Abstract
RNA levels in a cell are regulated by the relative rates of RNA synthesis and decay. We recently developed a new approach for measuring both RNA synthesis and decay in a single experimental setting by biosynthetic labeling of newly transcribed RNA. Here, we show that this provides measurements of RNA half-lives from microarray data with a so far unreached accuracy. Based on such measurements of RNA half-lives for human B-cells and mouse fibroblasts, we identified conserved regulatory principles for a large number of biological processes. We show that different regulatory patterns between functionally similar proteins are characterized by differences in the half-life of the corresponding transcripts and can be identified by measuring RNA half-life. We identify more than 100 protein families which show such differential regulatory patterns in both species. Additionally, we provide strong evidence that the activity of protein complexes consisting of subunits with overall long transcript half-lives can be regulated by transcriptional regulation of individual key subunits with short-lived transcripts. Based on this observation, we predict more than 100 key regulatory subunits for human complexes of which 28% could be confirmed in mice (P < 10−9). Therefore, this atlas of transcript half-lives provides new fundamental insights into many cellular processes.
INTRODUCTION
mRNA levels in a cell are determined by the relative rates of RNA synthesis by polymerases and degradation by nucleases. Constant transcript levels reflect an equilibrium of RNA synthesis and decay while changes in transcript levels may be caused by alterations in either of them (1). State-of-the-art gene expression profiling allows precise measurements of total transcript abundance on whole transcriptome level but cannot distinguish whether changes in total mRNA are due to alterations in de novo transcription or in decay. RNA decay rates have previously been determined by blocking transcription, e.g. using actinomycin D (act-D), and subsequently monitoring ongoing RNA decay over time (2–8). If RNA decay continues at the same rate after inhibition of transcription, decay rates for thousands of transcripts can be obtained. However, transcriptional arrest induces a major stress response in the cell. This influences key regulatory mechanisms governing RNA decay which leads to substantial stabilization of individual transcripts (9–12).
De novo transcription can be measured in a non-disruptive way by introducing 4-thiouridine (4sU) into newly transcribed RNA utilizing nucleoside salvage pathways (13) followed by thiol-mediated isolation of newly transcribed RNA from total RNA (13–18). By combining this technique with standard microarray techniques, newly transcribed RNA can be directly measured for thousands of genes at the same time (14,15,18). Furthermore, with the integrative approach we developed recently total cellular RNA can be separated into both newly transcribed, labeled RNA and pre-existing, unlabeled RNA with high specificity (14). RNA half-lives can then be determined based on both newly transcribed RNA/total RNA ratios as well as pre-existing RNA/total RNA ratios.
In this article, we demonstrate that half-life measurements based on RNA decay, e.g. after blocking transcription, are inherently imprecise for medium- to long-lived transcripts. In contrast, RNA half-lives determined from newly transcribed/total RNA ratios are precise independent of transcript half-life. In this study, we present the first atlas of RNA half-lives in human B-cells and murine fibroblasts determined with this superior precision. This atlas was used to identify patterns in transcript half-life conserved in mammals across species and cell types. A transcriptome-wide comparison between species revealed that transcript half-lives are conserved and specifically correlated to gene function. This enabled us to better characterize the regulation of important genes and identify conserved regulatory principles for a broad range of biological processes.
We show that differences in regulation between functionally similar proteins are reflected in differences in the corresponding transcript half-lives. As a consequence, such differences in transcript half-lives can be used to detect differential regulatory patterns for these genes. Most importantly, we provide strong evidence that the activity of protein complexes consisting of multiple subunits with overall long transcript half-lives can be both fast and efficiently regulated by transcriptional regulation of individual key subunits characterized by very short-lived transcripts. Based on this concept, we could identify more than 100 potential key regulators for protein complexes in human for which at least 28% were confirmed in mice. Accordingly, analysis of RNA half-life provides fundamental new insights into conserved regulatory mechanisms of many biological processes. Therefore, the atlas of transcript half-lives we provide in this study will be valuable for further studies on a large variety of biological processes.
MATERIALS AND METHODS
Sample preparation for microarray experiments
Newly transcribed RNA was labeled in human B-cells (BL41) and murine NIH-3T3 fibroblasts by culturing cells in the presence of 4-thiouridine (4sU) for 1 h. Total cellular RNA was isolated, thiol-specifically biotinylated and separated into labeled, newly transcribed RNA and unlabeled, pre-existing RNA using streptavidin coated magnetic beads as described (14) (see also Supplementary Data for details). In addition, RNA decay rates were obtained for NIH-3T3 cells by blocking RNA synthesis for 1, 2 and 3 h using actinomycin D at a final concentration of 5 µg/ml. Three biological replicates per condition were analyzed using Affymetrix HG U133 Plus 2.0 arrays (human) and MG 430 2.0 arrays (mouse). For murine fibroblasts, an additional three replicates were performed for newly transcribed and total RNA to assess the reproducibility of our approach.
