235–2247. doi: 10.1093/genetics/165.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ismail AM, Hall AE, Close TJ. Allelic variation of a dehydrin gene cosegregates with chilling tolerance during seedling emergence. Proc Natl Acad Sci USA. 1999;96:13566–13570. doi: 10.1073/pnas.96.23.13566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mould MJR, Xu T, Barbara M, Iscove NN, Heath MLC. cDNAS generated from individual epidermal cells reveal that differential gene expression predicting subsequent resistance or susceptibility to rust fungal infection occurs prior to the fungus entering the cell lumen. Mol Plant-Microbe Interact. 2003;16:835–845. doi: 10.1094/MPMI.2003.16.9.835. [DOI] [PubMed] [Google Scholar]
- 17.Simões-Araújo JL, et al. Identification of differentially expressed genes by cDNA-AFLP technique during heat stress in cowpea nodules. FEBS Lett. 2002;515:44–50. doi: 10.1016/s0014-5793(02)02416-x. [DOI] [PubMed] [Google Scholar]
- 18.Kosambi DD. The estimation of map distance from recombination values. Ann Eugen. 1944;12:172–175. [Google Scholar]
- 19.Voorrips RE. MapChart: Software for the graphical presentation of linkage maps and QTLs. J Heredity. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.