Summary
Polycomb group (PcG) proteins exert essential functions in the most disparate biological processes. The contribution of PcG proteins to cell commitment and differentiation relates to their ability to repress transcription of developmental regulators in embryonic stem (ES) cells and in committed cell lineages, including skeletal muscle cells (SMC). PcG proteins are preferentially removed from transcribed regions but the underlying mechanisms remain unclear. Here, PcG proteins are found to occupy and repress transcription from an intronic region containing the microRNA miR-214 in undifferentiated SMC. Cell differentiation coincides with PcG disengagement, recruitment of the developmental regulators MyoD and myogenin, and activation of miR-214 transcription. Once transcribed, miR-214 negatively feeds back on PcG by targeting the Ezh2 3′UTR, the catalytic subunit of the PRC2 complex. miR-214-mediated Ezh2 protein reduction accelerates SMC differentiation and promotes unscheduled transcription of developmental regulators in ES cells. Thus, miR-214 and Ezh2 establish a regulatory loop controlling PcG-dependent gene expression during cell differentiation.
Introduction
Histone H3K27 trimethylation (H3K27me3) is mediated by Ezh2, the catalytic subunit of Polycomb group (PcG) proteins that, with Suz12 and Eed, constitutes the Polycomb Repressive Complex 2 (PRC2) (Cao et al., 2002). The PRC1 complex includes Bmi1, Ring1A and B and can function independently of PRC2 (Schoeftner et al., 2006), (Pasini et al., 2007). However, often H3K27me3 serves as a docking site for PRC1 recruitment, which promotes chromatin remodeling and condensation to ensure gene silencing (Francis et al., 2001). PcG proteins regulate developmental gene expression patterns by repressing transcription of key developmental regulators in ES cells (Bernstein et al., 2006; Boyer et al., 2006; Bracken et al., 2006; Lee et al., 2006). Indeed, Suz12 and Eed-null ES cells express PcG-target genes that are normally repressed (Boyer et al., 2006; Chamberlain et al., 2008; Lee et al., 2006; Pasini et al., 2007). Ezh2-deficient ES cells could not be established (O'Carroll et al., 2001), leading to the suggestion that Ezh2 may be crucial for the propagation of the pluripotent state (Erhardt et al., 2003). However, the role of PcG proteins in the maintenance of ES pluripotency remains controversial (Silva and Smith, 2008). In addition to ES cells, PcG proteins may regulate differentiation of a wide range of cell lineages. For instance, decreased H3K27me3 and PcG binding at certain neuron-specific genes occur during ES cell differentiation into neural precursor cells (Boyer et al., 2006) and during neuronal differentiation (Bracken et al., 2006), at glial-specific genes in neural progenitor cells (Mikkelsen et al., 2007), and at epidermal genes during keratinocyte maturation (Sen et al., 2008). Moreover, conditional Ezh2 ablation has revealed its critical role in controlling B cell development and immunoglobulin gene rearrangement (Su et al., 2003), and gene expression in epidermal basal cells (Ezhkova et al., 2009). Whilst PcG binding and H3K27me3 marks are lost from most transcribed chromatin regions, the underlying mechanisms remain unclear.
Skeletal myogenesis provides a useful biological context within which to examine PcG regulation. The locus for the muscle developmental regulator MyoD is occupied by PcG proteins, bivalently marked by H3K4me3 and H3K27me3, and silent in ES cells (Lee et al., 2006). In skeletal muscle cells (SMC), PcG proteins and H3K27me3 no longer occupy the MyoD locus, allowing for its transcriptional activation. Although MyoD is expressed in undifferentiated SMC, certain MyoD-target genes remain occupied by PcG proteins, marked by H3K27me3, and not expressed. It is only after additional signals initiate the complete myogenic program that PcG binding and H3K27me3 are lost at MyoD-target loci, resulting in appropriate muscle gene expression and SMC differentiation (Caretti et al., 2004). Here, we describe a mechanism for PcG regulation. A genomic region containing the miR-214 transcriptional unit is occupied and repressed by PcG proteins in undifferentiated SMC. During differentiation, PcG proteins are disengaged and the developmental regulators MyoD and myogenin are recruited at the miR-214 region, resulting in its transcriptional activation. Once transcribed, miR-214 loops back to target the Ezh2 3′UTR, thus reducing Ezh2 protein accumulation. Indicating the Ezh2 3′UTR is a bona fide target and is accessible to miR-214 in other cell types, miR-214 expression decreases Ezh2 protein levels also in mouse embryonic fibroblasts (MEFs) and mouse ES cells. miR-214-induced Ezh2 protein reduction is accompanied by accelerated differentiation of SMC and derepression of developmental regulators that are PcG targets in ES cells. Thus, miR-214 can impact transcription controlled by PcG by regulating Ezh2 protein levels.
Results
The Ezh2 mRNA-But Not Its Corresponding Protein-is Detected in Adult Skeletal Muscle
Ezh2 mRNA expression decreases during skeletal muscle development (Laible et al., 1997) (Caretti et al., 2004). However, when amplified by RT-PCR, Ezh2 transcripts were detected in both liver and skeletal muscle of adult mice (Figure 1A). In skeletal muscle, Ezh2 mRNA was not effectively translated, as surmised by the absence of Ezh2 protein (Figure 1B). This observation suggested that Ezh2 mRNA might undergo post-transcriptional regulation in skeletal muscle.
Figure 1. Ezh2 mRNA and Protein Levels in Mouse Adult Liver and Skeletal Muscle and microRNAs Expression in SMC.
