Abstract
The endogenous human telomerase reverse transcriptase (hTERT) gene is repressed in somatic cells. To study the mechanisms of its repression, we developed a strategy of retrovirus-directed Cre recombinase-mediated BAC targeting, or RMBT, to generate single-copy integrations of BAC at pre-engineered chromosomal sites. This technique involved retroviral transduction of acceptor loci, containing an HSV thymidine kinase marker, and subsequent integration of BAC constructs into the acceptor sites, utilizing the loxP and lox511 sites present in the vector backbones. The BAC reporter, with a Renilla luciferase cassette inserted downstream of the hTERT promoter, was retrofitted with a puromycin marker. Through puromycin selection and ganciclovir counter-selection, a targeting efficiency of over 50% was achieved. We demonstrated that the activity and chromatin structures of the hTERT promoter in chromosomally integrated BAC reporter recapitulated its endogenous counterpart of the host cells. Therefore, we have established a genetically amendable platform to study chromatin and epigenetic regulation of the hTERT gene. The highly efficient and versatile RMBT technique has general applicability for studying largely unexplored chromatin-dependent mechanisms of promoter regulation of various genes.
INTRODUCTION
Transcriptional regulation of gene expression is central to development, cell differentiation, and disease progression (1,2). Cumulative evidence has indicated that epigenetic and chromatin-dependent mechanisms play critical roles in regulating gene activities (3,4). For example, in addition to transcription factors that directly bind DNA, compositions and covalent modifications of histones, organization of nucleosomal structures, and DNA methylation are all playing vital roles in gene regulation.
Currently, transient transfection of small plasmid reporters is the most commonly used approach for analyzing cis-regulatory elements. Using this method, a large body of data has been accumulated on the structural and functional studies of cis-elements in the regulation of promoter activities. However, nucleoprotein complexes formed by transiently transfected plasmids were different from nucleosomes at the native loci (5). They had altered histone H1 stoichiometry and lacked nucleosome positioning (6). In addition, the transient transfection method could yield erroneous results, due to frequent formation of concatamers between cotransfected plasmid DNAs through non-homologous end-joining ligation (7), further complicating the interpretation of results from experiments involving cotransfection of a control reporter plasmid, a commonly used strategy.
Many cis-elements regulating local chromatin environment have been identified (8,9). Transient transfection assays are unsuitable for studying such elements. At present, methods to examine these chromatin elements are limited. For example, chromatin boundary elements or insulators are often taken out of their normal chromosomal contexts and tested in small plasmids for their ability to generate drug-resistant colonies (10). In many cases, the roles of such elements in their native chromatin context remain to be verified.
The hTERT gene, which encodes human telomerase reverse transcriptase, is silenced in most somatic cells but activated in immortal cell lines and cancer cells. We and others have previously reported that transiently transfected plasmid reporters containing the hTERT promoter did not recapitulate the endogenous promoter in several fibroblast lines, suggesting that distal-regulatory elements and/or native chromatin environment were required for its regulation (11–13). Ideally, one would like to directly analyze the endogenous hTERT locus to delineate cis-regulatory elements. However, any modifications of a native gene are technically arduous, if not impossible, by known methods, including homologous recombination that occurs at an extremely low efficiency in human somatic cells. Thus, it would be important to develop an hTERT reporter system that recapitulates its native chromatin context. First, such a reporter system should contain a large segment of genomic sequence including distal regulatory elements that are often dispersed in regions exceeding 100 kb. A large DNA construct, such as a bacterial artificial chromosome (BAC) that encompasses several loci, likely contains sufficient information to establish a chromatin environment similar to the native loci upon chromosomal integration. Second, this reporter should be integrated, preferably as a single-copy, into a pre-determined chromosomal acceptor site, allowing its mutant derivatives to be analyzed with identical genomic contexts. A number of recombination-based technologies are now available, such as BAC recombineering and heterologous site-specific recombination, making it feasible to develop such a system (14–18). In particular, heterologous site-specific recombinase (SSR, e.g. Cre, FLP and øC31) could catalyze efficient exchange of sequence cassettes at targeted chromosomal loci, a process known as recombinase-mediated cassette exchange (RMCE) (19). Indeed, several groups have succeeded in targeted insertion of various large DNA constructs, including BAC constructs, into pre-selected chromosomal sites in mammalian cell lines by using SSRs (20–24). This type of precise chromosomal integration would allow detailed mutagenesis studies of a chromatinized reporter at specific chromosomal sites.
To study the mechanisms of hTERT gene regulation in a relevant chromatin context, we developed a retrovirus-directed and recombination-mediated BAC targeting (RMBT) strategy to specifically integrate a modified BAC construct into pre-engineered chromosomal sites. We demonstrated that the targeting process was highly efficient despite that large BAC constructs were used. Upon chromosomal integration, the BAC construct containing the hTERT locus adopted a chromatin configuration similar to its native counterpart. The expression of integrated hTERT promoter closely resembled that of the endogenous promoter in the host cells. The minor variations of hTERT-promoter activity among independently targeted BAC reporter clones at the same integration sites were eliminated by a transient treatment of 5-azacytidine, an inhibitor of DNA methyltransferases. Therefore, our data revealed, for the first time, that the chromatin-dependent regulation of the hTERT promoter was re-established by inserting a large segment of human genomic DNA containing the hTERT locus into heterologous chromosomal sites in human cells.
MATERIALS AND METHODS
Plasmids and BACs
The construction of acceptor plasmid pML-2 is described in Supplementary Table S1. BAC reporter constructs 117B23cFtRvSVP (Figure 1) and 183M22cFtRvSVP were generated from the original BAC clones RPCI11-117B23 and RP24-183M22 (BACPAC Resources Center at Children's Hospital Oakland Research Institute, Oakland, CA). These reporters were constructed through a two-step recombination procedure (25), which was based on a BAC recombineering protocol from the Copeland lab (26). In this procedure, an rpsL+-kanamycin marker was first inserted into the site of modification by selecting for kanamycin resistance and subsequently replaced with a luciferase cassette by selecting against streptomycin sensitivity (Supplementary Figure S1), resulting in precise modifications without any unwanted sequences. The puromycin resistant marker was inserted into the vector backbone using a kanamycin selection marker, which was deleted via FLP-induced recombination in bacterial host EL350. Following each recombineering step, the modified BAC constructs were examined by digestion with at least four different restriction enzymes to ensure that only intended recombinations occurred, which were further confirmed by sequencing the junctions. For transfection, high quality BAC DNAs were prepared using a Genopure Plasmid Maxi kit (Roche) and examined by pulse field gel electrophoresis to ensure the integrity.
