Abstract
ChIP-Seq technology, which combines chromatin immunoprecipitation (ChIP) with massively parallel sequencing, is rapidly replacing ChIP-on-chip for the genome-wide identification of transcription factor binding events. Identifying bound regions from the large number of sequence tags produced by ChIP-Seq is a challenging task. Here, we present GLITR (GLobal Identifier of Target Regions), which accurately identifies enriched regions in target data by calculating a fold-change based on random samples of control (input chromatin) data. GLITR uses a classification method to identify regions in ChIP data that have a peak height and fold-change which do not resemble regions in an input sample. We compare GLITR to several recent methods and show that GLITR has improved sensitivity for identifying bound regions closely matching the consensus sequence of a given transcription factor, and can detect bona fide transcription factor targets missed by other programs. We also use GLITR to address the issue of sequencing depth, and show that sequencing biological replicates identifies far more binding regions than re-sequencing the same sample.
INTRODUCTION
Chromatin immunoprecipitation, or ‘ChIP’, allows for the capture of the binding events between transcription factors or other DNA binding proteins and their targets in vivo at the moment of biochemical cross-linking. With the development of ‘ChIP-on-chip’ technology, the near genome-wide location analysis of binding sites for transcription factors became a reality (1). While this technology has greatly improved our understanding of transcriptional regulation in mammals, it is limited by the type of microarray platform used for the hybridization, in terms of spatial resolution and genomic regions that can be covered (1–3). ChIP-Seq technology addresses these issues, providing sequences for target regions anywhere in the genome with dramatically improved spatial resolution (2–9).
While ChIP-Seq technology offers many advantages over ChIP-on-chip, the large amount of data produced from each run (>1 Gb of sequence) poses a challenge for the accurate identification of transcription factor binding sites (3,8). Over the last few months, a number of new methods have been released which attempt to address these challenges (8–17). Many initial approaches did not employ control (i.e. input derived) datasets to eliminate falsely called binding regions that occur due to sequencing biases (6,10,11,15). More recent methods enable the user to specify a control data set to eliminate false positive regions that result from these biases (8,9,12–14,16,17). For instance, the CisGenome software system uses a conditional binomial model to identify enriched regions when a control data set is provided and includes an option for incorporating sequence strand information (16). MACS (Model-based Analysis of ChIP-Seq) uses the control dataset to model the tag distribution across the genome using the Poisson distribution (λBG) (14). After identifying candidate peaks that are significantly enriched over λBG, a local λ is estimated using windows around each peak to eliminate local biases (14). The PeakSeq algorithm is a two-step process that first identifies regions enriched compared to a null background model, and then returns regions that are statistically significant after taking ‘genome-mappability’ and control data into account (17). QuEST (Quantitative Enrichment of Sequence Tags) employs control data sets to eliminate false positive regions, and also to estimate a false discovery rate (FDR) (13). QuEST first calculates a ‘peak shift’ based on profiles generated from forward and reverse sequence tags reads. Once the shift is estimated, profiles are combined and peaks are called based on the enrichment of ChIP sequence tags to control sequence tags in the same region. The default parameter settings for QuEST are very stringent, yielding very small numbers of targets if the antibody used for ChIP provides only weak to moderately enriched regions. The SISSRS (Site Identification from Short Sequence Reads) algorithm utilizes sequence strand information to identify binding sites, which eliminates false positives. However, this approach may be too stringent for some applications, as only the strongest binding sites will contain sufficient sequence tags to fit the SISSRS model. When ChIP-Seq is used to identify binding regions for a transcription factor that has not been well studied, adjusting parameters without knowing the expected number of binding sites and without knowing the affinity of the antibody can be a difficult task.
Here, we introduce a novel method, termed GLITR (GLobal Identifier of Target Regions), to address some of the important issues with ChIP-Seq analysis. GLITR randomly samples sets of control sequence tags to accurately estimate a fold-change for each region identified in a target dataset. Following fold-change calculation, GLITR uses a classification method that incorporates two values, peak height and fold-change, to identify regions that are enriched above a specified FDR, which is calculated by comparing ChIP classification results to pseudo-ChIP (a sample of control sequence tags) classification results. By combining two attributes of a region GLITR greatly improves the ability to distinguish signal from noise in ChIP-Seq data. This is important because solely using peak height to identify targets leads to inclusion of multiple false-positives corresponding to regions that are also sequenced in control samples. Likewise, relying only on fold-change values is problematic, because a high-fold change cutoff eliminates many targets while a low fold-change, which is common in pseudo-ChIP data, drastically increases the number of false positives. After discussing the importance of using control DNA in ChIP-Seq experiments, we establish that sequencing input DNA from different tissues yields comparable results. We then compare the ability of GLITR to identify binding regions in ChIP-Seq data, obtained from sequencing Foxa2 ChIP material from adult mouse liver, to current published methods. We show that while all methods are able to identify regions that have the strongest Foxa2-binding sites, when moving deeper into a target list only GLITR continues to discover regions with a strong match to the Foxa2 consensus. Additionally, we show that the experimental design used to obtain sufficient sequencing tags greatly influences the regions identified as occupied by a transcription factor.