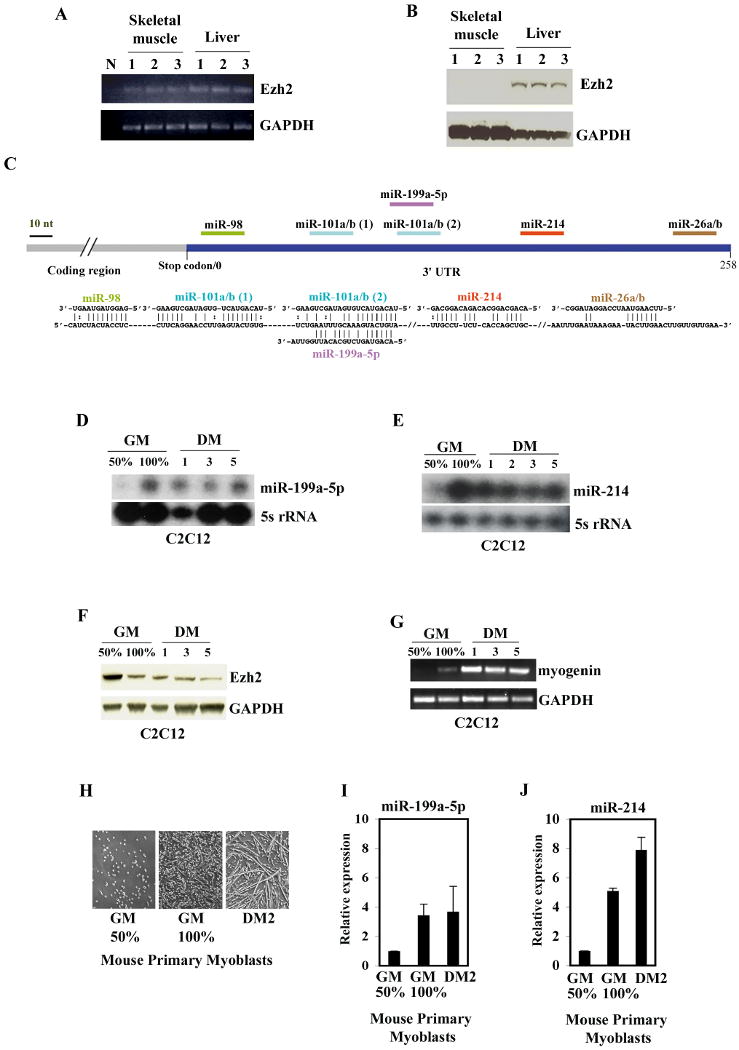
(A) Ezh2 and GAPDH mRNA in the skeletal muscles and livers from three (lanes 1-3) adult mice. N, negative control without cDNA template. (B) Ezh2 and GAPDH proteins in the same tissue samples isolated in (A). (C) The putative targeting sites of miRNAs within the Ezh2 3′UTR are shown in the upper panel. The alignment of each Ezh2 3′UTR/miRNA hybrid is shown in the lower panel. (D) Expression of miR-199a-5p by Ribonuclease Protection Assay (RPA) in subconfluent (50% GM, growth medium), confluent (100% GM), or 1, 3, or 5 days differentiated C2C12 cells (DM, differentiation medium). 5s rRNA was used as a loading control. (E) Expression of miR-214 and 5s rRNA by RPA in C2C12 cells cultured as described in (D). (F) Immunoblot of Ezh2 and GAPDH, and (G) Myogenin mRNA expression. (H) Light microscopic images of mouse primary myoblasts cultured in growth medium (GM, 50% and 100% confluency) or in differentiation medium for 2 days. (I) miR-199a-5p and (J) miR-214 expression determined by real-time PCR in mouse primary myoblasts cultured as described in (H). Error bars represent standard deviations (n=3).
miR-199/214 Are Expressed in Differentiating Skeletal Muscle Cells
Alignment of the Ezh2 3′UTRs of several species revealed a high degree of evolutionary conservation (Figure S1A), and the presence of putative sites for several miRNAs (Figure 1C), including those for miR-98, miR-101, miR-199, miR-214, and miR-26. miR-101, a microRNA that regulates Ezh2 in cancer (Varambally et al., 2008) (Friedman et al., 2009) was barely detectable and its expression, like that of miR-98, was not developmentally modulated in C2C12 cells (Figure S1B, C). Thus, we focused on miR-199 and 214. Both miRNAs were hardly detected in undifferentiated, sub-confluent C2C12 myoblasts (Figure 1D, E, GM, growth medium 50%), but clearly expressed when cells reached full confluency (100% GM) and underwent differentiation (DM, differentiation medium). Genome-wide expression profiling also indicates that the primary miR-199/214 transcript is up-regulated during C2C12 cell differentiation (Caretti et al., 2006) (GEO, GDS2515 record, Mm.29567, http://www.ncbi.nlm.nih.gov). Increased miR-199 and 214 accumulation in fully confluent cells coincided with initial Ezh2 protein reduction (Figure 1F, 100% GM) and cell differentiation, as indicated by the appearance of myogenin transcripts (Figure 1G, 100% GM). Similar expression profiles of miR-199 and 214 were observed in mouse primary myoblasts (Figure 1H-J). In developing mouse embryos (9.5 d.p.c), miR-214 was expressed in several anatomical structures, including the somites, where muscle precursors are initially specified and differentiate (Figure S1F). Consistent with its non-muscle-restricted expression in the embryo, mir-214 was expressed in confluent CH310T1/2 mouse fibroblasts, and coincided with reduced Ezh2 protein accumulation (Figure S1D). miR-26a may be relevant at later stages of differentiation, as its expression occurs in terminally differentiated C2C12 cells (Wong and Tellam, 2008), well after the initial Ezh2 protein decrease.
miR-214 Targets the Ezh2 3′ UTR in Skeletal Muscle Cells
Grafting the Ezh2 3′UTR to a luciferase reporter construct reduced luciferase activity in differentiating C2C12 cells (Figure 2A, B, p<0.0005, Figure S2A). The luciferase-Ezh2 3′UTR construct was less active- or more repressed-in differentiating than in proliferating C2C12 cells (Figure 2B). We tested whether luciferase repression may be ascribed to miR-199, miR-214, or both by modifying their presumptive sites (Figure 2A). Mutations of these sites were predicted to reduce pairing with their corresponding miRNAs, as suggested by increased free energy hybridization values (ΔG) (Figure 2A). Luciferase activity of wild-type (WT) or miR-199 mutant (199Mut) Ezh2 3′ UTR construct was comparably reduced (Figure S2B). These data suggest that miR-199 has no significant role in Ezh2 3′UTR regulation. Conversely, luciferase activity of the single (214Mut) or double mutants (199/214Mut) was similar to that of control vector (Figure S2A,B, and Figure 2B), indicating that the presumptive miR-214 site is required to mediate repression. To further assess the relevance of miR-214 to Ezh2 3′UTR regulation, WT or 214Mut Ezh2 3′UTR reporter construct was introduced in C2C12 cells expressing either wild-type or mutant miR-214 (Figure S2C). Cells expressing miR-214-wt, but not miR-214-mut, further suppressed the luciferase activity of WT but not 214Mut construct (Figure 2C). The repression of WT construct observed in cells expressing miR-214-mut (Figure 2C, right panel) was similar to that in control cells (Figure 2C, left panel), and is likely due to endogenous miR-214. Next, we depleted endogenous miR-214 in C2C12 cells by transfecting chemically modified, single-stranded anti-miR-214 oligomers (Figure S2D). MiR-214 depletion relieved the repression exerted by the Ezh2 3′UTR on luciferase activity (Figure 2D). Overall, these results are consistent with a direct targeting of Ezh2 3′UTR by miR-214.
Figure 2. miR-214 Down-Regulates Ezh2 by Direct Targeting Its 3′UTR.