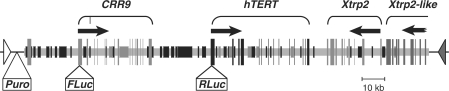
Figure 1.
A schematic illustration of the BAC reporter 117B23cFtRvSVP containing the hTERT, CRR9 and Xtrp2 loci. Exons are designated as long vertical bars and lines. Short black rectangles represent repetitive elements and short gray rectangles are mini-satellite sequences. Horizontal arrows indicate the directions of transcription. Fluc and Rluc, the Firefly and codo-optimized Renilla luciferase expression cassettes, respectively. Puro, the puromycin-resistant gene controlled by the SV40 promoter. The open and closed triangles show the positions and orientations of loxP and lox511 sites, respectively.
Cell culture and transfections
Immortal human fibroblast cell lines (13) were cultured in minimal essential medium containing 10% fetal bovine serum. Transfections were performed using FuGene 6 or FuGene HD reagent according to the manufacturer's recommendations (Roche). For stable clones, cells were selected first with 0.5 µg/ml puromycin for 3 days, followed by selection in 25 µM ganciclovir for 48 h. For 5-azacytidine treatment, cells were plated in 6-well plates at 30% confluence and treated with 0.5 µM 5-azacytidine (5-AC) (Sigma) the next day continuously for 9 days. Luciferase assays were performed before, at 9 days of treatment, 12 days and 20 days after the completion of treatment.
Gene expression analyses
For qRT–PCR, RNA isolation and cDNA synthesis were performed as described previously (27). Real-time PCR was performed in triplicates using an ABI StepOnePlus system. PCR primers and Taqman probes are summarized in Supplementary Table S2. Luciferase assays were performed as described previously (28). Luciferase activities were normalized to cell viability, as determined by thiazolyl blue tetrazolium bromide (MTT) assays (29).
Southern blotting and DNase I sensitivity assays
For characterization of acceptor loci and targeted BAC integrations, 3–5 µg genomic DNAs were digested with various restriction enzymes and Southern blots were hybridized to chicken β-globin insulator sequence cHS4 isolated from pUC19-cHS4 (Supplementary Table S1). For DNase I assays, nuclei preparation and genomic DNA extraction were performed as previously described (13,27).
RESULTS
Construction of a BAC reporter for monitoring transcriptional regulation of the hTERT gene
It has been previously shown that transiently transfected small plasmid reporters containing the hTERT promoter failed to recapitulate the endogenous hTERT gene expression (11,13). In fact, we have shown that chromatin environment played an important role in the regulation of the hTERT gene (27,30). In order to further study its regulatory mechanisms in depth, we constructed a reporter from a BAC clone (RPCI11-117B23) containing 160 kb of human genomic sequence, including the entire hTERT gene, the upstream CRR9 locus, and the downstream Xtrp2 locus (Figure 1). The CRR9 gene is constitutively expressed in all cell types examined, whereas expression of the Xtrp2 gene is only detected in kidney-derived cells (27,31). Because both upstream and downstream loci have very different expression patterns compared to the hTERT gene, this BAC presumably contains all the essential cis-regulatory elements for the correct regulation of hTERT gene.
For convenient and quantitative measurement of the hTERT promoter activity, a codon-optimized Renilla (Rluc) and a Firefly (Fluc) luciferase expression cassette were inserted into the initiation codons of the hTERT and CRR9 genes, respectively (Figure 1 and Supplementary Figure S1). The Fluc transcribed from the constitutive CRR9 promoter was intended to be an internal control for determining the relative level of hTERT promoter activity. For selection of BAC containing cells, a cassette containing a puromycin marker driven by the SV40 promoter and a kanamycin resistance gene surrounded by two FRT sites, was placed within the pBACe3.6 vector backbone immediately downstream of the lox511 site (Figure 1 and Supplementary Figure S1). The kanamycin marker was subsequently removed via FLP recombinase-mediated recombination (26). The final BAC reporter construct, 117B23cFtRvSVP, is shown in Figure 1.
Experimental strategy of targeted BAC integration
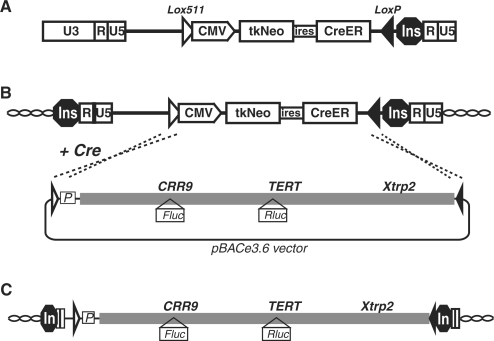
To develop a controlled BAC integration method, we designed a two-step BAC targeting scheme, RMBT, involving the establishment of pre-determined acceptor cell lines and subsequent site-directed chromosomal integrations of BAC reporter constructs (Figure 2). pBACe3.6 and pTARBAC1, the vector backbones used in the majority of currently available BAC libraries, had two lox sites, a lox511 and a loxP site (32). To make use of these sites for chromosomal integration, an acceptor vector, pML-2, was constructed (Figure 2A and Supplementary Table S1). pML-2 is a retroviral vector that contained a tkNeo ORF, a fusion protein of the HSV-thymidine kinase and neomycin resistance gene (33), and a CreER ORF, a fusion protein of Cre recombinase and the hormone-binding domain of human estrogen receptor. These two ORFs, separated by an internal ribosomal entry site (IRES), were transcribed from a CMV promoter. The selection cassette was ‘floxed’ by lox511 and loxP sites so that their recombinations with the lox sites in a BAC would lead to the replacement of the selection cassette by the entire insert of the BAC construct (Figure 2B). Cre recombinase catalyzes efficient homo-specific recombination of both loxP and lox511 sites with no observable in vivo hetero-specific recombination between them (34). Finally, an 1.2-kb insulator, containing the cHS4 element of the chicken β-globin locus (35), was inserted into the enhancer-less U3 region of the 3′ LTR. During retroviral transduction, the 3′LTR was copied to the 5′ end, inactivating the retroviral promoter and surrounding the entire vector by insulators, and thus shielding the acceptor locus from possible chromosomal positional effects [Figure 2C and (36)].
Figure 2.
Strategy of retrovirus-directed recombinase-mediated targeted BAC targeting (RMBT). (A) Acceptor retroviral vector pML-2. The 5′ LTR contains U3, R, and U5 elements, whereas U3 in the 3′ LTR was replaced by a 1.2-kb chicken β-globin insulator, cHS4 (Ins). (B) Cre-lox mediated recombination between a proviral acceptor locus and a BAC reporter. Following retroviral replication, the 5′ LTR is replaced by the 3′ LTR. (C) The integrated BAC reporter. Open and closed triangles mark the positions and orientations of lox511 and loxP sites, respectively. CMV, the cytomegalovirus major promoter; tkNeo, a fusion protein of herpes simplex virus thymidine kinase and neomycin-resistant protein; ires, internal ribosomal entry site; CreER, a fusion protein of Cre-recombinase and the hormone-binding domain of human estrogen receptor; P, puromycin-resistant marker. Thick gray lines represent the human sequences in the BAC construct; wavy lines, endogenous chromosomal sequences.