Normalization of microarray data
Microarray data were pre-processed with R and Bioconductor (19,20). A first normalization incorporated background correction, normalization and probe-level summarization by GCRMA. As these standard methods assume equal overall intensities for all arrays, a second normalization step is required to compensate for the different amounts of template mRNA present in newly transcribed RNA, pre-existing RNA and total RNA samples. In previous studies based on blocking transcription (e.g. using actinomycin D) this was either done by using reference genes (8) or by fitting the exponential decay model to time series measurements (5). We performed this normalization based on the combined analysis of total, newly transcribed and unlabeled pre-existing RNA from a single RNA sample (14). Since total RNA (N) is quantitatively separated into labeled, newly transcribed (L) and unlabeled, pre-existing RNA (U), normalized ratios of newly transcribed/total RNA and pre-existing/total RNA should add up to 100% and normalization factors can be obtained by a simple linear regression analysis (see Supplementary Data).
Calculation of RNA half-life
RNA decay has been shown to follow first-order kinetics (21) with
where λ is the decay rate for a given transcript. The transcript half-life then is t1/2 = ln 2/λ. At the beginning of labeling, U(0) = N(0). We assume that total RNA at time t is a multiple or fraction of the original concentration, i.e. N(t) = α (t)N(0) with α (t) a function of time. The RNA half-life of a specific probe set is then calculated as (see Supplementary Data for details)
where L(t)/N(t) and U(t)/N(t) are the normalized ratios of newly transcribed/total RNA and pre-existing/total RNA. The proportionality factor α (t) can be defined in different ways to model different scenarios: α (t) = 1 for the steady state and α (t) = 2t/CCL to model cell growth and division where CCL is the cell-cycle length of the cell. In this study, we used steady-state assumptions as reproducibility between replicates was higher than in the cell division model which amplifies measurement errors for long-lived transcripts (Supplementary Figure S1).
Transcript uracil number
To calculate the number of uracils in the spliced transcript for each gene, cDNA sequences for human and mouse were downloaded from the Ensembl site (release 54, May 2009) (22). The uracil number was then calculated as the number of thymines in the cDNA sequence for each gene. For genes with alternatively spliced transcripts uracil numbers were averaged.
Probe set quality score
As genes may be represented by more than one probe set on the array, we defined a probe set quality score PQS based on the difference between 1 and the sum of normalized pre-existing/total RNA ratios and newly transcribed/total RNA ratios (for details see Supplementary Data):
Gene half-life for a gene g is determined using the probe set with the maximum PQS for gene g.
RNA half-life ratio
Differences in transcript half-life between two genes gi and gj were calculated as the RNA half-life ratio
where t1/2 (gi) is the RNA half-life for gene gi.
Functional analysis
To identify functional groups significantly over-represented among short- or long-lived transcripts, we compared the overall distribution of RNA half-lives against the distribution of RNA half-lives for specific functional categories. For this purpose the functional categories of the Gene Ontology (GO) (23) were used and GO annotations were taken from the GO website. Only GO terms were analyzed with at least 10 annotated genes. Significance of differences in the distributions was determined with the Kolmogorov–Smirnov test (K–S test) in R (19). P-values were corrected for multiple testing using the method by Benjamini and Yekutieli (24), a more conservative version of the method of Benjamini and Hochberg (25), which controls the false discovery rate (FDR) and does not require the tests to be independent. Correction of P-values was performed for all ontologies taken together and statistically significant results were determined at a significance level of 0.001.
Analysis of protein families
Protein family annotations were taken from the Pfam database (26) (downloaded 1 August 2008). In total, we obtained 3170 families for human and 3031 families for mouse. Using orthology mappings from the mouse genome database (MGD) (27), protein family members were mapped between species. After removing redundant families, we obtained a final list of 738 families consisting of at least two members with half-lives in both human and mouse. Average RNA half-life ratios for members of the same family or subunits of the same protein complex (see below) were compared against results for randomized half-lives or families/complexes (10 000 randomizations each). P-values were calculated as the fraction of randomizations with lower or equal average half-life ratios than the observed average. Transcripts were defined as fast-decaying (slow-decaying) if the corresponding half-life was among the 20% shortest (longest) half-lives in at least one cell line and the 40% shortest (longest) ones in the other cell line. Functional similarity between members of the same family were calculated using the relevance similarity measure defined by Schlicker et al. (28) which determines the similarity between GO annotations of two proteins. For our purposes, similarity was calculated separately for the molecular function and biological process ontologies of the GO.
Analysis of protein complexes
Protein complexes for human and mouse (1185 and 285 protein complexes, respectively) were taken from the CORUM database (29) (downloaded 4 June 2008). Complexes were mapped between species and redundant complexes identical to another complex for either species were eliminated. This resulted in a large set containing 1434 non-redundant but partially overlapping protein complexes on which all analysis for both human and mouse were based.