(A) Schematic representation of luciferase reporter constructs. The predicted structure of each base-paired Ezh2 3′ UTR/wild-type or mutated miRNA hybrid is diagrammed, and the calculated free energy (ΔG) in kilocalories per mole of the 5′ seed region of each hybrid is shown on the right. The top strand in each diagram represents 5′ to 3′ Ezh2 3′ UTR WT and mutants (nucleotides in color), and the bottom strand represents the microRNAs. (B) Luciferase reporters shown in (A) were transfected into C2C12 myoblasts (50%, 100% confluent, or differentiated in DM for 24 hrs) and luciferase activity was determined 48 hrs after transfection. The ratio of reporter (Firefly Luciferase, FL) to control plasmid (Renilla Luciferase, RL) in relative luminescence units was normalized for each reporter to the buffer control and plotted as fold increase or decrease of the control value. Error bars represent standard deviations from 3 independent experiments done in triplicate (*** p<0.0005; ** p<0.005). (C) Control luciferase reporter (pGL3), pGL3 with wild-type Ezh2 3′UTR (WT), or with Ezh2 3′ UTR mutated miR-214 site (Mut) were transfected into C2C12 cells overexpressing empty vector (left panel), wild-type miR-214 (miR-214wt, middle panel), or mutated miR-214 (miR-214-mut, right panel). The sequences of miR-214-wt and the mutations introduced into miR-214-mut are indicated on the bottom. Luciferase activity was determined as described in (B). *** p<0.0005; ** p<0.005. (D) C2C12 cells were co-transfected with either control or anti-miR-214 oligomers, pGL3 (control vector), pGL3-Ezh2 3′ UTR (WT), or pGL3-Ezh2 3′ UTR mutated miR-214 site (Mut) reporter constructs. Luciferase activity was determined as described in (B). **** p<10-6. (E) Schematic representation of RNA-immnuoprecipitation (upper panel) and miRNA pull-down experiments (lower panel). (F) Cell extracts were prepared from 24 hours differentiated C2C12 cells overexpressing empty vector, wild-type miR-214, mutated miR-214 or miR-22 and immnuoprecipitated with either Ago2 antibody or IgG and blotted with Ago2 antibody (top panel). Asterisk indicates 55 kDa IgG heavy chains. The precipitated RNA was converted to cDNA and amplified by real-time PCR using Ezh2 primers (lower panel). Error bars represent standard deviations (n=3), * p< 0.05, ** p< 0.005. (G) C2C12 cell extracts were prepared from cells transfected with biotinylated double-strand RNA miR-214 or miR-214 mutant and subjected to either immunoblot (left panel) or pull-down assay with streptavidin beads. The pulled-down RNA was converted to cDNA and amplified by real-time PCR using Ezh2 or SIRT1 primers. Error bars represent standard deviations from 3 independent experiments. * p < 0.05, ** p< 0.005. (H) Cell extracts from C2C12 cells transfected with control (pGL3), WT Ezh2 3′UTR (WT), or 214Mut Ezh2 3′ UTR (Mut) luciferase constructs were immunoprecipitated with either Ago2 antibody or unrelated IgG. The precipitated RNA was converted to cDNA and amplified by real-time PCR using primers spanning the luciferase and Ezh2 3′UTR junction. The values were corrected for the amount of total luciferase –Ezh2 3′ UTR RNAs (either WT or Mut), measured by real-time PCR. Error bars represent standard deviations from 3 independent experiments, * p< 0.05.
The putative miR-214 site within the Ezh2 3′UTR is atypical, as it does not conform to the canonical seed rule (pairing to the 5′ region of the miRNA centered on nucleotides 2-7). Therefore, we felt it necessary to rigorously probe for miR-214 pairing to Ezh2 mRNA. To this end, the Argonaute-2 (Ago2) protein –an integral component of the RISC complex- was immunoprecipitated from extracts of C2C12 cells (Figure 2E, top panel; Figure 2F, top panel) expressing either exogenous wild-type, mutated miR-214, or control miR-22. The immunoprecipitated materials –along with control reactions- were processed to detect Ezh2 mRNA. Endogenous Ezh2 mRNA was significantly enriched in Ago2-containing protein complexes derived from wild-type -but not control, miR-22, or mutated -miR-214- C2C12 expressing cells (Figure 2F, lower panel). We confirmed the presence of miR-214 in the Ago2-containing complex (Figure S2E). In a second set of experiments, C2C12 cells were transfected with biotinylated wild-type or mutated miR-214 RNA duplexes, respectively. The transfected RNA duplexes were captured by incubating C2C12 cell extracts on streptavidin-coated agarose beads (Figure 2E, bottom panel), and the purified materials processed for Ezh2 mRNA detection. Ezh2 mRNA was enriched in extracts from cells transfected with wild-type- but not mutated-miR-214 (Figure 2G). Target specificity was indicated by lack of amplification of SIRT1- an mRNA not affected by miR-214 (Figures 2G and S4C). Finally, to address the role of the predicted, but atypical, miR-214 site within the Ezh2 3′UTR, extracts derived from C2C12 cells transfected with control, WT or 214Mut Ezh2 3′UTR-luciferase reporter constructs were immunoprecipitated with Ago2 antibodies or unrelated IgG. The immunoprecipitated materials were processed to detect the luciferase-Ezh2 3′UTR hybrid mRNA. A significantly higher mRNA enrichment was observed in Ago2-immunoprecipitated extracts of cell transfected with the WT Ezh2 3′UTR-luciferase reporter compared to those of 214Mut Ezh2 3′UTR-luciferase reporter-transfected cells (Figure 2H). Overall, the results of these experiments indicate that a miR-214-programmed RISC complex can specifically target the Ezh2 3′UTR in C2C12 cells.
Epigenetic and Functional Characterization of the miR-199/214 Genomic Region in Undifferentiated and Differentiated Skeletal Muscle Cells
miR-199/214 are co-transcribed from a conserved antisense intronic transcript at the dynamin3 (Dnm3) locus (Loebel et al., 2005). Both sense Dnm3 and antisense (Dnm3os) transcripts are expressed in skeletal muscle (Loebel et al., 2005). Since PcG proteins occupy muscle-specific genes in undifferentiated C2C12 cells (Caretti et al., 2004) and because of the miR-214 regulated expression during differentiation (Figure 1E), we analyzed the genomic region (∼ 10 Kb) surrounding and encompassing the miR-199/214 transcript by chromatin immunoprecipitation coupled to DNA microarray analysis (ChIP-chip).
In undifferentiated C2C12 myoblasts, when miR-214 is not expressed, the miR-199/214 region was occupied by Suz12, Eed (PRC2), and Bmi1 (PRC1) proteins (Figure 3A). Consistent with H3K27me3 being a mark for PRC1 recruitment, the region bound by Bmi1 displayed enrichment for H3K27me3. The initiating (hypophosphorylated) form of RNA polymerase II (RNAP2) and H3K4me3 were also detected at the miR-199/214 locus in MB (Figure 3B). H3K4me3 is restricted to the sites of transcription initiation and has been successfully employed to identify the location of miRNA promoters (Marson et al., 2008). The data indicate that this genomic region has the characteristics of a miRNA promoter where transcriptional pausing may be mediated by PcG proteins (Breiling et al., 2001; Guenther et al., 2007; Saurin et al., 2001; Sessa et al., 2007; Stock et al., 2007).