Establishment of acceptor lines
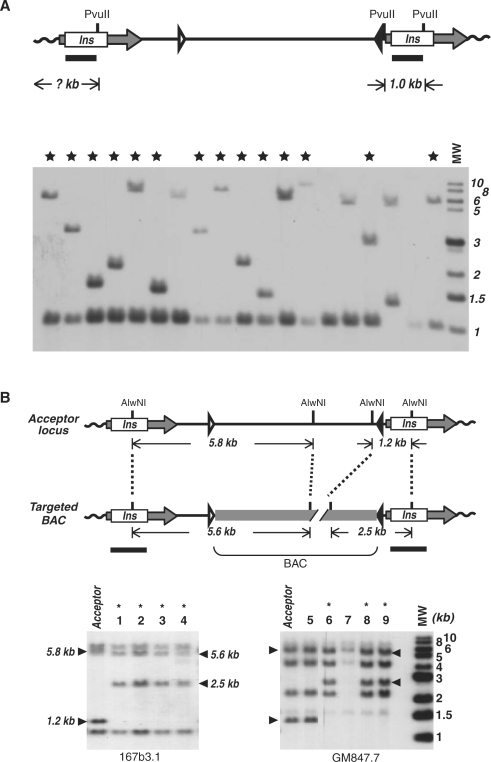
To test the RMBT strategy, we chose to use a pair of immortal fibroblast lines, telomerase-positive (tel+) 3C167b and telomerase-negative (tel−) 3C166a, which were derived from a common parental pre-crisis strain 3C (13). Another pair of cell lines, the tel+ GM639 and tel− GM847, were also used (37). Retroviruses derived from pML-2 were used to infect immortal cell lines at a low MOI (multiplicity of infection), minimizing multiple proviral insertions within a single cell. Stable neomycin-resistant clones were isolated and the copy number of proviral integrations in each clone was determined by Southern analysis (Figure 3A), in which Pvu II-digested genomic DNA fragments were hybridized to a portion of chicken β-globin insulator sequence. In more than half of the isolated clones, single proviral integrations occurred, which resulted in an internal DNA band of 1.0 kb and an external band of variable sizes. Two to three clones with single insertions of pML-2 provirus were utilized for the subsequent BAC targeting experiments.
Figure 3.
Southern analyses of acceptor loci and targeted BAC integrations. (A) Analysis of proviral acceptor loci. Upper panel is a diagram of a proviral acceptor locus following retroviral transduction. Horizontal arrows indicate defective LTR elements in which the U3 regions are replaced by the insulator cHS4 (Ins). Lox511 and loxP sites are shown as open and closed triangles, respectively. Lower panel shows a representative Southern analysis of acceptor loci. Genomic DNAs were digested with Pvu II and hybridized to a 32P-labeled Pvu II-Hind III fragment of cHS4. MW, molecular weight markers in kilo base pairs. Stars mark the acceptor clones with a single-copy provirus. (B) Analysis of integrated BAC constructs. Upper panel illustrates an acceptor locus before and after targeted BAC integration. The integrated BAC is shown as a gray thick line. Sizes of internal genomic bands recognized by the insulator probe are also indicated. Lower panels show representative Southern analyses of BAC constructs integrated into acceptor loci in 167b3.1 (left) and GM847.7 (right) cells. Genomic DNAs were digested with AlwN I and hybridized to a full-length insulator probe. As indicated by arrowheads, the 5.8-kb and 1.2-kb internal bands changed to 5.6 kb and 2.5 kb, respectively, upon integration of the BAC, whereas the two external bands specific to each acceptor locus were not affected. The positions of the Southern probe are shown as horizontal bars below the diagram. Asterisk indicates clones with targeted integration of the BAC reporter.
Chromosomal targeting of BAC reporter construct
To target the hTERT BAC reporter 117B23cFtRvSVP into an acceptor site, the circular BAC DNA was transfected into the acceptor cells. Although tamoxifen (4-HT) was able to induce Cre recombinase activity in acceptor cells (data not shown), we found that 4-HT treatment of transfected cells caused significant cell death in some of the host cell lines. We therefore opted to use a cotransfected Cre-expressing plasmid pCBM to supply the Cre recombinase. Transfected cells were initially selected with puromycin for three days, followed by recovery in drug-free medium until visible colonies were formed. About one week after transfection, ganciclovir (GCV) was added to the cells for 48 h to select for recombinant clones that had lost the tkNeo marker. Many of the initial puromycin-resistant colonies died upon GCV treatment and therefore contained randomly integrated BACs. The colonies that survived both puromycin and GCV selections were isolated, which presumably had undergone specific recombination events at the acceptor locus. About 80% of these dual drug resistant clones expressed both Fluc and Rluc, indicating that they likely contained the entire BAC reporter sequences (Table 1).
Table 1.
Chromosomal targeting of BAC vector and BAC reporter constructs
| Cell lines | GM639.3 |
GM639.6 |
167b3.1 |
GM847.7 |
166a11.1 |
Overall BAC | |||
|---|---|---|---|---|---|---|---|---|---|
| Input DNAs | pBACa SVpuroF | 117B23 cFtRvSVP | 117B23 cFtRvSVP | 117B23 cFtRvSVP | 183M22 cFhRvSVP | 117B23 cFtRvSVP | 183M22 cFtRvSVP | 117B23 cFtRvSVP | targeting |
| Vector backbone | pBACe3.6 | pBACe3.6 | pBACe3.6 | pBACe3.6 | pTARBAC1 | pBACe3.6 | pTARBAC1 | pBACe3.6 | |
| Total numbers of isolated clones | 30 | 9 | 20 | 29 | 40 | 18 | 33 | 53 | 202 |
| No. of clones with dual luciferases | ∼ | 9 | 20 | 22 | 37 | 18 | 32 | 22 | 160 |
| No. of clones examined by Southern analysisb | 5 | 9 | 14 | 16 | 16 | 13 | 16 | 12 | 96 |
| No. of clones with targeted integration | 5 | 8 | 9 | 15 | 14 | 8 | 11 | 5 | 70 |
| Targeting efficiencies among luciferase-positive clonesc | ∼ | 89% | 64% | 94% | 88% | 62% | 69% | 42% | 73% |
| Targeting efficiencies among all clonesd | 100% | 89% | 64% | 71% | 81% | 62% | 67% | 17% | 58% |
aA plasmid of 13.4 kb.
bOnly a subset of luciferase-expressing clones were examined by Southern analysis.
cTargeting efficiencies = numbers of targeted integration/numbers of clones examined by Southern analysis.
dTargeting efficiencies = efficiencies among luciferase-positive clones × number of clones with luciferases/total number of isolated clones.
