Abstract
Statins are compounds prescribed to lower blood cholesterol in millions of patients worldwide. They act by inhibiting HMG-CoA reductase, the rate-limiting enzyme in the mevalonate pathway that leads to the synthesis of farnesyl pyrophosphate, a precursor for cholesterol synthesis and the source of lipid moieties for protein prenylation. The nematode Caenorhabditis elegans possesses a mevalonate pathway that lacks the branch leading to cholesterol synthesis, and thus represents an ideal organism to specifically study the noncholesterol roles of the pathway. Inhibiting HMG-CoA reductase in C. elegans using statins or RNAi leads to developmental arrest and loss of membrane association of a GFP-based prenylation reporter. The unfolded protein response (UPR) is also strongly activated, suggesting that impaired prenylation of small GTPases leads to the accumulation of unfolded proteins and ER stress. UPR induction was also observed upon pharmacological inhibition of farnesyl transferases or RNAi inhibition of a specific isoprenoid transferase (M57.2) and found to be dependent on both ire-1 and xbp-1 but not on pek-1 or atf-6, which are all known regulators of the UPR. The lipid stores and fatty acid composition were unaffected in statin-treated worms, even though they showed reduced staining with Nile red. We conclude that inhibitors of HMG-CoA reductase or of farnesyl transferases induce the UPR by inhibiting the prenylation of M57.2 substrates, resulting in developmental arrest in C. elegans. These results provide a mechanism for the pleiotropic effects of statins and suggest that statins could be used clinically where UPR activation may be of therapeutic benefit.
Keywords: small GTPase, unfolded protein response (UPR), HMG-CoA reductase
Statins are widely prescribed cholesterol-lowering drugs that inhibit 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) reductase, the enzyme responsible for the rate-limiting step in the mevalonate pathway (Fig. S1). This pathway splits into two branches: one leading to cholesterol synthesis and the other to the synthesis of farnesyl pyrophosphate, the source of the lipid moiety that is attached to proteins upon prenylation (1). While the effects of statins on plasma lipid levels are well described, the actual mechanisms by which these and other effects are obtained are not always clear (2). Statins have pleiotropic effects at the physiological level (e.g., depression, insomnia, neuropathy, anti-inflammatory, and antiatherogenic effects) as well as at the cellular (e.g., interference with lysosomal functions) and biochemical levels (e.g., reduced isoprenylation of proteins and inhibition of GTPases) (3–6). Furthermore, statins are increasingly being considered as anti-cancer drugs because of their effects on the RAS superfamily of GTPases (1) and have recently proven protective against heart failure in patients where cholesterol levels are not elevated (7).
The connection between statins and RAS-type GTPases provides the best explanation for the noncholesterol effects of statins. Following their synthesis, RAS-type GTPases, including RAS, RHO, RHEB, and RAB GTPases, must be lipid-modified to become membrane-associated, itself a requirement for their activity. Typically, the lipid moiety is either a farnesyl group (for RAS GTPases) or a geranylgeranyl group (for RHO and RAB GTPases) (1, 8, 9). In either case, the modification is accomplished by specialized prenyltransferases, i.e., farnesyl transferases or geranylgeranyl transferases, that typically recognize a CAAX motif at the C terminus of their target GTPases (C stands for a cysteine, A for an aliphatic amino acid, and X for any amino acid). Since farnesyl-pyrophosphate and geranylgeranyl-pyrophosphate are both produced from the HMG-CoA reductase-dependent mevalonate pathway, statins have strong effects on GTPase prenylation, and hence are promising therapeutics for controlling tumors in which GTPase oncogenes are activated. Given the varied and important role of RAS-type GTPases, including organelle biogenesis and homeostasis, it is perhaps not surprising that their inhibition would activate cellular stress responses. For example, the unfolded protein response (UPR) is activated when misfolded proteins accumulate in the endoplasmic reticulum (ER), as would be expected when processes such as ER homeostasis or vesicle trafficking are impaired (10–12). Consistently, a recent study showed that statin treatment causes the induction of the UPR in cultured macrophages (13).
Studies on the cellular effects of statins are typically confounded by the dual roles of the mevalonate pathway: production of cholesterol on the one hand and production of lipid moieties to be used in protein prenylation on the other. The availability of an experimental system that could allow to specifically study the noncholesterol effects of statins would therefore be a definite boon for the field. Much of the mevalonate pathway is preserved in the free-living nematode Caenorhabditis elegans, but this organism lacks the branch of the pathway that leads to cholesterol synthesis: worms are unable to synthesize cholesterol and therefore need to have it supplied exogenously. C. elegans may therefore be an ideal model to elucidate the effects of statins that are not related to cholesterol biosynthesis, i.e., the effects on the branch of the mevalonate pathway that leads to farnesyl pyrophosphate and protein prenylation. To better understand the noncholesterol activities of statins, we have examined their effects on C. elegans and found that they cause growth arrest, reduced Nile red staining without affecting lipid stores, and induction of the UPR.