Figure 3. Epigenetic Characterization of the miR199a/214 Locus in Myoblasts and Myotubes.

ChIP-chip analyses of binding of the PRC2 subunits Suz12 and Eed, the PRC1 subunit Bmi1, and the H3K27 trimethyl mark (A) and the hypophosphorylated form of RNAP2 and the H3K4 trimethyl marker of transcriptional initiation (B) at the miR199a-2/214 locus in proliferating myoblasts; and the same factors in differentiated myotubes (C) along with analyses of the myogenic factors MyoD and Myog (D). (E) Real-time PCR analysis of Suz12 and Ezh2 mRNA (left panel) and immunoblot of Suz12 and tubulin (right panel) in C2C12 myoblasts transfected with siRNA against Suz12 (si-Suz12). (F) miR-214 and myogenin expression by real-time PCR in C2C12 cells transfected with si-Suz12. (G) Real-time PCR analysis of Suz12 and Ezh2 mRNA (left panel) and immunoblot of Ezh2 and tubulin (right panel) in C2C12 myoblasts transfected with siRNA against Ezh2 (si-Ezh2). (H) miR-214 and myogenin expression by real-time PCR in C2C12 myoblasts transfected with si-Ezh2. Error bars represent the standard deviations for n = 3 in (E, F, G, H), * p<0.05; ** p<0.005.
In differentiated C2C12 myotubes, Suz12, Eed, and Bmi1were reduced near the putative start site of miR-199/214 transcription (TSS) (Figure 3C). These modifications were accompanied by increased RNAP2 occupancy, maintenance of H3K4me3, and recruitment of MyoD and myogenin (Figure 3D). Sequence analysis of the region surrounding the MyoD/myogenin peaks revealed the presence of putative E-boxes, the canonical DNA binding sites for MyoD/myogenin. The sequences of three E-boxes display fairly strong evolutionary conservation, with one of these three (E3) being the closest to the peak of MyoD/myogenin binding (Figure S3A). Despite reduced recruitment of both PRC2 and PRC1 proteins near the miR-199/214 TSS, a new peak of H3K27me3 was observed in MT at a region within the putative miR-199/214 transcript, just upstream of miR-214 domain (Figure 3C). This shifted peak of H3K27me3 may reflect nucleosome remodeling that accompanies muscle differentiation and is reminiscent of recent observations in Drosophila where H3K27me3 was found to be enriched not only at Polycomb Responsive Elements but was broadly distributed in gene flanking regions (Schwartz et al., 2006). The results of the ChIP-chip experiments were confirmed by quantitative ChIP (Figure S3D). As control for specificity of the ChIP-chip data at the miR-214 locus, we analyzed H3K27me3 and Suz12 binding at the muscle creatine kinase (Ckm) (Caretti et al., 2004) and MyoD loci (Lee et al., 2006). In agreement with previous studies, the Ckm enhancer was H3K27me3-marked and occupied by Suz12 in undifferentiated C2C12 myoblasts but not differentiated myotubes (Figure S3B, D) (Caretti et al., 2004). Neither H3K27me3 nor Suz12 enrichment were observed at the MyoD locus in either myoblasts or myotubes (Figure S3C) (Lee et al., 2006). Suz12 enrichment at the PcG target Pax6 gene (Lee et al., 2006) in C2C12 myoblasts was comparable to that observed at miR-199/214 region (Figure S3E).
To test for the regulatory function of PcG proteins at the miR-199/214 region, either Suz12 or Ezh2 were depleted by siRNA in undifferentiated C2C12 myoblasts. Lowering either protein (Figure 3E, G) increased miR-214 expression (Figure 3F, H). Consistent with PcG binding to the myogenin promoter (Bracken et al., 2006) and the regulatory role exerted by miR-214 on myogenin expression (see below, Figure 4), either Suz12 or Ezh2-knock-down increased also myogenin expression (Figure 3F, H). These results indicate that PcG proteins occupy and repress transcription of the miR-199/214 region in undifferentiated C2C12 myoblasts. Moreover, they reveal that the miR-199/214 region is a direct target of MyoD and myogenin.
Figure 4. miR-214 Reduces Ezh2 Protein and Accelerates Muscle Cell Differentiation.

(A) miR-214 transcripts (left panel) and time course of protein expression of Ezh2, MHC, myogenin and tubulin (right panel) in empty vector or miR214-wt retrovirally-transduced C2C12 cells. Quantification of the band corresponding to Ezh2 protein was performed with the ImageJ software program. (B) Ezh2 mRNA in empty vector or miR-214-wt C2C12 cells. (C) MHC immunostaining of C2C12 cells transduced with empty vector, miR-214-wt, or mir-214-mut and differentiated for 24 hrs. The nuclei were visualized with DAPI. The fusion index was determined by counting of the number of nuclei present in MHC+ cells per number of total nuclei and expressed as percentage. Error bars represent the standard deviations. (D) Immunoblot of Ezh2, myogenin, and tubulin in C2C12 cells transduced with empty vector, miR-214-wt, or mir-214-mut and differentiated for 12 hrs. (E) Immunoblot of Ezh2, MHC, myogenin and tubulin in C2C12 cells transfected with Ezh2 siRNAs (left panel) and immunoblot of Ezh2, GAPDH, and myogenin mRNA in Ezh2 flox/flox primary myoblasts infected with adenoviruses expressing Cre recombinase (right panel). (F) Immunoblot of Ezh2, MHC, myogenin and tubulin in C2C12 myoblasts transfected without (-), or with scrambled or anti-miR-214 oligomers. Bands corresponding to Ezh2 protein were quantified using the NIH Image J software. (G) Immunoblot of Ezh2, Myc, myogenin and tubulin of extracts derived from C2C12-expressing wt or mutated miR-214 cells transfected with pCDNA3 (empty vector) or pCDNA3-Myc-tagged-Ezh2 vector, and differentiated for 12 hrs. The signal corresponding to the endogenous Ezh2 protein in C2C12 cells expressing exogenous Ezh2-myc and miR-214 WT was reduced by ∼ 40% when compared to control. The miR-214 mutant had no effect on endogenous Ezh2. Quantification was performed with NIH ImageJ software. (H) Light microscopic images of myofibers (left panels) and immunofluorescence staining of myogenin (red) and counterstained with DAPI (blue) in myofiber-derived MSCs (right panel) infected with either empty vector (upper panel) or miR-214 (lower panel) expressing lentivirus. (I) Myofiber-derived MSCs were assayed for myogenin expression and expressed as a percentage of total myofiber- derived MSCs analyzed (empty vector, blue bar, n=5, 1712 total cells; miR-214, red bar, n=3, 2318 total cells, * p<0.05). (J) Real-time PCR analysis of myogenin expression in myofiber-derived MSCs transduced with empty vector or miR-214. Data represents pooled total RNA from three independent infections and is presented as averaged replicates (**p<0.001). (K) Real time PCR analysis of miR-214 expression in myofiber-derived MSCs transduced with empty vector or miR-214. (L) Immunoblot of Ezh2, Su(fu), and tubulin in empty vector or miR-214-wt retroviral transduced C2C12 cells. (M) Immunoblot of Ezh2 and tubulin in MEFs transduced with empty vector, miR-214-wt, or mir-214-mut. 214-1 and 214-2 represent two different experiments conducted with miR-214-wt constructs. (N) Immunoblot of Ezh2 and tubulin in MEFs non-transfected, transfected with scrambled or anti-miR-214 oligomers. Bands corresponding to Ezh2 protein were quantified using the NIH Image J software. (O) Ezh2 mRNA expression in empty vector or miR-214-wt retrovirally-transduced MEFs.