To confirm that the BAC reporter was targeted into the acceptor loci, genomic DNAs of a subset of luciferase-expressing clones were digested with AlwN I and analyzed by Southern analysis using the chicken β-globin insulator as a probe. Shown in Figure 3B is a representative experiment of such analyses for acceptor lines 167b3.1 (left panel) and GM847.7 (right panel). In each case, four genomic fragments were detected from the acceptor loci, including two internal (1.2 and 5.8 kb) and two variable external fragments. Upon Cre-mediated recombination with the BAC reporter construct, the two internal bands changed to 2.5 kb and 5.6 kb, respectively, whereas the external bands remained the same. The integrity of integrated BAC reporters in a subset of these targeted clones was further verified by Southern analyses using multiple probes specific for different regions of the BAC reporter, including the hTERT and CRR9 promoter regions (data not shown). Although most puromycin and GCV resistant clones contained correctly targeted BAC reporter, some were not targeted. These non-targeted clones were likely escapers resulted from either epigenetic silencing of the CMV promoter in the acceptor locus (#5, Figure 3B) or loss of the acceptor locus (#7, Figure 3B), both leading to the loss of GCV sensitivity.
Successfully targeted integrations have been achieved in acceptor cell lines derived from tel+ 3C167b and GM639 and tel− 3C166a and GM847 cells (Table 1). For the pBACe3.6-derived plasmid pBACSVpuroF (13.4 kb), five out of five puromycin and GCV-resistant GM639.3 clones (100%) contained targeted integrations. The 173-kb BAC 117B23cFtRvSVP was successfully targeted into multiple acceptor loci from all these four cell lines. Since the vector pTARBAC1 was also used in many BAC libraries, we tested a pTARBAC1-derived BAC RP24-183M22, which contained 135-kb mouse genomic sequence encompassing the mTERT locus and the upstream mCRR9 and downstream mXtrp2 loci. This BAC clone was similarly modified to 183M22cFtRvSVP as the human BAC reporter. As shown in Table 1, the mTERT BAC reporter was also efficiently targeted into two acceptor loci in 3C167b and GM847 cells. The average BAC targeting efficiency was 58% overall, ranging from 17% to 89% in different acceptor lines, which was increased to 73%, ranging from 42% to 94%, when pre-screened for luciferase expressions. We also targeted several other BAC constructs and attained similar efficiencies (data not shown). The relatively low targeting efficiency of 166a11.1 cells seemed to be resulted from the nature of these tel− cells that utilized alternative lengthening of telomeres, the ALT mechanism. Continued dying of these cells during passaging made it difficult to recover and expand the isolated recombinant clones. Overall, with a combination of positive and negative selections, single-copy large BAC constructs were efficiently targeted into acceptor loci of various cell types through Cre-mediated recombination.
Activity of the integrated hTERT promoter in targeted BAC reporter
To determine the reporter activities in the targeted BAC clones, transcription from the integrated hTERT or CRR9 promoter was measured using Rluc or Fluc as readouts. Two independent clones from tel− GM847.1 cells and five from other acceptor lines, including both tel+ and tel− cells, were examined (Table 2). Rluc activity expressed from the hTERT promoter was much higher in tel+ 3C167b and GM639 cells than in tel− 3C166a and GM847 cells. On the other hand, Fluc activity expressed from the CRR9 promoter was similarly high in all cells, consistent with the constitutive expression pattern of the CRR9 gene. Therefore, the ratio of Rluc to Fluc activity was a reliable assessment of the hTERT promoter activity in the integrated BAC construct. As shown in Table 2, the ratio of Rluc to Fluc was higher in 167b3.1 clones than in GM639.3 and GM639.6 clones, consistent with the higher level of endogenous hTERT mRNA in 3C167b cells than in GM639 cells, as determined by real-time RT–PCR analysis. In contrast, 3C166a and GM847 cells expressed very little hTERT mRNA and consistently, the Rluc to Fluc ratio was significantly lower in these tel− cells. Furthermore, BAC reporters integrated in different acceptor loci of the same cell lines functioned essentially the same, indicating that the strong chicken β-globin insulator placed on both sides of the acceptor loci likely protected the integrated BAC reporter from potential chromosomal positional effects (38). We therefore conclude that the expression of integrated hTERT promoter in the targeted BAC reporter recapitulated the activity of the native promoter.
Table 2.