Results
Phenotypic Effects of Statin on C. elegans.
Previous publications reporting on high-throughput studies in C. elegans have shown that inhibition of HMG-CoA reductase using RNAi causes sterility and a reduction in Nile red staining in adults, as well as embryonic/larval developmental arrest (14–20). These effects were also obtained when growing C. elegans in the presence of statins; incubation of L1 larvae on fluvastatin at concentrations of 0.125 mM or higher caused larval developmental arrest (Fig. 1 A–C), while incubation of L4 larvae caused adult sterility. Furthermore, embryos from adults grown overnight on 1 mM fluvastatin arrested early during development. These effects were highly reproducible, affected 100% of the treated animals, and were fully rescued by supplying mevalonate exogenously at concentrations above 1.25 mM (Fig. 1 D–F). Similar results were also obtained with two other statins, namely atorvastatin and lovastatin, when used at 1 mM.
Fig. 1.
Fluvastatin causes developmental arrest and decreased Nile red staining, but only the development is rescued by mevalonate. Eggs were placed on plates containing various concentrations of fluvastatin: (A) 0 mM; (B) 0.125 mM; (C) 1 mM; or 1 mM fluvastatin plus various concentrations of mevalonate: (D) 10 mM; (E) 1.25 mM; (F) 0.156 mM; then allowed to develop and grow for 72 h, at which time they were photographed. Nile red staining in a control worm (G), in a worm grown overnight on 1 mM fluvastatin (H), or on 1 mM fluvastatin and 10 mM mevalonate (I).
Statins Inhibit Protein Prenylation.
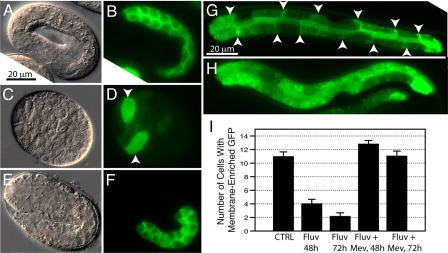
To verify that inhibition of the mevalonate pathway causes the expected reduction in protein prenylation, we used the plasmid pGLO-1P::GFP-CAAX, which expresses a green fluorescent protein (GFP) modified to carry the C terminus of the C. elegans ras-2 protein and expressed in the intestine from the glo-1 promoter. Control transgenic embryos or larvae carrying this reporter show enrichment of GFP labeling on the membranes of intestinal cells (Fig. 2A, B, and G). When young adults are placed on 1 mM fluvastatin and their progeny examined 48 or 72 h later, the developmentally arrested embryos show a diffused staining (Fig. 2 C and D). Similarly, viable statin-treated transgenic L1/L2 progeny exhibit much more diffuse staining with significantly reduced membrane enrichment by 48 and 72 h (Fig. 2H). The reduced membrane localization of the prenylation reporter was highly significant, as determined by counting the number of intestinal cells with distinct membrane-enriched GFP in the various treatments (Fig. 2I). Note that even after 72 h of culture on 1 mM fluvastatin, there are still some cells in which the GFP prenylation reporter is membrane-enriched, suggesting either that fluvastatin does not inhibit completely all production of mevalonate, that some mevalonate is present on the culture plates, or that the turnover of prenylated GFP is a slow process. Inclusion of 10 mM mevalonate on the 1 mM statin plates completely rescued the membrane localization of the GFP prenylation reporter (Fig. 2 E–G and I).
Fig. 2.
Fluvastatin inhibits protein prenylation. The worms shown are transgenic for the prenylation reporter pGLO-1P::GFP-CAAX. In control embryos (A and B), the reporter is clearly membrane localized. Embryos from parents grown on 1 mM fluvastatin plates for 48 h (C and D) often arrest with diffused GFP localization (note the diffused expression in the two strongly GFP positive cells indicated by arrowheads). Embryos from parents grown on 1 mM fluvastatin plus 10 mM mevalonate plates for 48 h (E and F) all show strong membrane association of the GFP signal. (G) Shows an L2 larva from parents grown on 1 mM fluvastatin plus 10 mM mevalonate plates for 48 h; note the membrane enrichment of the GFP signal (indicated by arrowheads); this profile is identical to larvae grown on control plates. (H) Shows L2 larvae progeny from parents grown on 1 mM fluvastatin plates for 48 h; note the diffuse distribution of the GFP signal. (I) Shows the results of counting the number of intestinal cells with a clear membrane enrichment of the GFP signal in L1/L2 worms grown under various conditions. The bars show the average ± SEM (n > 35).