miR-214 Expression Reduces Ezh2 Protein, Accelerates Muscle Gene Expression, and Promotes Muscle Differentiation
We evaluated the functional significance of our findings by analyzing C2C12 cells transduced with a retrovirus expressing miR-214 (Figure 4A, left panel). These cells had reduced Ezh2 protein (Figure 4A, right panel), but similar Ezh2 mRNA levels (Figure 4B). Mir-214-transduced cells initiated premature expression of myogenin and myosin heavy chain (MHC) (Figure 4A). Moreover, miR-214-expressing cells contained a higher percentage of MHC-positive cells (Figure 4C). Cells transduced with either miR-214 mutant (Figure 4C, D) or miR-22 (Figure S4A,B) -for which there are no putative sites within the Ezh2 3′UTR- behaved as control cells.
To further probe the concept that miR-214 affects muscle gene expression by modulating Ezh2, we reduced Ezh2 in C2C12 cells by siRNA, or in primary myoblasts derived from mice harboring floxed Ezh2 alleles by adenoviral-mediated expression of the Cre recombinase. In siRNA Ezh2-depleted C2C12 cells, expression of both myogenin and MHC was prematurely initiated. Similarly, reducing Ezh2 in mouse primary myoblasts resulted in increased myogenin expression (Figure 4E).
In addition to Ezh2, miR-214 is likely to affect other targets involved in regulating muscle gene expression and differentiation. A bioinformatics search identified the 3′ UTR of the deacetylases SIRT1 and HDAC9 (Figure S4C), both regulators of muscle differentiation (Fulco et al., 2003) (Haberland et al., 2007), as potential miR-214 targets. However, neither protein was influenced by miR-214 expression (Figure S4C). To continue testing for the miR-214-Ezh2 link, we blocked endogenous miR-214 with chemically modified, single-stranded anti-miR-214 oligomers. C2C12 cells receiving anti-miR-214 oligomers and prompted to differentiate had increased Ezh2 and reduced myogenin and MHC protein levels (Figure 4F). Finally, we employed an Ezh2 version - devoid of its 3′UTR region- expected to escape miR-214-dependent regulation. Indeed, endogenous Ezh2 protein-but not transfected 3′UTR-less Myc-tagged Ezh2- was reduced in miR-214-expressing C2C12 cells (Figure 4G). Moreover, anticipated myogenin expression induced by miR-214 was effectively counteracted by 3′UTR-less Ezh2 (Figure 4G). Overall, the results support the contention that Ezh2 is a specific and functionally relevant target of miR-214 in skeletal muscle cells.
Next, we wished to evaluate the role of mir-214 on differentiation of mouse primary myoblasts. Individual myofibers isolated from the soleus muscle of 8-week-old mice were plated (Figure 4H, left panels) and transduced with control or miR-214-expressing lentivirus. Cultures transduced with miR-214 lentivirus contained a higher percentage of myogenin-positive cells, compared to those of control transduced myofibers (∼50% increase of myogenin-positive cells, p<0.05) (Figure H-K). Effects of comparable magnitude were noted for miR-233 in hematopoietic progenitor cells (Chen et al., 2004). Thus, miR-214 promotes differentiation not only of an established skeletal muscle cell line (i.e., C2C12) but also of myofiber-derived myoblasts. In zebrafish, miR-214 influences formation of slow muscle cell types by targeting Su(fu), a negative regulator of Hedgehog signaling (Flynt et al., 2007). We aligned zebrafish with mammalian Su(fu) 3′UTRs and found no conservation of these regions (data not shown). Accordingly, the levels of mammalian Su(fu) protein were not decreased in miR-214-expressing C2C12 cells (Figure 4L). Since Ezh2 is transcriptionally down-regulated in differentiating C2C12 cells (Caretti et al., 2004) (Figure S1E), it could be argued that miR-214 overexpression may indirectly promote muscle cell differentiation without necessarily targeting Ezh2. To address this issue, we evaluated how miR-214 affects Ezh2 protein in MEFs, which do not undergo cell lineage commitment and differentiation. MEFs were transduced with the miR-214 retrovirus and Ezh2 mRNA and protein evaluated. Ezh2 protein was reduced in miR-214-wt, but not miR-214-mut expressing MEFs (Figure 4M), whereas MEFs transfected with chemically modified, single-stranded anti-miR-214 oligomers had increased Ezh2 protein (Figure 4N). miR-214 overexpression did not reduce the Ezh2 mRNA levels (Figure 4O). Overall, these results suggest that miR-214 directly targets Ezh2's 3′UTR in both C2C12 cells and MEFs and regulates skeletal muscle differentiation.
miR-214 Targets Ezh2's 3′UTR During Retinoic Acid-Induced Differentiation and Modifies Transcription of Developmental Regulators in Pluripotent ES Cells
Given the regulatory role exerted by PcG on gene transcription in ES cells, we extended our studies to these cells. Depending on the culture conditions, ES cells can be differentiated into various cell types. Retinoic acid (RA) promotes repression of the pluripotent markers Oct-4, Sox2, and Nanog (Chambers and Smith, 2004), and activation of the Hox genes (Langston et al., 1997), Sox1(Zhao et al., 2004), the Wnt antagonist Dkk1 (Verani et al., 2007), and of the neuronal marker nestin (Okabe et al., 1996). Pluripotent ES cells formed rounded, tightly packed colonies with well-defined margins and expressed Oct-4 and Sox2 (Figure S5A,B). Once exposed to RA, ES cells changed their morphology to assume a flattened neuronal-like phenotype (Figure S5A), down-regulated expression of Oct-4 and Sox2, initiated expression of Sox1, Hoxa1, Hoxb1, and nestin (Figure S5B,C).