Transcription from integrated and native hTERT promoters
| Acceptor lines (targeted clones) | Rluca | Fluca | Rluc/Fluc | Endogenous hTERT mRNAb |
|
|---|---|---|---|---|---|
| hTERT/18S | hTERT/CRR9 | ||||
| 166a11.1 (tel−)c | 13.2 ± 7.8 | 404 ± 137 | 0.04 ± 0.03 | 0.02 ± 0.01 | 0.07 ± 0.04 |
| #8 | 14.4 ± 0.4 | 187 ± 3 | 0.08 ± 0.00 | ||
| #15 | 9.5 ± 0.4 | 392 ± 1 | 0.02 ± 0.00 | ||
| #26 | 3.8 ± 0.7 | 401 ± 2 | 0.01 ± 0.00 | ||
| #27 | 13.1 ± 0.3 | 547 ± 10 | 0.02 ± 0.00 | ||
| #47 | 25.1 ± 0.4 | 492 ± 6 | 0.05 ± 0.00 | ||
| 167b3.1 (tel+)c | 944 ± 575 | 559 ± 112 | 1.63 ± 0.71 | 104 ± 10 | 720 ± 71 |
| #1 | 720 ± 47 | 520 ± 38 | 1.39 ± 0.09 | ||
| #2 | 1968 ± 40 | 687 ± 1 | 2.86 ± 0.06 | ||
| #3 | 589 ± 13 | 483 ± 13 | 1.22 ± 0.03 | ||
| #4 | 697 ± 39 | 439 ± 25 | 1.59 ± 0.09 | ||
| #10 | 746 ± 27 | 668 ± 33 | 1.12 ± 0.04 | ||
| GM847.1 (tel−)c | 5.9 ± 2.7 | 484 ± 129 | 0.01 ± 0.00 | 0.02 ± 0.01 | 0.03 ± 0.01 |
| #26 | 4.0 ± 0.8 | 392 ± 3 | 0.01 ± 0.00 | ||
| #27 | 7.8 ± 2.1 | 576 ± 15 | 0.01 ± 0.00 | ||
| GM847.7 (tel−)c | 19 ± 5 | 476 ± 107 | 0.04 ± 0.02 | ||
| #5 | 12 ± 1 | 628 ± 40 | 0.02 ± 0.00 | ||
| #6 | 21 ± 2 | 437 ± 13 | 0.05 ± 0.01 | ||
| #7 | 24 ± 2 | 338 ± 10 | 0.07 ± 0.01 | ||
| #18 | 16 ± 1 | 519 ± 28 | 0.03 ± 0.00 | ||
| #19 | 20 ± 4 | 458 ± 5 | 0.04 ± 0.01 | ||
| GM639.3 (tel+)c | 93 ± 35 | 279 ± 48 | 0.33 ± 0.10 | 62 ± 3 | 75 ± 4 |
| #1 | 67 ± 7 | 241 ± 22 | 0.28 ± 0.03 | ||
| #2 | 69 ± 6 | 221 ± 6 | 0.31 ± 0.03 | ||
| #4 | 119 ± 2 | 281 ± 13 | 0.42 ± 0.01 | ||
| #5 | 67 ± 12 | 331 ± 12 | 0.20 ± 0.04 | ||
| #8 | 142 ± 8 | 318 ± 17 | 0.45 ± 0.02 | ||
| GM639.6 (tel+)c | 233 ± 96 | 791 ± 55 | 0.29 ± 0.12 | ||
| #9 | 387 ± 17 | 834 ± 25 | 0.46 ± 0.02 | ||
| #10 | 194 ± 33 | 790 ± 126 | 0.25 ± 0.04 | ||
| #11 | 145 ± 17 | 857 ± 44 | 0.17 ± 0.02 | ||
| #25 | 176 ± 8 | 744 ± 24 | 0.24 ± 0.01 | ||
| #26 | 261 ± 10 | 730 ± 45 | 0.36 ± 0.01 | ||
The bold numbers in these rows are averages of the numbers below.
aRelative levels of luciferase activities were normalized to amounts of live cells as determined by MTT assays.
bRelative levels of endogenous hTERT mRNA in parental immortal lines were determined by reverse transcription and quantitative PCR, normalized to either 18S ribosomal RNA or CRR9 mRNA.
cAverage luciferase activities in targeted clones at the same acceptor line.
Chromatin structures of the hTERT promoter in targeted BAC reporters
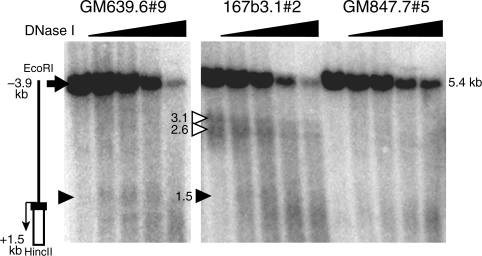
To determine the chromatin structures of the targeted BAC reporter, DNase I assays were performed to examine the integrated hTERT promoter. Nuclei from targeted clones 167b3.1#2, GM639.6#9 and GM847.7#5 were treated with DNase I and genomic DNA fragments were analyzed by Southern analysis for the presence of DNase I hypersensitive sites (DHSs) using a Rluc probe, which was specific for the integrated hTERT promoter. As shown in Figure 4, the integrated hTERT promoter coincided with a DHS (1.5 kb) in both 167b3.1#2 and GM639.6#9 cells, consistent with active transcription from the hTERT promoter in these tel+ cells. In contrast, the hTERT promoter was not sensitive to DNase I in tel− GM847.7#5 cells. Furthermore, two extra DHS bands (2.6 and 3.1 kb) were also identified in 167b3.1#2 cells, corresponding to DHSs at 1.1-kb and 1.6-kb upstream of the hTERT promoter, respectively. A closer examination of our previously published data indicated the presence of these two DHSs at the endogenous hTERT locus in 3C167b cells [Figure 4 in (13)]. Since these upstream DHSs were not present in GM639 and GM847 cells, they might contain binding sites for transcription activators and contributed to the higher level of hTERT transcription in 3C167b cells. Taken together, our experiments indicated that the hTERT promoter in integrated BAC recapitulated its endogenous promoter in host cells at the chromatin level.
Figure 4.
DNase I sensitivity of the hTERT promoter in integrated BAC. Nuclei were treated with 0, 2, 4, 8 and 16 units/ml DNase I for at 37°C for 20 min. Genomic DNAs were isolated, digested with EcoR I and Hinc II, analyzed by Southern analysis using the Rluc sequence as the probe. A diagram of the integrated hTERT promoter is shown on the left, in which the black rectangle represents a portion of the first hTERT exon. The white rectangle shows part of the Rluc sequence, and the thin arrow shows the transcription start site (TSS) and direction. The thick arrow designates the 5.4-kb EcoR I-Hinc II full-length genomic band. The positions of restriction sites are relative to the TSS. The closed triangles denote the 1.5-kb DHS bands at the hTERT promoter. The two open triangles indicate the two additional hypersensitive bands (2.6 kb and 3.1 kb) in 167b3.1 cells, corresponding to DHSs that are 1.1-kb and 1.6-kb upstream of the hTERT promoter. The sizes of DNase I hypersensitive bands are indicated by numbers in kb.
Effects of inhibition of DNA methyltransferases on the integrated hTERT promoter activity
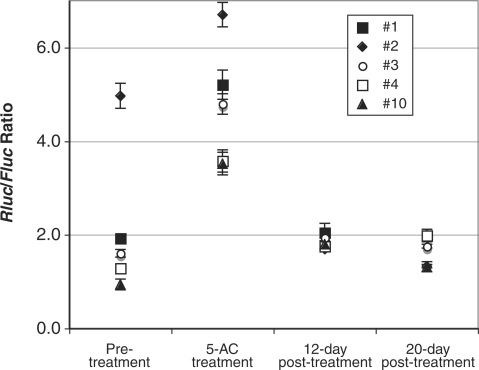
Independent clones with the hTERT BAC reporter targeted at the same acceptor site had identical genomic contexts and were thus expected to have the same promoter activity. Although differences of Rluc activity among clones derived from the same acceptor lines were relatively small, as compared to the disparities between tel+ and tel− cells (Table 2), such differences were reproducible in repeated experiments (data not shown). In fact, we observed a tendency of increase in clonal variations upon passaging cells, especially among clones of tel+ 167b cells. To determine whether DNA methylation was a cause of such clonal variations, five-targeted clones derived from the 167b3.1 acceptor line were treated with 5-azacytidine (5-AC). 5-AC is a potent inhibitor of DNA methyltransferases DNMT1 and DNMT3a and known to induce demethylation and reactivation of a number of silenced genes (39,40). As shown in Figure 5, Rluc to Fluc ratio ranged 1–5 prior to 5-AC treatment. Immediately following treatment with 0.5 µM 5-AC for 9 days, the ratio of Rluc to Fluc increased significantly in all five clones and reached 3–7, suggesting that DNA methylation contributed to the suppression of integrated hTERT promoter in these clones. Interestingly, when these treated cells were cultured in the absence of the drug for 12 days, the Rluc to Fluc ratio reduced to about 2 in all clones. Further culturing of these cells led the Rluc/Fluc ratios to diverge again. Therefore, accumulated DNA methylation during cell proliferation was likely the cause of clonal variations among the BAC reporters integrated at the same chromosomal site.