MATERIALS AND METHODS
Software availability
The GLITR software and the data described in this study are available at http://web.me.com/kaestnerlab1/GLITR/.
ChIP-Seq library construction and sequencing
ChIP was performed as described previously (18). ChIP-Seq libraries were prepared for four ChIPs performed on four mouse livers. The libraries were prepared as per Illumina's instructions (http://www.illumina.com). Briefly, ChIP sample DNA fragments were blunted, phosphorylated, and ligated to library adapters provided through Illumina. For input DNA preparation, 10 ng of starting material was used. Following ligation, size selection was performed by gel electrophoresis by excising DNA fragments at 200 ± 25 base pairs. Following gel purification, PCR amplification was performed [30 s at 98°C; (10 s at 98°C, 30 s at 65°C, 30 s at 72°C) × 18 cycles; 5 min at 72°C]. Amplified material was run on the Agilent 2100 bioanalyzer using the DNA 1000 Kit to ensure proper size selection, and was subsequently diluted to a concentration of 10 nM. These products were sequenced on the Illumina 1G Genome Analyzer at a concentration of 3–4 pM.
Data processing
Genome Analyzer sequencing output was analyzed using the Genome Analyzer Pipeline provided by Illumina. Sequence tags that aligned uniquely to the mouse genome build MM8 with zero, one, or two mismatches, according to the ELAND alignment algorithm, were used for further analysis.
GLITR algorithm
GLITR was developed using the framework of the ChIPSeq Peak Finder (http://woldlab.caltech.edu/html/chipseq_peak_finder versions 0.9 and 1.6) (5). While the method used for binding region identification is significantly different from the method used by ChIPSeq Peak Finder, a small set of variable names remain unchanged. All Perl and Python scripts needed to run GLITR are called from the main script, GLITR.pl. As input, GLITR requires a file containing the chromosome, start coordinate and strand for every uniquely aligned sequence tag in a ChIP data set and a control data set. GLITR extends sequence tags to the expected fragment length, as specified by the user. GLITR then chooses a set of pseudo-ChIP tags from the control data, which has the same number of tags as ChIP data. These tags are removed from the control set, which is then used as background. A directory is created for ChIP data as well as pseudo-ChIP data, and a Perl script is run to separate files by chromosome to increase efficiency. Overlapping sets of tags, where the number of tags must be at least three, are grouped into regions, and regions are split and trimmed if there is only one tag at any base in the region. Following region identification in ChIP and pseudo-ChIP data, a script is called that randomly samples background tags into datasets that are the same size as the ChIP data. For each of these background samples, the fold-change is calculated for every ChIP and pseudo-ChIP region. The fold-change reported is the average fold-change for the entire region, and is calculated by taking the average of the number of ChIP (or pseudo-ChIP) tags divided by the number of tags in the sampled background set at each position in the region. The median fold-change over each background sample is used for further analysis.
FDR calculation
Each ChIP region and pseudo-ChIP region is assigned a pair of coordinates, which correspond to the log2 of peak height and median fold-change value. Before calculating the Euclidean distance between coordinates, peak height values are variance-normalized by dividing them by the standard deviation of all values. Fold-change values are also variance-normalized. For every ChIP region, the k-nearest neighbors, whether a ChIP or pseudo-ChIP point are identified. A region is considered bound if n out of k nearest neighbors are also ChIP points. These bound points are considered true positives. Using the same n, every pseudo-ChIP point is ‘swapped’ for a random ChIP point, and is considered bound, or a false positive, if n out of its k nearest neighbors are ChIP points. The FDR is then calculated as the false positive proportion divided by the true positive proportion.
Background sequence generation
For de novo motif analysis, as well as generation of receiver operating characteristic (ROC) curves, background sequences are required. To generate background sequence sets, target sequences are binned in a two dimensional array by floor (log2) of both GC% and length of the region. For every target sequence in every bin, three background sequences that fall into the same bin are selected randomly from the genome.
De novo motif analysis
When performing de novo motif analysis, a background sequence set specific to the target sequence set was generated as described above. For all de novo motif analysis, Bioprospector was run 100 times, and a boot-strapping approach was employed using the top scoring motif to generate the positional weight matrix (PWM) used in scanning. For a position to be used in the final PWM, it was required that Bioprospector report it in 90 out of 100 runs.