RA-induced ES differentiation was accompanied by increased expression of both primary and mature miR-214 and reduced Ezh2 protein levels (Figure 5A,B). To causally link miR-214 to RA-mediated Ezh2 down-regulation, ES cells were transfected with either the WT Ezh2-3′UTR or 214Mut reporter construct. ES cells exposed to RA suppressed luciferase activity of WT but not 214Mut construct, indicating that, as observed in SMC, miR-214 targets the Ezh2 3′UTR also in ES cells (Figure 5C). To further study the role of miR-214, ES cells were transduced with control, miR-214-wt or miR-214-mut-expressing lentivirus, respectively (Figure 5D). MiR-214-wt, but not miR-214-mut, reduced the Ezh2 protein without affecting the mRNA levels (Figure 5D, middle and right panels). ES cells transduced with miR-214-wt and exposed to RA displayed increased Dkk1 and nestin expression, when compared to RA-treated control cells (Fig.5E). Of interest, Dkk1 is occupied by PcG and upregulated in Suz12-null ES cells (Boyer et al., 2006).
Figure 5. miR-214 Modulates Gene Expression in ES Cells by Regulating Ezh2 Protein Levels.

(A) Real-time PCR of primary (left panel) and mature (right panel) miR-214 in ES cells kept in medium containing LIF or induced to differentiate by removing LIF and adding RA for 3 d. (B) Immnuoblot of Ezh2 and GAPDH in ES cells cultured in conditions described in (A). (C) Control luciferase reporter (pGL3), pGL3- Ezh2 3′UTR (WT), or Ezh2 3′ UTR mutated miR-214 site (Mut) were transfected into ES cells and the luciferase activity was determined after 3 d treatment with (right panel) or without (left panel) RA. Error bars represent standard deviations from 2 independent experiments done in triplicate (* p<0.05). (D) Real-time PCR of primary miR-214 transcripts (left panel), protein expression of Ezh2, Oct-4, and GAPDH (middle panel), and Ezh2 mRNA expression in empty vector, miR-214-wt or miR-214-mut lentiviral-transduced ES cells (right panel). (E) Nestin and Dkk1 mRNA expression in 3 day RA-induced lentiviral transduced ES cells with empty vector, miR-214-wt or miR-214-mut. (F) Gata-4, Sox17, and Pax7 mRNA expression in ES cells transduced with empty vector, miR-214-wt, or miR-214-mut lentivirus. MyoD mRNA expression was evaluated by Taqman real-time PCR. (G) Alkaline phosphatase assay in empty vector, miR-214-wt or miR-214-mut lentiviral-transduced ES cells. Differentiated percentage was determined by counting of colonies with reduced AP staining per number of total colonies. Error bars represent standard deviations from values of colonies counted in 2 independent experiments (** p<0.005, * p<0.05).
MiR-214 expression in ES cells kept in their pluripotent state repressed expression of the pluripotent marker Oct-4 (Figure 5D, middle panel) and was sufficient to de-repress transcription of the developmental regulators Gata-4, Sox17, and Pax7 (Figure 5F), three PcG targets whose expression is increased in Suz12- and Eed-null ES cells (Boyer et al., 2006) (Lee et al., 2006). Since Oct-4 expression is not reduced in Ezh2-/- ES cells (Shen et al., 2008), miR-214 may have additional, Ezh2-independent, targets. Alternatively, it is possible that a subpopulation of cultured Ezh2-/- ES had reduced levels of Oct-4, and, as a consequence, may have lost cell identity, pluripotency, and self-renewal potential (Niwa et al., 2000). MyoD transcription was not activated by miR-214 (Fig.5F, right panel) (see Discussion). MiR-214-expressing ES cells assumed a flattened phenotype and displayed reduced staining for alkaline phosphatase, a marker of pluripotency (Figure 5G). Overall, these results indicate that miR-214 can reduce Ezh2 and derepress transcription of developmental regulators in ES cells.
Discussion
During cell lineage commitment and differentiation, H3K27me3 is reduced at many genes, including those encoding for developmental regulators. This phenomenon often coincides with transcriptional activation. The important role played by PcG proteins in ES cell biology has been reinforced by the discovery that H3K27me3 marks are reestablished at genes for developmental regulators during reprogramming of differentiated cells into induced pluripotent cells (iPS) (Maherali et al., 2007) (Mikkelsen et al., 2008; Wernig et al., 2007). Because of the important functions exerted, decreased H3K27me3 at selected genes during cell commitment and differentiation may be robustly regulated by independent and coherent mechanisms. Reduced deposition and active removal of the H3K27me3 mark may account for it. Indeed, PcG expression is developmentally regulated (Caretti et al., 2004) (de la Cruz et al., 2007) (Silva et al., 2003) and H3K27me3 can be erased by the UTX/JMJD3 demethylase complex (Cloos et al., 2008).
Our findings provide evidence that miR-214 introduces an additional level of PcG regulation in different cell types via a posttranscriptional mechanism. miRNAs have emerged as central regulators of biological processes. The repressive effect of a given miRNA on individual proteins is relatively small (Selbach et al., 2008), rarely exceeding 1.5-2 fold (Baek et al., 2008). Perhaps because of their limited impact on protein accumulation, miRNAs often act in concert with other regulatory processes, such as reduced transcription (Figure S1E). For instance, an upstream event may oppositely regulate transcription of the miRNAs and their targets mRNAs (Alon, 2007b; Tsang et al., 2007). With the additive nature of these two regulatory mechanisms, the repressive effect of miRNAs on target expression can be modest yet functionally meaningful. The results of the experiments reported here indicate that miR-214-dependent regulation of Ezh2 protein is relevant not only for cell lineage-specific differentiation (i.e., SMC) but also influence ES cell commitment. Pluripotent ES cells, which express very low levels of miR-214 (Marson et al., 2008), readily activate miR-214 transcription when induced to differentiate, a process that is accompanied by reduction of the Ezh2 protein levels. Moreover, miR-214 expression in pluripotent ES cells is sufficient to derepress transcription of the developmental regulators Gata-4, Sox17, and Pax7. These regulators are PcG-targets whose expression is also up regulated in both Suz12-null (Pasini et al., 2007) (Lee et al., 2006) and Eed-null ES cells (Boyer et al., 2006). In addition, Pax7 plays a central role in muscle development (Seale et al., 2000) and, following transcriptional activation, its locus shows reduced levels of Suz12 occupancy in muscle cells relative to ES cells (Lee et al., 2006). MyoD's locus is occupied by PcG proteins and H3K27me3 in ES cells (Lee et al., 2006). Nonetheless, in contrast to other PcG targets, MyoD expression is not activated in either Suz12-null (Lee et al., 2006) or Eed-null ES cells (Boyer et al., 2006), nor was it induced by miR-214 in ES cells (Figure 5F), indicating that different PcG-targets undergo distinct regulation in ES cells. For some of them, such as MyoD, simple transcriptional derepression may be insufficient to activate transcription in the absence of positive regulators.