Statin Treatment Causes Reduced Nile Red Staining Without Affecting Lipid Stores.
Others have reported that RNAi against HMG-CoA reductase causes a decrease in Nile red staining in C. elegans, which was taken as an indication that lipid stores were reduced in the treated animals (14). We could partially reproduce this result and found that statins are even more effective; growing L4 larvae overnight in the presence of 1 mM fluvastatin caused a strong decrease in Nile red staining (see Fig. S2 for dose-response, kinetics, controls, and similar results using two other statins). Unexpectedly, mevalonate did not visibly reduce the effects of fluvastatin on Nile red staining, suggesting that this effect is caused by the action of statins on other targets (Fig. 1 G–I).
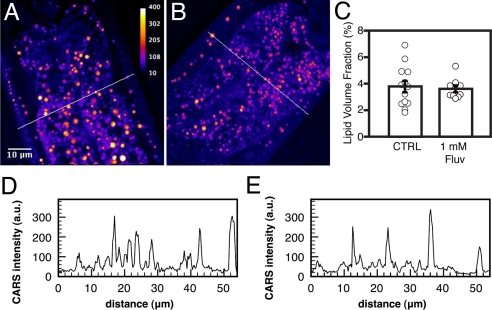
We used coherent anti-stoke raman scattering (CARS) microscopy to determine whether the reduced Nile red staining was indicative of changes in lipid stores. CARS microscopy images fatty acid molecules directly by exciting specifically the C-H bonds in which they are rich and is therefore a dye-independent method to quantitatively visualize lipid droplets with great spatial resolution (21). CARS microscopy could detect no difference between fluvastatin-treated worms or control worms; the fraction of the sampled volume occupied by lipid droplets did not vary between controls and statin-treated worms (Fig. 3 A–C), nor did the average intensity and volume of these droplets (Fig. 3 D and E). In the worms that we observed using both methods, there was generally poor co-localization between Nile red-stained structures and those detected by CARS; at best, Nile red stained only a subset of the CARS-detected structures (Fig. S3). Sudan black staining and gas chromatography also revealed no changes in lipid stores in the statin-treated worms (Fig. S3). Schroeder and coworkers have previously shown that Nile red stains lysosome-like granules rather than lipid droplets within the C. elegans intestine (22). However, our efforts to document defects in lysosomes has so far been negative (Fig. S4). We conclude that the decreased Nile red staining in the statin-treated worms does not reflect changes in the composition of the fat stores, but may instead reflect minor, rather than major, changes in the intestinal lysosome-like granules.
Fig. 3.
CARS imaging shows that lipid content usually does not correlate well with Nile red staining and is not affected by statin treatment. (A and B, respectively) Control worm and a fluvastatin-treated worm imaged by CARS microscopy; an intensity scale is provided in panel A. (C) Percentage of the sampled volume occupied by lipid droplets as determined by CARS microscopy on 13 individuals grown on Nile red plates (average % ± SEM: 3.79 ± 0.44), nine grown on 1 mM fluvastatin and Nile red (3.53 ± 0.30). (D and E) Intensity profiles expressed in absolute units over the area indicated by lines in panels A and B, respectively.
Inhibition of Farnesyl Diphosphate Synthase Causes Larval Arrest.
Bisphosphonates were used to test if inhibition of enzymes downstream of HMG-CoA reductase could uncouple larval arrest from the decrease in Nile red staining observed in statin-treated worms. Bisphosphonates are known inhibitors of farnesyl diphosphate synthase, and two different compounds of this group were tested, namely ibandronate and pamidronate. Eggs allowed to hatch and develop on plates containing 1 mM ibandronate arrested as larvae, just as they did when placed on fluvastatin, but L4 worms treated for 24 h showed no significant decrease in Nile red staining (Fig. S5). This result is consistent with the interpretation that statins inhibit growth at this or a downstream step, but cause reduced Nile red staining via off-target effects. Pamidronate had no visible effects on growth or Nile red staining.
Statins and HMG-CoA Knock-Down Induce the UPR.