Quantifying PWM enrichment
PWMs generated through de novo motif analysis were scanned as described previously (18). Scanning was performed on target regions as well as background regions, generated as described above. Enrichment of PWMs was quantified by calculating the area under the ROC curve (AUC), as described previously (18). An AUC of 0.5 corresponds to no enrichment.
Comparison of ChIP-Seq analysis methods
To compare GLITR to other methods, we used approximately a 1.5% FDR, which corresponds to 4051 target regions. To rank these regions by peak height and median fold-change simultaneously, we used a 2D rotation across the best-fit linear regression line of these points, and then sorted by the transformed x-coordinates. We ran MACS using default cutoff parameters, and sorted regions according to the FDR. Because the default cutoff parameters in SISSRS returns 2040 regions, we increased the E-value to 1000 and p-value to 0.5, to obtain 7662 regions. These regions were sorted by p-value, and only the top 4000 were used for comparative analysis. Because the default cutoff parameter settings for QuEST returns only 34 regions, we decreased the ChIP threshold from 0.61 to 0.08. Again, only the top 4000 were used for comparative analysis and QuEST regions were sorted by the reported ‘locmax’ value. CisGenome was run using default cutoff parameters, and were already in sorted format. Default cutoff parameters for PeakSeq returned 2994 regions, and in order to obtain enough regions for comparative analysis we set a p-value of 0.095. PeakSeq regions were sorted by q-value.
Quantitative RT-PCR
Real-time PCR reactions were assembled using SYBR GreenER (Invitrogen). Reactions were performed using the Mx3000 PCR System (Stratagene). The enrichment of target genes was calculated using the 28S rRNA locus as a reference for non-specific DNA, and was calculated by comparing input (sheared genomic DNA) to ChIP material. Primer sequences are provided in Supplementary Table 1.
RESULTS
Analysis method: the GLITR algorithm
We performed ChIP-Seq using a Foxa2-specific antibody on four adult mouse livers. After library preparation, cluster generation and sequencing, we used only those sequence reads that mapped uniquely to the genome for further analysis. After pooling data from all four replicates, we obtained 12 190 018 tags as our ChIP data set. Similar to QuEST, GLITR utilizes a large number of control tags (input DNA), which are sequences from sheared genomic DNA carried through the library preparation protocol. We pooled input sequencing reads from several runs to obtain 48 867 305 tags as the control data set.
The first step of the GLITR algorithm is to filter each dataset such that each start coordinate is represented once, to reduce the effect of tags that are sequenced repeatedly because of sequencing biases caused by amplification or non-random shearing of DNA (8,9,14). This reduced our ChIP data set to 9 627 691 tags and our control data set to 44 344 055 tags. Following data set reduction, GLITR creates a ‘pseudo-ChIP’ sample by randomly sampling the same number of tags from the filtered control set as are contained in the filtered ChIP dataset. The remaining control tags are used as background to estimate the fold-change for candidate regions as well as to estimate the FDR. All tags are extended based on the average fragment length (typically around 108 bp), which is estimated from the chromatin fragment size used in the preparation of the library sequenced minus the adaptor length. Both the ChIP and pseudo-ChIP datasets are grouped into ‘regions’, which are chains of overlapping extended tags. Each region is assigned a peak height, which is the maximum number of overlapping tags in the region. Then random samples of background tags, each with the same number of tags as the ChIP dataset, are used to calculate the fold-change of the ChIP and pseudo-ChIP regions (Figure 1A). By default, GLITR performs the fold-change calculation using 100 samples of background tags, to ensure an accurate median fold-change value (Supplementary Figure 1). To reduce computation time, the number of samples may be reduced. Using the background data allows for elimination of regions that are amplified due to sequencing bias or sequencing errors, and random sampling of this data prevents losing regions where by chance one particular background sample contains many input tags in a bound region (Figure 1B). In addition, the large set of background data allows for a model-free analysis. As described previously, model-free approaches will remain robust as additional experimental and biological factors that effect ChIP-Seq sequence data are uncovered (13).
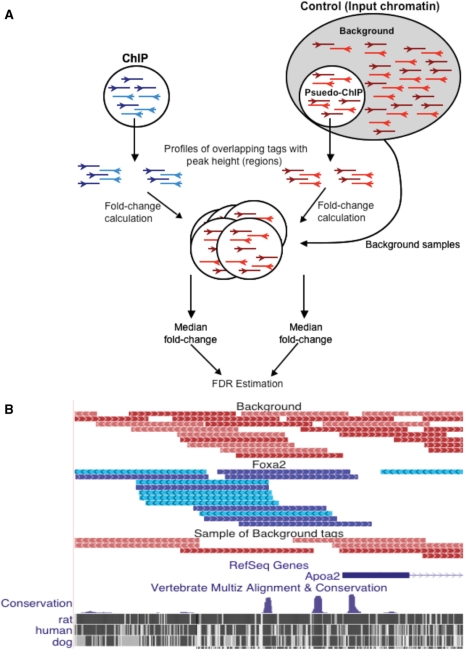
Figure 1.