The miR-214 site within the Ezh2 3′UTR does not obey the canonical seed rule (Bartel, 2009). In this respect, it is of interest to note that a recent study found that ∼ 30% of Ago-mRNA clusters have no predicted seed matches among the top 20 Ago-miRNA families (Chi et al., 2009). These findings indicate that miRNA pairing may, under circumstances yet to be defined, tolerate single mismatch, wobble or bulge nucleotides (Vella et al., 2004) (Didiano and Hobert, 2006) (Tay et al., 2008).
Mice with experimental ablation of the miR-199/214 locus display developmental defects, including skeletal muscle hypoplasia, suggesting a relevant role of these microRNAs in skeletal myogenesis (Watanabe et al., 2008). Several miRNAs influence myogenesis (Williams et al., 2009). In particular, miR-26a regulates Ezh2 in SMC and promote their differentiation (Wong and Tellam, 2008). It is likely that miR-214 and miR-26a affect Ezh2 at distinct developmental steps. Indeed, miR-214 accumulation is evident at the very initial stages of cells differentiation –coinciding with the initial and most evident Ezh2 protein reduction (Figure 1E,F)- whereas miR-26a could not be detected until SMC had completed their terminal differentiation (Wong and Tellam, 2008).
The link between mir-214 and PcG proteins in muscle cells may be rationalized in a working model where Ezh2 transcriptional down-regulation occurring at the initial phases of muscle differentiation (Caretti et al., 2004) (Figure 6A) would promote, along with MyoD and/or myogenin recruitment, expression of miR-214, which in turn would negatively feedback on Ezh2 by inhibiting translation of its mRNA. This relation is best described as a two-node bistable feedback loop (Alon, 2007a) (Figure 6B), a network motif that, by providing a lock-on-mechanism, is conducive to epigenetic inheritance and, as such, often recurs in developmental transcription networks. This network may have been selected to increase robustness of the system to effectively and rapidly reduce Ezh2 availability at critical stages, such as those regulating SMC differentiation.
Figure 6. PcG-miR-214 feedback loop model.
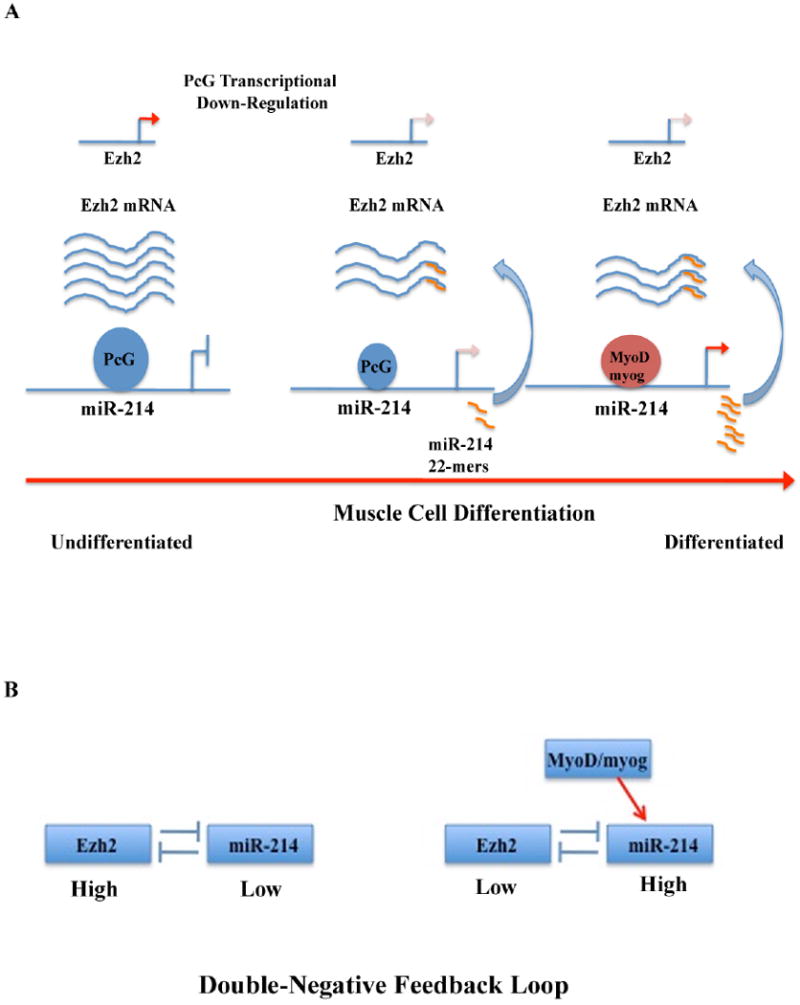
(A) Ezh2 is highly expressed in undifferentiated myoblasts (left part of the model). The initial phase of cell differentiation (middle part) is characterized by reduced Ezh2 expression and consequent de-repression of the miR-214 locus. miR-214 feeds back on Ezh2, reducing its mRNA translation. MyoD/myogenin binding (right part) actively promotes miR-214 transcription, further inhibiting Ezh2 mRNA translation. (B) Double negative feedback loop. MyoD/myogenin acts to switch the steady state. In undifferentiated myoblasts, Ezh2 is high and represses miR-214 (left part of the panel). Upon differentiation, MyoD/myogenin expression is activated, miR-214 is produced and Ezh2 is repressed (right part of the panel).
Experimental Procedures
Cells, Retroviral, Adenoviral, and Lentiviral Constructs, Viral Transduction and Transient Transfection
A detailed description of the reagents and methods is reported in Supplemental Experimental Procedures.
Myofiber Isolation, Culture, and Lentiviral Infection
Mouse soleus muscles from 8 weeks-old C57BL mice were isolated and incubated in DMEM with 0.2% (w/v) collagenase I. Individual myofibers were serially diluted and subsequently manually isolated in GM. For infection, myofibers were cultured for 36 hrs post-isolation prior to infection. Isolated myofibers were incubated with viral supernatants diluted 1:1 in GM, supplemented with polybrene (4μg/ml). Infected cells were incubated for an additional 72 hrs, and then analyzed by immunofluorescence microscopy and quantitative PCR.
Reporter Constructs, Mutagenesis, and Luciferase Reporter Assay
A detailed description of the reagents and methods is reported in Supplemental Experimental Procedures.