Figure 5.
Effects of inhibition of DNA methylasetransferases on hTERT promoter reporter activities in integrated BACs. Five 3C167b3.1 clones containing the BAC reporter at the same acceptor locus were treated with 0.5 µM 5-AC for 9 days and luciferase activities were determined. Pre-treatment, 5-AC treatment, 12-day, and 20-day post-treatment: immediately before and after the 9-day treatment with 5-AC, 12 and 20 days after the completion of treatment, respectively.
DISCUSSION
Chromatin structures and epigenetic marks are ultimately determined by the underlining primary DNA sequences and the differentiation status of host cells. It has become increasingly evident that such chromatin and epigenetic mechanisms are critical for the regulation of gene expression. We have previously shown that randomly integrated BAC reporter of the hTERT gene, similar to the reporter used in this study, was able to undergo silencing upon osteogenic differentiation of mouse embryonic stem cells (28). However, uncontrolled random reporter integration was inadequate for analyzing cis-regulatory elements, because each integration event differed in chromosomal position effects, copy numbers, and structural integrities, making it difficult to further dissect the regulatory sequences via deletions and site-directed mutagenesis. To establish a system for such mechanistic studies, we developed the strategy of retrovirus-directed recombinase-mediated BAC targeting or RMBT. First, clones with a single-copy acceptor locus were created by retroviral transduction and neomycin selection. Second, large BAC constructs, upon transfection into cells, integrated into the acceptor sites via Cre-lox mediated recombination and a puromycin/ganciclovir selection scheme. This targeted integration procedure was highly efficient, as the majority of isolated clones contained the entire BAC reporter at the acceptor site. This efficiency was further increased with additional screening for the luciferase activities. We compared luciferase expression of the BAC reporters targeted at six acceptor loci in four different cell lines with their endogenous counterparts. Rluc expression from the integrated hTERT promoter largely correlated with the endogenous hTERT mRNA levels in host cells, whereas Fluc expression from the CRR9 promoter was similarly high in all cell lines. The ratio of Rluc to Fluc, a reliable assessment of the integrated hTERT promoter activity, thereby paralleled the endogenous hTERT mRNA level. The luciferase expression data, together with the formation of multiple DHSs at and upstream of the integrated hTERT promoter, indicated that expression of the endogenous hTERT promoter in host cells was recapitulated by that of the transgenic BAC reporter, whereas all previous attempts to model the endogenous hTERT expression by transiently transfected reporters were unsuccessful (11,13).
During initial experimental designing, we included a hormone inducible Cre recombinase (CreER) in the acceptor cassette as the source of recombinase activity. While the Cre activity was induced by tamoxifen in acceptor lines, the hormone treatment also caused significant cell death in a subset of cell lines, especially tel− ALT cells. Thus, a transiently transfected Cre-expressing plasmid was used to supply the recombinase activity. Nevertheless, compared to their parental cells, the pre-targeted acceptor cell lines did not show any change in the rate of cell proliferation in the absence of hormone. Besides, the entire cassette was excised upon targeted integration of the BAC constructs. Therefore, although the CreER ORF will be removed in future experiments, we reasoned that the presence of CreER sequence in the pre-targeted acceptor loci would not have any effects on the interpretation of the data from current experiments.
Although an integrated BAC reporter at the same acceptor site would be expected to adopt the same chromatin configuration in independent clones, we did detect, albeit relatively small, differences in reporter activities among these clones. As shown in Figure 5, these clonal variations were largely eliminated by a transient inhibition of DNA methyltransferases, suggesting that the variations were caused by DNA methylation accumulated during cell proliferation.
It has been reported that the hTERT promoter subjected to methylation in cultured cells and many cancer-derived cell lines, but the reported effects of DNA methylation on hTERT transcription were variable. Whereas some studies showed that methylation of the hTERT promoter had negative or no effect on hTERT regulation (41,42), another study demonstrated that hypermethylation of the hTERT promoter correlated with higher telomerase expression level in cancer cells (43). Although our previous data showed that transcriptional silencing of the hTERT gene did not require CpG methylation at its promoter (28), our current study suggested that DNA methylation was likely a cause of variations in hTERT promoter activation in tel+ cells. For example, Rluc expression in the 3C167b#2 clone was initially higher than in its sibling clones. Interestingly, following the 5-AC treatment and recovery, its Rluc activity returned to a level very similar to those of its sibling clones. This result indicated that stochastic methylation of specific cis-elements in the BAC reporter, such as repressors of the hTERT promoter, might lead to an increase of the hTERT reporter promoter activity in 3C167b3.1#2 cells. Alternatively, DNA methylation could inactivate a negative trans regulator and thus indirectly activated the hTERT promoter. However, the endogenous hTERT mRNA level in 3C167b3.1#2 cells was similar to that in other 3C167b clones (data not shown), suggesting that the second possibility was unlikely.
Despite the minor variations, analysis of gene expression using RMBT provides many advantages over conventional methods. First, cis-acting elements, such as enhancers, chromatin barriers/insulators, and locus control regions, may be dispersed over hundreds of kilobases from the promoter (44). Small plasmid reporters are therefore not suited for studying the structures and functions of these chromatin elements in their native contexts. BAC constructs contain up to 300 kb of genomic sequences and likely include all of such distal chromatin elements. Second, integration of the BAC reporters allows the DNA to be assembled as part of the chromosomes. BAC constructs often contain sufficient sequences to establish a chromatin environment that is similar, if not completely identical, to their endogenous counterparts. Third, a single copy of the BAC reporter is integrated at the acceptor locus. Thus, the presence of integrated sequence unlikely alters the stoichiometry of sequence-specific DNA-binding factors. Single-copy integrations also allow easier detection of unintended rearrangements within the integrated reporter and ensure its structural integrity. Most importantly, modified BAC constructs, containing deletions or other mutations, can be reproducibly integrated into the same acceptor sites and evaluated in the same genetic and chromatin context.