The GLITR algorithm for identifying binding regions in ChIP-Seq data. (A) A pseudo-ChIP sample, which contains the same number of tags as the ChIP-Seq sample, is randomly selected from a large number of control tags obtained from multiple sequencing runs of sheared input chromatin. Overlapping regions of tags are identified in the ChIP and pseudo-ChIP samples and a median fold-change is then calculated for each of these regions, based on the fold-change to several random samplings of background tags. (B) Example of FoxA2 ChIP-Seq data in the Apoa2 promoter region. FoxA2 ChIP-Seq tags (blue) align over a known Foxa2 binding site in Apoa2 promoter. Since one sequencing lane of input is not enough to cover the entire genome, the background tags that are sequenced could by chance be in the region of a Foxa2 binding site. Sampling a large background set for the same number of tags used in the ChIP sample prevents artificial compression of fold-changes in these regions by more accurately estimating the background rate in each area of the genome. The complete set of background tags, as well as one random sample of these tags, is shown in red.
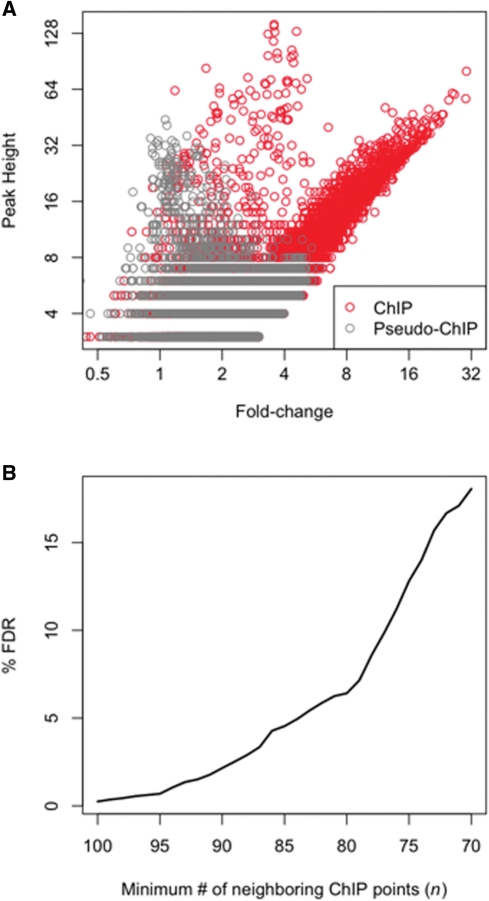
A scatter plot of the median fold-change and peak height for all of the ChIP and pseudo-ChIP regions yielded insight into the problem with using each attribute separately. First, many regions had a high peak height, but low fold-change in both the ChIP and pseudo-ChIP samples (Figure 2A). Additionally, the high density of pseudo-ChIP regions with low fold-change and low peak height are valuable for identifying ChIP regions that are above a background signal. By using both the fold-change and peak height as a signature for each region, we could better distinguish a truly bound region from one that is likely to occur by chance in background data. We also note a small set of points in the ChIP data that have very high peak heights and medium fold-changes. Specifically, 38 points have a peak height greater than fifteen and a fold-change between three and four. Ninty-five percent of these regions are within a satellite repeat or rRNA repeat and are not enriched for the Foxa2 consensus site, which indicates they are likely false-positives resulting from the ChIP procedure. GLITR employs a k-nearest neighbors classification method to make the distinction between truly bound regions versus those that are likely false-positives. For every ChIP region GLITR identifies the k (default = 100) closest points in the peak height versus fold-change plot. A region is considered bound if at least n of the k neighboring points are also ChIP regions. To estimate the FDR for a given n, the process is repeated for the pseudo-ChIP data points, treating each pseudo-ChIP point as a ChIP point. If at least n of the nearest neighbors are ChIP points, then the pseudo-ChIP point is considered a false-positive. Thus, for any given n, GLITR calculates the ratio of the proportion of points that are falsely called bound in the pseudo-ChIP set to the proportion of points called as bound in the ChIP data set as an estimate of the FDR. To increase the FDR, the size of the minimum number of ChIP regions (n) needed among the k nearest neighbors may be decreased. GLITR reports the FDR for all values where N ≤ n ≤ k (Figure 2B). The exact choice of k is not critical, because the number of regions identified with different values of k but the same proportion of n/k is comparable (Supplementary Figure 2). When we employed a 1.5% FDR cutoff, which corresponded to n/k = 92/100, we obtained 4051 regions that are bound by Foxa2. We also ran GLITR after disabling the steps used to filter data such that each start coordinate is represented once in order to assess the importance of this first step of the algorithm. Supplementary Figure 3 compares regions identified at a 1.5% FDR with and without filtering data for unique start coordinates, and demonstrates that the filtering step is critical in eliminating thousands of false-positive regions.