The metabolic steps occurring downstream of farnesyl disphosphate synthase lead to the prenylation of several GTPases important for essential processes, including organelle homeostasis (23–25). A recent study demonstrated that the UPR is induced in macrophages cultivated in the presence of statins or RNAi against HMG-CoA reductase (13). Another recent study coupled GTPases of the RAS superfamily with induction of the UPR in C. elegans (26). These studies led us to hypothesize that statins may induce the UPR by inhibiting RAS-type GTPases, which require farnesylation to become membrane-bound and active.
We examined whether inhibition of the mevalonate pathway induces the UPR in C. elegans by using a phsp-4::GFP transgene as a reporter of UPR activation; hsp-4 encodes a worm homolog of the mammalian endoplasmic reticulum chaperone BiP that is a component of the UPR (26–28). We found that C. elegans grown on 1 mM fluvastatin induced the expression of hsp-4::GFP, although to levels clearly lower than those obtained by treating worms with 5 μg/mL tunicamycin, a protein glycosylation inhibitor and a strong UPR activator (Fig. 4 A–D). Progeny from tunicamycin-treated worms arrested as embryos, just as they did in the fluvastatin treatment (Fig. 4 E–G).
Fig. 4.
Fluvastatin induces the UPR. (A–C) A hsp-4::gfp reporter was used to monitor UPR induction in controls or worms grown overnight on 5 μg/mL tunicamycin or 1 mM fluvastatin; worms with average fluorescence for each treatment are shown. The average fluorescence for each treatment is provided in panel D, where error bars indicate the SEM; n > 20 worms. (E–G) The progeny from treated worms were photographed 72 h after treatment initiation. Note the healthy control adult that has begun to lay new eggs, and the arrested embryos (arrowheads) and larvae (arrows) in the tunicamycin and fluvastatin samples.
In C. elegans and other organisms, the UPR response is regulated by the transmembrane inositol-requiring 1 protein kinase (ire-1 in C. elegans), which activates the xbp-1 mRNA by cleaving it, leading to the synthesis of XBP-1 protein which in turn positively regulates UPR (27). The hsp-4::gfp reporter was not activated by fluvastatin or HMG-CoA reductase RNAi in the ire-1 or xbp-1 mutant backgrounds (Fig. 5 A–E), indicating that their effects occur upstream of ire-1 and xbp-1. pek-1 and atf-6 are members of a distinct UPR activation pathway (29); their inhibition by RNAi had no effects on the ability of statins to induce the UPR (Fig. S6), suggesting that statins act specifically through ire-1 and xbp-1.
Fig. 5.
Fluvastatin and RNAi against HMG-CoA reductase induce the UPR via ire-1 and xbp-1. For panels A and B, worms of the strain SJ4005, which harbor the hsp-4::gfp transgene, were picked at the L3 stage and placed on control plates (A) or plates containing 1 mM fluvastatin (B) then photographed 24 h later. For panels C–E, worms of the strains SJ4005 (C), SJ17, which harbor the hsp-4::gfp transgene in a xbp-1 mutant background (D), and SJ30, which harbor the hsp-4::gfp transgene in a ire-1 mutant background (E) were grown for two generations on bacteria producing dsRNA against HMG-CoA reductase, then photographed at the L4 stage. In a separate experiment (lower row), RNAi against the M57.2 (annotated in Wormbase as a protein geranylgeranyl transferase type II, α subunit) caused the activation of the UPR reporter in strain SJ4005 (G), and this could not be rescued by supplementing the plates with mevalonate (H), farnesyl-pp (I), or geranylgeranyl-pp (J).
UPR Activation by Suppression of Prenylation Enzymes and Rescue with Prenylation Substrates.
UPR activation did not occur when fluvastatin- or HMG-CoA RNAi-treated worms were provided with exogenous mevalonate or farnesyl-pyrophosphate, but still occurred when geranylgeranyl-pyrophosphate was supplied (Fig. S7). These results demonstrate that fluvastatin and HMG-CoA reductase RNAi exert their effects on the UPR via the mevalonate pathway and specifically implicate the steps that immediately precede protein farnesylation, i.e., the addition of farnesyl at the C terminus of proteins, such as RAS-type GTPases, but exclude any critical role for geranylgeranyl modification, as occurs on RAB-type GTPases. Consistently, inhibition of the RAS farnesyl-transferase using manumycin A induced the UPR and caused growth retardation (Fig. S8). As a control to establish that manumycin A does indeed inhibit the RAS farnesyl-transferase in C. elegans, we successfully used it to rescue the multivulva phenotype of the let-60(n1046) allele, which corresponds to a constitutively active form of RAS (Table S1). That manumycin A recapitulates the effects of statins suggests that small GTPases of the RAS class may be critical downstream molecular targets of statin treatment in C. elegans.