Recently, several other groups have reported methods for site-specific integration of large DNA constructs. For example, Dafhnis-Calas et al. reported the first site-specific integration of BAC constructs by RMCE using øC31 and Cre recombinases (24). For studying gene regulation, Wallace et al. reported the replacement of an 85-kb mouse genomic fragment containing the α-globin gene cluster with a 117-kb human synthetic sequence by recombinase-mediated genomic replacement, or RMGR (22). RMGR involved two gene-targeting procedures to insert lox sites at both ends of a genomic region, which was subsequently replaced by a pre-engineered BAC sequence via Cre-mediated recombination. Prosser et al. introduced an acceptor containing loxP and lox511 sites into the Hprt locus in a mouse embryonic stem cell line, in which a BAC containing myosin VIIa gene was subsequently integrated (21). Similar to RMBT, these techniques offered well-controlled experimental approaches to study the genetic and epigenetic regulation of a chromosomal segment. However, these strategies were not readily adaptable for studies of other genomic loci in a large variety of cell types.
In this regard, RMBT is a novel approach and possesses several advantages in its efficiency and general applicability. First, retroviral transduction of acceptor sites is a much simpler procedure and can be used in different cell types, including those where homologous recombination is difficult to achieve. Second, RMBT takes advantage of the existing loxP/lox511 sites in the BAC vector backbones. Retrofitting most BAC clones for RMBT requires only a single recombineering step. Third, a pair of cHS4 insulators was designed to surround the entire acceptor locus to minimize chromosomal position effects. Due to the scope of this study, we did not provide the data supporting that these insulators functioned to reduce chromosomal position effects on the targeted BAC loci. However, numerous previous reports have demonstrated that similarly positioned cHS4 insulators protected retrovirus vectors from chromosomal position effects and increased the probability of retroviral expression at various chromosomal integration sites (36,45). Finally, compared to previously published methods, the targeting efficiency of RMBT was very high. In fact, all transfections for each specific targeting described in this study were performed at small scales (105 cells and 1–2 µg of BAC DNA in 6-well plates), and thus handling a relatively large number of different BAC constructs in several cell types simultaneously is feasible.
In conclusion, the RMBT strategy is highly efficient and versatile for targeted single-copy integration of large BAC constructs. Combined with mutagenesis of BAC constructs using recombineering (14), the method would offer a genetically amendable system for dissecting the chromatin-dependent and epigenetic mechanisms of hTERT gene regulation as well as the regulation of many other genes in their native genomic contexts. In addition, the strategy can also be adapted to study regulatory elements for DNA replication and DNA damage repair in vivo, and to generate transgenic animals by inserting large BAC constructs into defined chromosomal loci.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM071725). Funding for open access charge: National Institutes of Health (GM071725).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Frank Buchholz for the generous gifts of pACYC and pTNFUS69 plasmids and Dr Robert Paulson for his critical comments on the manuscript. We also thank Anne Stanley, David Stanford and Joe Bednarczyk of the Macromolecular and Molecular Genetics Core Facilities at Penn State College of Medicine for their excellent services.
REFERENCES
- 1.Jones KA. Transcription strategies in terminally differentiated cells: shaken to the core. [comment] Genes Dev. 2007;21:2113–2117. doi: 10.1101/gad.1598007. [DOI] [PubMed] [Google Scholar]
- 2.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 3.Wolffe AP, Matzke MA. Epigenetics: regulation through repression. Science. 1999;286:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- 4.Higgs DR, Vernimmen D, Hughes J, Gibbons R. Using genomics to study how chromatin influences gene expression. Annu. Rev. Genom. Hum. Genet. 2007;8:299–325. doi: 10.1146/annurev.genom.8.080706.092323. [DOI] [PubMed] [Google Scholar]
- 5.Jeong S, Stein A. Micrococcal nuclease digestion of nuclei reveals extended nucleosome ladders having anomalous DNA lengths for chromatin assembled on non-replicating plasmids in transfected cells. Nucleic Acids Res. 1994;22:370–375. doi: 10.1093/nar/22.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hebbar PB, Archer TK. Altered histone H1 stoichiometry and an absence of nucleosome positioning on transfected DNA. J. Biol. Chem. 2008;283:4595–4601. doi: 10.1074/jbc.M709121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishikawa T, Lee EJ, Jameson JL. Nonhomologous end-joining ligation transfers DNA regulatory elements between cointroduced plasmids. Mol. Cell Biol. 2004;24:8323–8331. doi: 10.1128/MCB.24.19.8323-8331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saitoh N, Bell AC, Recillas-Targa F, West AG, Simpson M, Pikaart M, Felsenfeld G. Structural and functional conservation at the boundaries of the chicken beta-globin domain. EMBO J. 2000;19:2315–2322. doi: 10.1093/emboj/19.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Harju S, Peterson KR. Locus control regions: coming of age at a decade plus. Trends Genet. 1999;15:403–408. doi: 10.1016/s0168-9525(99)01780-1. [DOI] [PubMed] [Google Scholar]
- 10.Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 11.Ducrest AL, Amacker M, Mathieu YD, Cuthbert AP, Trott DA, Newbold RF, Nabholz M, Lingner J. Regulation of human telomerase activity: repression by normal chromosome 3 abolishes nuclear telomerase reverse transcriptase transcripts but does not affect c-Myc activity. Cancer Res. 2001;61:7594–7602. [PubMed] [Google Scholar]
- 12.Kirch HC, Ruschen S, Brockmann D, Esche H, Horikawa I, Barrett JC, Opalka B, Hengge UR. Tumor-specific activation of hTERT-derived promoters by tumor suppressive E1A-mutants involves recruitment of p300/CBP/HAT and suppression of HDAC-1 and defines a combined tumor targeting and suppression system. Oncogene. 2002;21:7991–8000. doi: 10.1038/sj.onc.1205965. [DOI] [PubMed] [Google Scholar]