Figure 2.
FDR estimation. (A) A plot of peak height versus fold-change (pseudo-ChIP tags—grey and FoxA2 ChIP-Seq tags—red) shows that both variables provide valuable information for determining if a region is likely to be bound. Using a height cutoff alone leads to inclusion of multiple false positives corresponding to regions that are also sequenced in control samples. Likewise, use of a high fold-change cutoff alone may be too stringent, whereas use of a low fold-change would increase the number of false positives. (B) The k-nearest neighbor classification method is used to determine if a region is considered bound, where a region is considered bound if a majority of at least n of the k closest regions are ChIP regions. The method is repeated on pseudo-ChIP data in order to estimate a FDR. Here, k is 100 and as the majority threshold (n) used in classification decreases, the FDR increases.
Input DNA as a control
Because input DNA covers all of the mappable portions of the genome, a large number of sequence tags are necessary to obtain an accurate distribution of tags across the genome. GLITR utilizes the large set of tags by sampling sets that are equal in size to the ChIP data set, as described above. Our starting set of control tags had approximately four times the number of ChIP tags. To assess the sensitivity of the results to the number of control tags used, we ran GLITR five times using different background datasets that have three times the number of ChIP tags (3× control), and five times using different background datasets that have four times the number of ChIP tags (4× control). Plotting the average number of regions across all five runs identified for various values of n (described above) shows that at higher values of n (corresponding to lower FDR's), which are more typically used in identifying truly bound regions, the use of more control tags will aid in identifying more regions (Supplementary Figure 4). We further analyzed regions at a 1.5% FDR, and found that 307 regions were identified in all five runs using the 4× control sets that were not found in all of the runs using the 3× control sets. These regions were also enriched for the Foxa2 consensus site, but represent an increase in target number of <10% (Supplementary Figure 4). We recommend running GLITR with a control set that contains at least three times the number of ChIP tags, to ensure that a pseudo-ChIP data set can be selected, and several samples may be used for fold-change calculations.
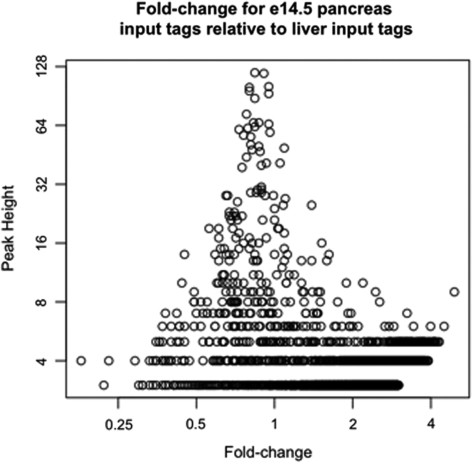
It has been reported that separate controls are needed for different cellular conditions (17), and in contrast that a control must only be sequenced once for the same organism as long as fragmentation conditions for each library are similar (8,17). To address this issue, we compared input libraries generated from adult mouse liver chromatin, adult mouse islet chromatin, and chromatin isolated from the mouse pancreas at embryonic day 14.5. Chromatin from each tissue was isolated on different days, from different mice, using the same fragmentation conditions. In one comparison, we identified regions in the e14.5 pancreas input data, and then calculated the fold-change for each region relative to liver input tags. Plotting the peak-height versus fold-change shows that all fold-change values are low, including those for regions that have a high peak height (Figure 3). The same is true when comparing islets to liver, and embryonic pancreas to islets (Supplementary Figure 5). Since we had far more liver input tags compared with islet or embryonic pancreas, we ran GLITR using the liver input tag set as our control and the islet input or embryonic pancreas input data sets as the ‘ChIP’ dataset. In both cases, GLITR did not identify any significant regions. It is apparent from our data that large peaks arising because of sequencing bias, chromatin preparation, or IP protocol are the same in all tissues investigated. Similarity of smaller peaks may not be addressed until full coverage of the mappable genome is achieved with different tissues. However, in our FDR calculation, the input region data is treated as a cloud of points and the exact location of the small peaks is not relevant for this purpose. Our pooled input data set is available on the GLITR website, and may be used for any experiment using mouse DNA as long as fragmentation conditions are similar to those described in the ‘Materials and Methods’ section.
Figure 3.