Finally, we tested RNAi against three genes annotated within Wormbase as subunits of protein prenyltransferases, namely M57.2 (annotated as a protein geranylgeranyl transferase type II, α subunit), F23D3.5 (annotated as a farnesyl transferase α subunit; also named fnta-1), and R02D3.5 (annotated as a farnesyl transferase β subunit; also named fntb-1). Of these, only M57.2 induced the UPR when inhibited by RNAi (Fig. 5 F and G). Importantly, the UPR induced by RNAi against M57.2 could not be rescued by mevalonate, farnesyl-pp, or geranylgeranyl-pp (Fig. 5 H–J). We conclude that inhibition of the mevalonate pathway causes activation of the UPR by limiting the prenylation of one or more M57.2 substrate(s).
Discussion
We have shown that statins can be used in C. elegans to replicate the effects of RNAi against HMG-CoA reductase. These effects include embryonic lethality, larval developmental arrest, adult sterility, and reduced Nile red staining. Because the mevalonate pathway lacks the branch leading to cholesterol synthesis in C. elegans, statins are likely to have their end effects on protein prenylation in this organism. Farnesyl pyrophosphate is near the end of the mevalonate pathway, which is inhibited by statins (30), and can be attached to RAS-type GTPases or converted to geranylgeranyl pyrophosphate, another lipid moiety that can be added to proteins during (iso)prenylation. Prenylation of GTPases has been documented in C. elegans (31), and many prenylated small GTPases are essential for developmental processes. It is therefore not surprising that statins caused developmental arrest in C. elegans. That the UPR was triggered in statin-treated worms is also consistent with inhibition of GTPase prenylation, as this would impair the activity of these regulators of organelle biogenesis and homeostasis, resulting in ER defects and accumulation of misfolded protein, as has been reported for statin-treated macrophages (13). RNAi against M57.2 induced the UPR and partly phenocopied the growth inhibitory effects of statins, suggesting that preventing prenylation of its substrates accounts for some or many of the statin effects. Our results are consistent with the published effects of RNAi against M57.2, which include embryonic lethality and larval arrest (18, 19). At the time of writing, M57.2 is annotated in Wormbase as a RAB geranylgeranyl transferase, although this has not been verified experimentally; its homology is with the α subunits of protein prenyltransferases, and it could well act as a farnesyl transferase. It will be very interesting to identify the substrates of M57.2 and to examine their possible roles in ER homeostasis or protein folding.
Medically, the range of indications for which statins may be used is likely to expand as its cellular effects are better understood. In particular, given that activation of small GTPases such as RAS contributes to about 20% of human cancers, the ability of statins to inhibit GTPases holds the potential for their use as anti-cancer therapeutics (1, 32). Another example is the in vivo use of statins to induce the UPR in a rat model where it had neuroprotective effects under ischemic conditions (33). Indeed, induction of the UPR is likely to be protective in several disease contexts, including Parkinson disease (34) and diabetes (35), where misfolded proteins or increased protein production impose a burden on the ER. Note that the potential of statins as inducers of beneficial effects via preventive UPR will clearly depend on whether they can induce UPR directly and not via accumulation of deleterious levels of unfolded proteins.
Our results also show that cultivation of C. elegans in the presence of 1 mM statin interferes with Nile red staining, but has no significant effects on lipid content or composition. It is possible that statins interfere with uptake processes and thus cause the reduced Nile red staining; for example, a screen for genes important for receptor-mediated endocytosis found that HMG-CoA reductase is important for that process (36). The fact that mevalonate rescued the growth and UPR but not the Nile red effects of statins, and that the bisphononate ibandronate caused growth arrest but not a reduction in Nile red staining, suggest that the effects of statins on Nile red staining are unrelated to the mevalonate pathway and are likely off-target effects. The discrepancy between the Nile red staining and the three other methods used to monitor lipid stores, i.e., CARS microscopy, Sudan black, and gas chromatography, point to some limitation of the Nile red staining method as an indicator of lipid store status in C. elegans. Indeed, others have shown that Nile red merely stains lysosome-related compartments in C. elegans (22, 37), and our results also show that Nile red allows the visualization of only a subset of the lipid droplets. There are obviously a variety of gut granules types in the C. elegans intestine, of which only a subset is stained by Nile red, and staining of this subset by Nile red does not always correlate with lipid content.