- 13.Wang S, Zhu J. Evidence for a relief of repression mechanism for activation of the human telomerase reverse transcriptase promoter. J. Biol. Chem. 2003;278:18842–18850. doi: 10.1074/jbc.M209544200. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 15.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 16.Muyrers JP, Zhang Y, Stewart AF. Techniques: recombinogenic engineering—new options for cloning and manipulating DNA. Trends Biochem. Sci. 2001;26:325–331. doi: 10.1016/s0968-0004(00)01757-6. [DOI] [PubMed] [Google Scholar]
- 17.Bouhassira EE, Westerman K, Leboulch P. Transcriptional behavior of LCR enhancer elements integrated at the same chromosomal locus by recombinase-mediated cassette exchange. Blood. 1997;90:3332–3344. [PubMed] [Google Scholar]
- 18.Wirth D, Gama-Norton L, Riemer P, Sandhu U, Schucht R, Hauser H. Road to precision: recombinase-based targeting technologies for genome engineering. Curr. Opin. Biotechnol. 2007;18:411–419. doi: 10.1016/j.copbio.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 19.Baer A, Bode J. Coping with kinetic and thermodynamic barriers: RMCE, an efficient strategy for the targeted integration of transgenes. Curr. Opin. Biotechnol. 2001;12:473–480. doi: 10.1016/s0958-1669(00)00248-2. [DOI] [PubMed] [Google Scholar]
- 20.Testa G, Zhang Y, Vintersten K, Benes V, Pijnappel WW, Chambers I, Smith AJ, Smith AG, Stewart AF. Engineering the mouse genome with bacterial artificial chromosomes to create multipurpose alleles. Nat. Biotechnol. 2003;21:443–447. doi: 10.1038/nbt804. [DOI] [PubMed] [Google Scholar]
- 21.Prosser HM, Rzadzinska AK, Steel KP, Bradley A. Mosaic complementation demonstrates a regulatory role for myosin VIIa in actin dynamics of stereocilia. Mol. Cell Biol. 2008;28:1702–1712. doi: 10.1128/MCB.01282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wallace HA, Marques-Kranc F, Richardson M, Luna-Crespo F, Sharpe JA, Hughes J, Wood WG, Higgs DR, Smith AJ. Manipulating the mouse genome to engineer precise functional syntenic replacements with human sequence. Cell. 2007;128:197–209. doi: 10.1016/j.cell.2006.11.044. [DOI] [PubMed] [Google Scholar]
- 23.Pscherer A, Schliwka J, Wildenberger K, Mincheva A, Schwaenen C, Dohner H, Stilgenbauer S, Lichter P. Antagonizing inactivated tumor suppressor genes and activated oncogenes by a versatile transgenesis system: application in mantle cell lymphoma. FASEB J. 2006;20:1188–1190. doi: 10.1096/fj.05-4854fje. [DOI] [PubMed] [Google Scholar]
- 24.Dafhnis-Calas F, Xu Z, Haines S, Malla SK, Smith MC, Brown WR. Iterative in vivo assembly of large and complex transgenes by combining the activities of phiC31 integrase and Cre recombinase. Nucleic Acids Res. 2005;33:e189. doi: 10.1093/nar/gni192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S, Zhao Y, Leiby M, Zhu J. A new positive/negative selection scheme for precise BAC recombineering. Mol. Biotechnol. 2009;42:110–116. doi: 10.1007/s12033-009-9142-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 27.Wang S, Zhu J. The hTERT gene is embedded in a nuclease-resistant chromatin domain. J. Biol. Chem. 2004;279:55401–55410. doi: 10.1074/jbc.M411352200. [DOI] [PubMed] [Google Scholar]
- 28.Wang S, Hu C, Zhu J. Transcriptional silencing of a novel hTERT reporter locus during in vitro differentiation of mouse embryonic stem cells. Mol. Biol. Cell. 2007;18:669–677. doi: 10.1091/mbc.E06-09-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 30.Wang S, Zhao Y, Hu C, Zhu J. Differential repression of human and mouse TERT genes during cell differentiation. Nucleic Acids Res. 2009;37:2618–2629. doi: 10.1093/nar/gkp125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S, Robertson GP, Zhu J. A novel human homologue of Drosophila polycomblike gene is up-regulated in multiple cancers. Gene. 2004;343:69–78. doi: 10.1016/j.gene.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 32.Frengen E, Weichenhan D, Zhao B, Osoegawa K, van Geel M, de Jong PJ. A modular, positive selection bacterial artificial chromosome vector with multiple cloning sites. Genomics. 1999;58:250–253. doi: 10.1006/geno.1998.5693. [DOI] [PubMed] [Google Scholar]
- 33.Chan MF, van Amerongen R, Nijjar T, Cuppen E, Jones PA, Laird PW. Reduced rates of gene loss, gene silencing, and gene mutation in Dnmt1- deficient embryonic stem cells. Mol. Cell Biol. 2001;21:7587–7600. doi: 10.1128/MCB.21.22.7587-7600.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soukharev S, Miller JL, Sauer B. Segmental genomic replacement in embryonic stem cells by double lox targeting. Nucleic Acids Res. 1999;27:e21. doi: 10.1093/nar/27.18.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 36.Emery DW, Yannaki E, Tubb J, Nishino T, Li Q, Stamatoyannopoulos G. Development of virus vectors for gene therapy of beta chain hemoglobinopathies: flanking with a chromatin insulator reduces gamma- globin gene silencing in vivo. Blood. 2002;100:2012–2019. doi: 10.1182/blood-2002-01-0219. [DOI] [PubMed] [Google Scholar]
- 37.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bell AC, West AG, Felsenfeld G. Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- 39.Laird PW, Jaenisch R. The role of DNA methylation in cancer genetic and epigenetics. Annu. Rev. Genet. 1996;30:441–464. doi: 10.1146/annurev.genet.30.1.441. [DOI] [PubMed] [Google Scholar]
- 40.Liang G, Gonzales FA, Jones PA, Orntoft TF, Thykjaer T. Analysis of gene induction in human fibroblasts and bladder cancer cells exposed to the methylation inhibitor 5-aza-2′-deoxycytidine. Cancer Res. 2002;62:961–966. [PubMed] [Google Scholar]
- 41.Dessain SK, Yu H, Reddel RR, Beijersbergen RL, Weinberg RA. Methylation of the human telomerase gene CpG island. Cancer Res. 2000;60:537–541. [PubMed] [Google Scholar]
- 42.Devereux TR, Horikawa I, Anna CH, Annab LA, Afshari CA, Barrett JC. DNA methylation analysis of the promoter region of the human telomerase reverse transcriptase (hTERT) gene. Cancer Res. 1999;59:6087–6090. [PubMed] [Google Scholar]
- 43.Guilleret I, Yan P, Grange F, Braunschweig R, Bosman FT, Benhattar J. Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase activity. Int. J. Cancer. 2002;101:335–341. doi: 10.1002/ijc.10593. [DOI] [PubMed] [Google Scholar]
- 44.Heintz N. BAC to the future: the use of bac transgenic mice for neuroscience research. Nat. Rev. Neurosci. 2001;2:861–870. doi: 10.1038/35104049. [DOI] [PubMed] [Google Scholar]
- 45.Pannell D, Ellis J. Silencing of gene expression: implications for design of retrovirus vectors. Rev. Med. Virol. 2001;11:205–217. doi: 10.1002/rmv.316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.