Comparing input DNA from different tissues. Chromatin was isolated from e14.5 pancreas and adult mouse liver on different days, using the same fragmentation conditions. Regions were identified from e14.5 input sequence tags, and the peak height was plotted against the fold-change, which was calculated relative to liver input sequence tags. All regions have a low fold-change and are not considered significant by GLITR.
Comparison to other methods
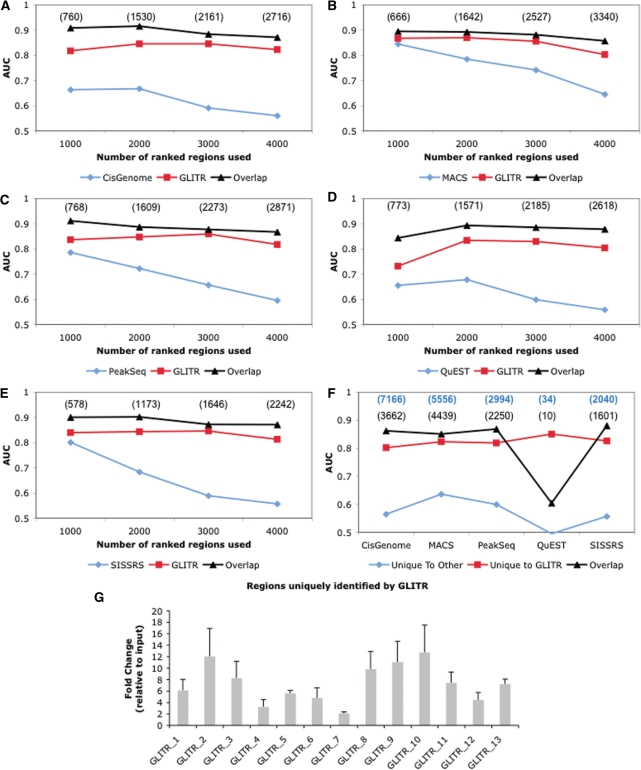
We compared GLITR to five recent programs that allow incorporation of an experimentally derived background model. Because each method calculates thresholds differently, we sorted the scored regions identified by each program and overlapped them with the GLITR-identified regions using sets of the top 1000, 2000, 3000 and 4000 regions. We used the enrichment of matches to the Foxa2 PWM as determined by the area under the ROC curve as our measure of peak quality. To determine the level of enrichment in the non-overlapping regions, we first performed de novo motif analysis using Bioprospector (19) on half of the overlapping regions. While de novo motif analysis returns similar PWMs in each of the analyses, we wanted to ensure that the PWM used for scanning was not biased towards a particular dataset. For each set of 1000 to 4000 regions, the PWM was scanned on the half of the overlapping regions that were not used in de novo motif analysis, as well as the regions that were unique to GLITR, or unique to CisGenome, MACS, PeakSeq, QuEST, or SISSRS when compared to GLITR. While regions unique to the other programs are enriched for the Foxa2 PWM when the top 1000 regions are analyzed, the enrichment decreases substantially as more regions are included, whereas enrichment remains high for regions unique to GLITR (Figure 4A–E). Several targets that were identified uniquely by GLITR were randomly selected and tested for occupancy by Foxa2 in vivo using ChIP followed by quantitative RT–PCR. Eleven of the thirteen sites tested showed an enrichment of more than four-fold in the ChIP sample compared to input, confirming that GLITR can identify bona fide transcription factor binding sites missed by the other programs (Figure 4G).
Figure 4.
Comparing GLITR to other methods for the analysis of ChIP-Seq data. Regions identified by CisGenome (A), MACS (B), PeakSeq (C), QuEST (D), SISSRS (E) and GLITR were ranked and analyzed in groups of the top 1000, 2000, 3000 and 4000 regions. For each of these groups, a training set was randomly selected from regions that overlap between each program and GLITR. The training set was used to derive a PWM which was then employed to scan the remaining regions that overlap, as well as the regions that were unique when comparing each program and GLITR. Regions unique to GLITR are more enriched for the Foxa2 PWM especially as more regions are incorporated into the analysis. The number of overlapping regions in each dataset is shown in parentheses. (F) Default regions returned by each program were overlapped with GLITR default regions (4051). Regions that were unique to each program and GLITR remained more enriched for the Foxa2 PWM. Bold blue numbers are the number of regions returned using default parameters for each program. The number of overlapping regions in each dataset is shown in black. (G) Quantitative RT-PCR confirmation of randomly selected Foxa2 target regions identified by GLITR but none of the other programs. Abundance of the target sequences was compared between Foxa2 ChIP from liver (n = 3) and liver input DNA.