The rather dramatic effects of statins on C. elegans should allow this organism to be used to screen for genes that act in the HMG-CoA reductase pathway. We plan to carry out forward genetics screens to identify mutations/RNAi that allow for the growth of C. elegans in the presence of statins. Such screens should help us to identify genes important for the statin effects and which may encode targets or regulators of protein prenylation and downstream effectors.
Materials and Methods
Nematode Strains and Maintenance.
Maintenance of worms were performed as described (38). The wild-type reference strain was the C. elegans Bristol variety strain, N2. In addition to the wild-type, the mutants ire-1(zc14) and xbp-1(zc12) were used. The following transgenic strains were also used and have been described elsewhere (27): SJ4005(zcIs4[phsp-4::gfp]), SJ17(xbp-1(zc12);zcIs4[phsp-4::gfp]), and SJ30(ire-1(zc14); zcIs4[phsp-4::gfp]). All strains were cultured at 20 °C.
Microinjection of dsRNA.
Bacteria containing the F08F8.2 RNAi clone (Geneservice Ltd.) were cultured overnight in 100 mL LB media containing tetracycline (12.5 μg/mL). The RNAi plasmid was prepared with a Qiagen midiprep kit (Qiagen) and cut with PciI or HindIII before vitro transcription of F08F8.2_α and F08F8.2_β using T7 polymerase (Promega) according to published protocols (39). Following annealing of the products, dsRNA was microinjected into adult wild-type worms.
Feeding RNAi.
The following ORFs (gene name; concise annotation) were inhibited by feeding RNAi using a published RNAi library (40): F08F8.2 (HMG-CoA reductase), F23B12.6 (fntb-1; farnesyl transferase β subunit), M57.2 (protein geranylgeranyl transferase type II α subunit), and R02D3.5 (fnta-1; farnesyl transferase α subunit). Bacterial clones were inoculated 18 h before seeding the RNAi-plates, and bacteria carrying the empty vector L4440 were used as controls. Ten L3/L4 worms were transferred to RNAi plates (1 day after seeding) containing the given RNAi bacteria and allowed to grow to young adults. These were then transferred to fresh RNAi plates and allowed to lay eggs for 24 h. The adults were then removed, and the progeny scored at the L4-stage using a Leica DMI6000 B microscope equipped with GFP filter and an Olympus DP71 camera. All pictures for a given experiment were taken with the same settings and exposure times.
Prenylation Assay.
To monitor protein prenylation in C. elegans, we adapted a GFP-based method previously described for mammalian cells (41). The plasmid pGLO-1P::GFP-CAAX carries the intestinal-specific promoter glo-1 (22, 42) to express a modified GFP fused to the last 12 aa of the C. elegans ras-2 gene, including the terminal prenylation motif sequence CLIS (see SI Materials and Methods). pGLO-1P::GFP-CAAX was microinjected together with the phenotypic marker rol-6 carried on plasmid pRF4 (43), and transgenic worms were maintained by picking individuals with the roller phenotype.
Statin Solutions and Plates with Additives.
Fluvastatin (brand Lescol; Novartis) or Lovastatin (brand Mevinolin; Sigma): for a 100 mM stock solution, 80 mg fluvastatin was dissolved in 1,844 μL dH2O. Insoluble components were spun down at 5,000 × g for 10 min, and the supernatent was filter-sterilized. Fluvastatin was added directly into NGM media (≈55 °C), with final concentrations being: 0.02, 0.04, 0.06, 0.08, 0.1, 0.2, 0.4, 0.6, 0.8, or 1 mM. Atorvastatin (brand Lipitor; Pfizer) stock solution and plates were prepared as for fluvastatin, but the drug was dissolved in methanol instead of dH2O.
Mevalonolactone (Sigma) was dissolved in dH2O to produce a 1 M stock solution of mevalonic acid. Mevalonic acid was added together with fluvastatin directly into NGM media (≈55 °C), with final concentrations of 1 mM fluvastatin plus 0.156, 0.313, 0.625, 1.25, 2.5, 5, or 10 mM mevalonic acid. Farnesylpyrophosphate (Sigma), geranylgeranylpyrophosphate (Sigma), manumycin A (dissolved in DMSO; Sigma), and tunicamycin (dissolved in DMSO; Sigma) were similarly used.
Nile Red Staining.
Nile red was dissolved in acetone (1 mg/mL), diluted in PBS, and added on top of NMG plates seeded with OP50 bacteria (without or containing various drugs, see above), to a final concentration of 1 ng/mL. Worms were placed on these plates as L4 larvae and allowed to grow overnight before mounting in a drop of 10 mM levamisole atop a 2% agarose pads laid on a microscopy glass slide then overlayed with a coverslip before fluorescence microscopy (rhodamine channel) using a Zeiss Axioplan microscope. All worms were photographed at a fixed exposure time.