Ribonuclease Protection Assay (RPA) and Northern Blot Hybridization
Total RNA was isolated from C2C12 cells using Trizol Reagent (Invitrogen). The expression of miR-199a-5p, miR-214, and miR-101b was determined by RPA using mirVana miRNA Detection Kit according to the manufacturer's instruction (Ambion). A detailed description of the reagents and methods is reported in Supplemental Experimental Procedures.
Sequence Alignments and microRNA Target Prediction
The methods are reported in Supplemental Experimental Procedures.
Whole Mount In Situ Hybridizations
Reagents and methods are reported in Supplemental Experimental Procedures.
Tiling of microRNA regions on DNA arrays, ChIP, and promoter-specific PCR
Sequences upstream of the miR-199-a2/214, Ckm, and MyoD loci were analyzed and probes were designed for the ∼10kb upstream by the methods described in (Boyer et al., 2005). Probes were spaced approximately ∼250 bp apart. Chromatin immunoprecipitation (ChIP) was performed as described in. Briefly, chromatin fragments were immunoprecipitated overnight at 4°C. After reversal of crosslinks, enriched fragments were amplified using a two-step ligation-mediated PCR (LM-PCR) protocol and fluorescently labeled with Cy5 dUTP (Amersham). Labeled fragments were hybridized to DNA arrays in Agilent hybridization chambers at 40° C for ∼40 hrs against a Cy3 dUTP labeled unenriched reference sample (H3K27me3 immunoprecipitations were hybridized against core H3 immunoprecipitations to normalize for total histone H3 density). Arrays were washed and scanned using an Agilent DNA microarray scanner. Unprocessed enrichment ratios from the DNA arrays were examined to look for evidence of binding.
RNA interference and anti-miR oligonucleotides transfections
Description of the oligonucleotides is provided in Supplemental Experimental Procedures.
Immunofluorescence and immunoblot
The methods and the antibodies employed are described in Supplemental Experimental Procedures.
Immnuoprecipitation and miRNA pull-down
The methods and the reagents are reported in Supplemental Experimental Procedures.
RT-PCR, Real-Time PCR, and microRNA TaqMan Real-Time PCR
Material and methods are provided in Supplemental Experimental Procedures.
Real-Time PCR Validation of ChIP-chip Data
Description of the material and methods is provided in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We would like to thank I-h. Su and A. Tarakhovsky for providing the Ezh2 floxed mice, Kambiz Mousavi for assistance with myofiber isolation and culture, and Rafael Casellas for sharing lentiviral constructs. This work was supported in part by the Intramural Research Program of the National Institute of Arthritis, Musculoskeletal, and Skin Diseases of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alon U. An Introduction to Systems Biology Design Principles of Biological Circuits. London: Chapman & Hall/CRC); 2007a. [Google Scholar]
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007b;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiling A, Turner BM, Bianchi ME, Orlando V. General transcription factors bind promoters repressed by Polycomb group proteins. Nature. 2001;412:651–655. doi: 10.1038/35088090. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caretti G, Schiltz RL, Dilworth FJ, Di Padova M, Zhao P, Ogryzko V, Fuller-Pace FV, Hoffman EP, Tapscott SJ, Sartorelli V. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev Cell. 2006;11:547–560. doi: 10.1016/j.devcel.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;23:7150–7160. doi: 10.1038/sj.onc.1207930. [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009 doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz CC, Kirmizis A, Simon MD, Isono K, Koseki H, Panning B. The polycomb group protein SUZ12 regulates histone H3 lysine 9 methylation and HP1 alpha distribution. Chromosome Res. 2007;15:299–314. doi: 10.1007/s10577-007-1126-1. [DOI] [PubMed] [Google Scholar]
- Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- Erhardt S, Su IH, Schneider R, Barton S, Bannister AJ, Perez-Burgos L, Jenuwein T, Kouzarides T, Tarakhovsky A, Surani MA. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development. 2003;130:4235–4248. doi: 10.1242/dev.00625. [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet. 2007;39:259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Saurin AJ, Shao Z, Kingston RE. Reconstitution of a functional core polycomb repressive complex. Mol Cell. 2001;8:545–556. doi: 10.1016/s1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, Zhou X, Jones PA. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Arnold MA, McAnally J, Phan D, Kim Y, Olson EN. Regulation of HDAC9 gene expression by MEF2 establishes a negative-feedback loop in the transcriptional circuitry of muscle differentiation. Mol Cell Biol. 2007;27:518–525. doi: 10.1128/MCB.01415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. Embo J. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston AW, Thompson JR, Gudas LJ. Retinoic acid-responsive enhancers located 3′ of the Hox A and Hox B homeobox gene clusters. Functional analysis. J Biol Chem. 1997;272:2167–2175. doi: 10.1074/jbc.272.4.2167. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebel DA, Tsoi B, Wong N, Tam PP. A conserved noncoding intronic transcript at the mouse Dnm3 locus. Genomics. 2005;85:782–789. doi: 10.1016/j.ygeno.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadfeld M, Yachechki R, Tchieu J, Jaenisch R, et al. Directly Reprogrammed Fibroblasts Show Global Epigenetic Remodeling and Widespread Tissue Contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008 doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe S, Forsberg-Nilsson K, Spiro AC, Segal M, McKay RD. Development of neuronal precursor cells and functional postmitotic neurons from embryonic stem cells in vitro. Mech Dev. 1996;59:89–102. doi: 10.1016/0925-4773(96)00572-2. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. Embo J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22:1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa L, Breiling A, Lavorgna G, Silvestri L, Casari G, Orlando V. Noncoding RNA synthesis and loss of Polycomb group repression accompanies the colinear activation of the human HOXA cluster. Rna. 2007;13:223–239. doi: 10.1261/rna.266707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Silva J, Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verani R, Cappuccio I, Spinsanti P, Gradini R, Caruso A, Magnotti MC, Motolese M, Nicoletti F, Melchiorri D. Expression of the Wnt inhibitor Dickkopf-1 is required for the induction of neural markers in mouse embryonic stem cells differentiating in response to retinoic acid. J Neurochem. 2007;100:242–250. doi: 10.1111/j.1471-4159.2006.04207.x. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Sato T, Amano T, Kawamura Y, Kawamura N, Kawaguchi H, Yamashita N, Kurihara H, Nakaoka T. Dnm3os, a non-coding RNA, is required for normal growth and skeletal development in mice. Dev Dyn. 2008;237:3738–3748. doi: 10.1002/dvdy.21787. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol. 2009;21:461–469. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CF, Tellam RL. microRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J Biol Chem. 2008 doi: 10.1074/jbc.M709614200. [DOI] [PubMed] [Google Scholar]
- Zhao S, Nichols J, Smith AG, Li M. SoxB transcription factors specify neuroectodermal lineage choice in ES cells. Mol Cell Neurosci. 2004;27:332–342. doi: 10.1016/j.mcn.2004.08.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