Because SISSRS and QuEST tend to return smaller region windows than GLITR, we extended the regions identified by each program by 108 base pairs (the expected fragment length) on each side from its center and then repeated the program comparisons. Because the method we used for choosing background sequences is based on region width, the trend in area under the curve (AUC) was similar to what was presented in Figure 4 (Supplementary Figure 6). We also derived the optimal motif from regions that were identified by all six programs. Scanning this motif against all datasets also produced similar results (Supplementary Figure 7).
Because some programs return more regions than GLITR and some return fewer using the default cutoff settings, we also compared GLITR to each of the datasets produced using default cutoff parameters specified by each program, as these are the cutoff settings with which the programs will most commonly be executed. Again, in each comparison, regions that are unique to GLITR are more strongly enriched for the Foxa2 consensus site than regions that are found uniquely by other programs (Figure 4F). We also carried out the ranking analysis described above, but rather than calculating the AUC until 4000 regions have been identified, which is close to the number returned by GLITR, we only calculated the AUC for 1000 region increments until the default value for the comparison program was reached (Supplementary Figure 8). In all cases, target regions identified by GLITR were more enriched for the Foxa2 consensus site than those of any other program. This indicates that GLITR is more sensitive in identifying regions strongly enriched for the Foxa2 consensus site when moving deeper into the dataset.
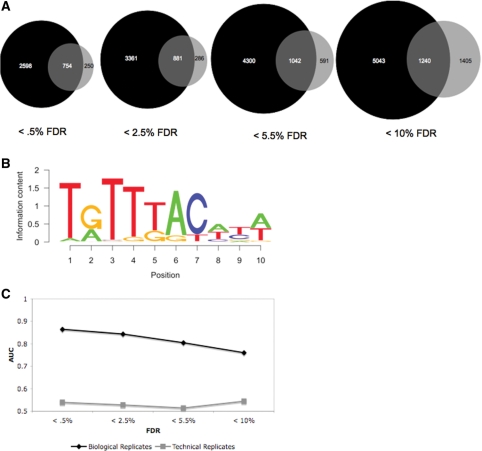
Technical replicates versus biological replicates
In order to identify enriched regions in ChIP-Seq data, sufficient sequencing reads must be obtained, and often one sequencing lane does not provide enough data for reliable peak detection. To acquire a large number of sequence tags, one can sequence the same ChIP sample several times (technical replicates), or sequence multiple samples once each (biological replicates). To determine which of these methods is more appropriate for the identification of transcription factor binding sites, we sequenced one technical replicate four times to compare to the dataset obtained from four biological replicates (starting at the earliest point in the process, i.e. by using livers taken from multiple animals) sequenced once each, described above. The total number of reads that align uniquely to the genome was comparable between the two datasets (12 190 018 tags for biological replicates and 15 521 648 tags for technical replicates). When GLITR filtered these datasets so that each start coordinate was represented only once, the biological replicate dataset had fewer tags removed (2 562 327 tags removed from biological replicates and 9 352 597 tags removed from technical replicates). This was expected, since when re-sequencing the same sample there is a higher chance of sequencing the same tag multiple times.
We ran GLITR on both the biological replicate and technical replicate datasets, and compared the enriched regions identified at several FDR thresholds. At the most stringent FDR, which is <0.5% for both datasets, the data set derived from four biological replicates identified 2348 more targets than the technical replicate dataset (Figure 5A). We performed de novo motif analysis on the regions that were common to both datasets to obtain the Foxa2 consensus motif (Figure 5B). We then calculated the enrichment of the match to the Foxa2 PWM in the regions that were unique to either the biological replicate or technical replicate data sets by computing the area under the Foxa2 ROC curve. Regions that were unique to the biological replicate dataset were much more enriched for the Foxa2 PWM than regions that were unique to the technical replicates dataset (Figure 5C). The trend was similar when GLITR was run using a subset of tags from the filtered biological replicate data set, which was equal to the number of tags in the filtered technical replicate data set (Supplementary Figure 9). This is likely due to sequencing additional tags in a region where the antibody bound to DNA non-specifically. Therefore, biological replicates are a more efficient use of sequencing capacity and are valuable for the identification of thousands of additional bona fide binding regions, as well as for the elimination of noise resulting from non-specific binding in one particular ChIP.
Figure 5.
Comparing biological replicates and technical replicates. (A) Running GLITR on biological replicate ChIP-Seq data and technical replicate ChIP-Seq data shows that thousands of additional binding sites are identified in the biological replicate dataset at several different FDR thresholds. (B) Foxa2 PWM derived from target regions identified in both the biological and technical replicate datasets, at an FDR of <0.5%. (C) When regions that are unique to either the biological replicate dataset or technical replicate dataset are scanned with the PWM shown in (B), enrichment for the PWM is consistently higher in regions that are only identified in the biological replicate data. An AUC of 0.5 indicates that a PWM is not enriched in the regions scanned.