Quantitation of Nile Red and hsp-4::gfp Levels.
Quantitative measurements of the Nile red staining was performed using the image processing program ImageJ (Rasband, WS. ImageJ, U.S. National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij/).
CARS Microscopy.
The CARS microscopy analysis was performed as previously described (21), with the modification that the area sampled corresponded to the first intestinal ring, just posterior to the pharynx, and the use of a slightly different setup (see SI Materials and Methods).
Supplementary Material
Acknowledgments.
We thank the C. elegans Genetics Center (funded by the National Institutes of Health Center for Research Resources) for the strains used in this study. This work was funded by the Åhlens Stiftelse, the Swedish Research Council (Vetenskaprådet), Cancerfonden, Magnus Bergvalls Stiftelse, the Danish Research Council for Strategic Research, the Swedish Society for Medical Research, and Sahlgrenska Center for Cardiovascular and Metabolic Research (no. A305:188), which is supported by the Swedish Strategic Foundation and the Novo-Nordic foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907117106/DCSupplemental.
References
- 1.Konstantinopoulos PA, Karamouzis MV, Papavassiliou AG. Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat Rev Drug Discov. 2007;6:541–555. doi: 10.1038/nrd2221. [DOI] [PubMed] [Google Scholar]
- 2.Wang CY, Liu PY, Liao JK. Pleiotropic effects of statin therapy: Molecular mechanisms and clinical results. Trends Mol Med. 2008;14:37–44. doi: 10.1016/j.molmed.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grosser N, et al. Rosuvastatin upregulates the antioxidant defense protein heme oxygenase-1. Biochem Biophys Res Commun. 2004;325:871–876. doi: 10.1016/j.bbrc.2004.10.123. [DOI] [PubMed] [Google Scholar]
- 4.Kumai T, et al. Pleiotropic effects of 3-hydroxy-3-methylglutaryl-coenzyme A reductase inhibitors: Candidate mechanisms for anti-lipid deposition in blood vessels. Curr Med Chem Cardiovasc Hematol Agents. 2005;3:195–201. doi: 10.2174/1568016054368223. [DOI] [PubMed] [Google Scholar]
- 5.Ostrowski SM, Wilkinson BL, Golde TE, Landreth G. Statins reduce amyloid-beta production through inhibition of protein isoprenylation. J Biol Chem. 2007;282:26832–26844. doi: 10.1074/jbc.M702640200. [DOI] [PubMed] [Google Scholar]
- 6.Yamagishi S, Nakamura K, Matsui T, Inoue H. A novel pleiotropic effect of atorvastatin on advanced glycation end product (AGE)-related disorders. Med Hypotheses. 2007;69:338–340. doi: 10.1016/j.mehy.2006.11.054. [DOI] [PubMed] [Google Scholar]
- 7.Ridker PM, et al. Rosuvastatin to prevent vascular events in men and women with elevated C-reactive protein. N Engl J Med. 2008;359:2195–2207. doi: 10.1056/NEJMoa0807646. [DOI] [PubMed] [Google Scholar]
- 8.Leung KF, Baron R, Seabra MC. Thematic review series: Lipid posttranslational modifications. Geranylgeranylation of Rab GTPases. J Lipid Res. 2006;47:467–475. doi: 10.1194/jlr.R500017-JLR200. [DOI] [PubMed] [Google Scholar]
- 9.Wright LP, Philips MR. Thematic review series: Lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J Lipid Res. 2006;47:883–891. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.van Anken E, Braakman I. Endoplasmic reticulum stress and the making of a professional secretory cell. Crit Rev Biochem Mol Biol. 2005;40:269–283. doi: 10.1080/10409230500315352. [DOI] [PubMed] [Google Scholar]
- 11.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 12.Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen JC, Wu ML, Huang KC, Lin WW. HMG-CoA reductase inhibitors activate the unfolded protein response and induce cytoprotective GRP78 expression. Cardiovasc Res. 2008;80:138–150. doi: 10.1093/cvr/cvn160. [DOI] [PubMed] [Google Scholar]