DISCUSSION
GLITR randomly samples sets of tags from a background set of sheared input chromatin to accurately identify enriched regions in the ChIP-Seq data. Each sequence tag is extended to the expected fragment length, and grouped into regions of overlapping tags. Each candidate region is assigned a peak height and a fold-change, based on the median fold-change to random samples of background tags. A classification method is then used which compares the peak height and fold change of the ChIP data set to the peak height and fold-change of a pseudo-ChIP set to distinguish regions that are likely to occur by chance from those where the transcription factor is actually bound.
The requirement of a large set of control tags can be daunting in terms of the time and cost necessary to sequence a ChIP-Seq library. We show that under similar fragmentation conditions, it is appropriate to combine input sequence tag data from different tissues, and thus reuse this information to identify bound regions for multiple ChIP-Seq libraries. It has been previously reported that in order to identify binding sites accurately, sufficient sequencing depth must be obtained (14). We used GLITR to identify Foxa2 targets in adult mouse liver and compare sequence data obtained from re-sequencing one ChIP sample four times to sequence data obtained from sequencing four biological replicates. We demonstrated that simply re-sequencing a single ChIP sample is not sufficient to capture all binding sites, and can even lead to the inclusion of a significant number of false positives. Additionally, we compared GLITR to other methods that incorporate background tags into binding site identification and show that GLITR more accurately identifies regions that contain a strong match to the consensus binding site of Foxa2. While strong binding sites are easily identified by all programs, GLITR brings us a step closer to obtaining a truly genome-wide list of binding sites, by identifying regions with lower tag counts that still have a strong match to the Foxa2 consensus site. It is important that these ‘weaker’ binding sites are included when biological inferences are made from ChIP-seq data sets.
While several programs have been released for the analysis of ChIP-Seq data, each method has pros and cons, and a standardized approach has not yet been established. As more data become available in different organisms and tissues, one can better assess the similarities between control data sets in different conditions. Also as these datasets become available, more experimental or biological factors that effect data analysis will be uncovered, which will allow for fine-tuning of all current methods and will result in a more accurate list of binding sites across the genome.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health, Training Grant in Computational Genomics (5-T32-HG000046-09 to G.T.), and National Institutes of Health (2-P01-DK49210 to K.H.K.). Funding for open access charge: National Institutes of Health (2-P01-DK49210).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Alan Fox, Ellen Tsai and Olga Smirnova for excellent technical support, Dr John Le Lay for use of the islet and embryonic pancreas input data sets, and Dr Sridhar Hannenhalli and Dr Shane T. Jensen for valuable discussions and suggestions.
Footnotes
Present address: Peter White, The Research Institute at Nationwide Children's Hospital, Columbus, OH 43205, USA.
REFERENCES
- 1.Bulyk ML. DNA microarray technologies for measuring protein-DNA interactions. Curr. Opin. Biotechnol. 2006;17:422–430. doi: 10.1016/j.copbio.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mardis ER. ChIP-seq: welcome to the new frontier. Nat. Methods. 2007;4:613–614. doi: 10.1038/nmeth0807-613. [DOI] [PubMed] [Google Scholar]
- 3.Wold B, Myers RM. Sequence census methods for functional genomics. Nat. Methods. 2008;5:19–21. doi: 10.1038/nmeth1157. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 5.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein–DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 6.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 7.Wederell ED, Bilenky M, Cullum R, Thiessen N, Dagpinar M, Delaney A, Varhol R, Zhao Y, Zeng T, Bernier B, et al. Global analysis of in vivo Foxa2-binding sites in mouse adult liver using massively parallel sequencing. Nucleic Acids Res. 2008;36:4549–4564. doi: 10.1093/nar/gkn382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barski A, Zhao K. Genomic location analysis by ChIP-Seq. J. Cell Biochem. 2009;107:11–18. doi: 10.1002/jcb.22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyle AP, Guinney J, Crawford GE, Furey TS. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics. 2008;24:2537–2538. doi: 10.1093/bioinformatics/btn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fejes AP, Robertson G, Bilenky M, Varhol R, Bainbridge M, Jones SJ. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 2008;24:1729–1730. doi: 10.1093/bioinformatics/btn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang ZD, Rozowsky J, Snyder M, Chang J, Gerstein M. Modeling ChIP sequencing in silico with applications. PLoS Comput. Biol. 2008;4:e1000158. doi: 10.1371/journal.pcbi.1000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat. Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuteja G, Jensen ST, White P, Kaestner KH. Cis-regulatory modules in the mammalian liver: composition depends on strength of Foxa2 consensus site. Nucleic Acids Res. 2008;36:4149–4157. doi: 10.1093/nar/gkn366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, Brutlag DL, Liu JS. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 2001;6:127–138. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.