- 14.Ashrafi K, et al. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- 15.Byrne AB, et al. A global analysis of genetic interactions in Caenorhabditis elegans. J Biol. 2007;6:8. doi: 10.1186/jbiol58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gönczy P, et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- 17.Hamamichi S, et al. Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson's disease model. Proc Natl Acad Sci USA. 2008;105:728–733. doi: 10.1073/pnas.0711018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol. 2001;11:171–176. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- 19.Simmer F, et al. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PloS Biol. 2003;1:e12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonnichsen B, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 21.Hellerer T, et al. Monitoring of lipid storage in Caenorhabditis elegans using coherent anti-stokes raman scattering (CARS) microscopy. Proc Natl Acad Sci USA. 2007;104:14658–14663. doi: 10.1073/pnas.0703594104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schroeder LK, et al. Function of the Caenorhabditis elegans ABC transporter PGP-2 in the biogenesis of a lysosome-related fat storage organelle. Mol Biol Cell. 2007;18:995–1008. doi: 10.1091/mbc.E06-08-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altan-Bonnet N, Sougrat R, Lippincott-Schwartz J. Molecular basis for Golgi maintenance and biogenesis. Curr Opin Cell Biol. 2004;16:364–372. doi: 10.1016/j.ceb.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Lazarow PB. Peroxisome biogenesis: Advances and conundrums. Curr Opin Cell Biol. 2003;15:489–497. doi: 10.1016/s0955-0674(03)00082-6. [DOI] [PubMed] [Google Scholar]
- 25.Quatela SE, Philips MR. Ras signaling on the Golgi. Curr Opin Cell Biol. 2006;18:162–167. doi: 10.1016/j.ceb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Caruso ME, et al. GTPase-mediated regulation of the unfolded protein response in Caenorhabditis elegans is dependent on the AAA+ ATPase CDC-48. Mol Cell Biol. 2008;28:4261–4274. doi: 10.1128/MCB.02252-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 28.Sasagawa Y, Yamanaka K, Ogura T. ER E3 ubiquitin ligase HRD-1 and its specific partner chaperone BiP play important roles in ERAD and developmental growth in Caenorhabditis elegans. Genes Cells. 2007;12:1063–1073. doi: 10.1111/j.1365-2443.2007.01108.x. [DOI] [PubMed] [Google Scholar]
- 29.Shen X, Ellis RE, Sakaki K, Kaufman RJ. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet. 2005;1:e37. doi: 10.1371/journal.pgen.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat Chem Biol. 2006;2:584–590. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 31.Aspbury RA, Prescott MC, Fisher MJ, Rees HH. Isoprenylation of polypeptides in the nematode Caenorhabditis elegans. Biochim Biophys Acta. 1998;1392:265–275. doi: 10.1016/s0005-2760(98)00040-x. [DOI] [PubMed] [Google Scholar]
- 32.Walker K, Olson MF. Targeting Ras and Rho GTPases as opportunities for cancer therapeutics. Curr Opin Genet Dev. 2005;15:62–68. doi: 10.1016/j.gde.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Urban P, et al. Molecular analysis of endoplasmic reticulum stress response after global forebrain ischemia/reperfusion in rats: Effect of neuroprotectant simvastatin. Cell Mol Neurobiol. 2009;29:181–192. doi: 10.1007/s10571-008-9309-7. [DOI] [PubMed] [Google Scholar]
- 34.Sado M, et al. Protective effect against Parkinson's disease-related insults through the activation of XBP1. Brain Res. 2009;1257:16–24. doi: 10.1016/j.brainres.2008.11.104. [DOI] [PubMed] [Google Scholar]
- 35.Scheuner D, Kaufman RJ. The unfolded protein response: A pathway that links insulin demand with beta-cell failure and diabetes. Endocr Rev. 2008;29:317–333. doi: 10.1210/er.2007-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balklava Z, Pant S, Fares H, Grant BD. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat Cell Biol. 2007;9:1066–1073. doi: 10.1038/ncb1627. [DOI] [PubMed] [Google Scholar]
- 37.Rabbitts BM, et al. glo-3, a novel Caenorhabditis elegans gene, is required for lysosome-related organelle biogenesis. Genetics. 2008;180:857–871. doi: 10.1534/genetics.108.093534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sulston JE, Hodgkin JA. Methods, in The Nematode Caernorhabditis elegans. In: Wood WB, editor. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. pp. 587–606. [Google Scholar]
- 39.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 40.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 41.Simonen M, et al. High-content assay to study protein prenylation. J Biomol Screen. 2008;13:456–467. doi: 10.1177/1087057108318757. [DOI] [PubMed] [Google Scholar]
- 42.Hermann GJ, et al. Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol Biol Cell. 2005;16:3273–3288. doi: 10.1091/mbc.E05-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.