Protein complex subunits with significantly shorter transcript half-life than the rest of the complex were identified by calculating for each subunit p in each complex C the difference ratio:
Here, hlr(p, C - p) is the average RNA half-life ratio between subunit p and all the other subunits of C and hlr(C - p, C - p) the average RNA half-life ratio between these other subunits. A subunit p was predicted as a regulatory subunit for a complex C if the following conditions were fulfilled: (i) the average RNA half-life ratio for p to the remaining subunits of C was at least 40% higher than the average half-life ratio between these subunits [dr(p, C) > 1.4]. (ii) RNA half-life was shorter than the median half-life in human B-cells and murine fibroblasts (∼5 h) and shorter than the median transcript half-life in complex C.
RESULTS
RNA half-life measurements in human B-cells and murine fibroblasts
To investigate RNA turnover rates in mouse and human, we analyzed RNA half-lives in human B-cells (BL41) and murine NIH-3T3 fibroblasts (see ‘Materials and Methods’ section). The murine measurements have recently been published (14). Human B-cells were chosen to compare the data from murine fibroblasts with a cell line of both a different species and cell type to identify conserved regulatory patterns. RNA half-lives were obtained based on both newly transcribed/total RNA and pre-existing/total RNA ratios (Figure 1A and B, and Supplementary Figure S2). In addition, we performed microarray measurements on total RNA following 1, 2 and 3 h of transcriptional arrest by actinomycin D (act-D) in murine fibroblasts (Supplementary Figure S3) in order to compare our new approach with this standard method used to determine RNA decay rates (2–8). While all three approaches provided highly reproducible data for short-lived transcripts (t½ < 1–2 h) only newly transcribed/total RNA ratios yielded reliable data for medium- to long-lived transcripts. Although reproducibility of half-lives increased with longer act-D treatment, differences between 2 and 3 h act-D treatment were still considerable (Supplementary Figure S3) indicating that transcriptional arrest would have to be prolonged by another several hours to obtain accurate transcript half-lives. By simulating the effect of noise on RNA half-life determination (Supplementary Figure S4), we confirmed that this is not a problem of the individual measurements but an inherent feature of RNA half-life measurements based on monitoring the decay of transcripts.
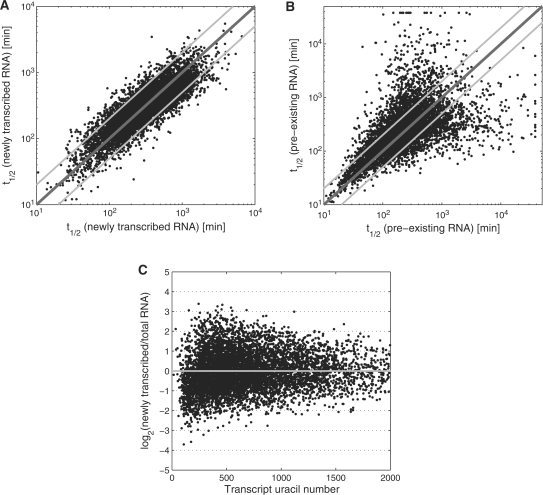
Figure 1.
Comparison of RNA half-lives determined in human B-cells based on newly transcribed/total RNA ratios (A) and pre-existing/total RNA ratios (B) following 1 h of 4sU labeling. RNA half-lives were determined independently for two replicates. The x-axis shows the half-life of each gene for the first replicate and the y-axis the corresponding half-life for the second replicate. Dark grey diagonals indicate equal RNA half-lives and light grey lines a deviation by a factor of 2. (C) Comparison of transcript uracil number (number of uracils in spliced transcript) against the logarithm of newly transcribed/total RNA ratios for human B-cells (correlation coefficient = −0.014). Here, expression levels before the second normalization step were used. The grey line indicates no correlation, i.e. no transcript length bias.
Efficient capture of nascent transcripts is dependent on the incorporation of sufficient amounts of 4sU. This is particularly important for short transcripts with low uracil content. Previously, efficiency of capture of nascent transcripts by the streptavidin-coated magnetic beads was shown to be very high as 80–90% of radioactively labeled nascent RNA could be recovered. Although so far this was not specifically evaluated for short transcripts, reduced capture rates of small transcripts may create a substantial bias when calculating RNA half-lives based on newly transcribed/total RNA ratios (30). To check for this kind of bias, we compared the uracil number for transcripts with the measured ratios of newly transcribed/total RNA for our microarray data. No significant correlation for human B-cells (Figure 1C) and only a weak correlation for murine fibroblasts (Supplementary Figure S2) was observed. Thus, the employed concentrations of 4sU were sufficient for highly efficient capture even of transcripts with low uracil content.
Increasing the accuracy of half-life measurements by using probe set quality scores
Accuracy and reproducibility of half-lives determined from newly transcribed/total RNA ratios can be further increased by assessing probe set quality based on the relationship between newly transcribed, pre-existing and total RNA. On both the Affymetrix MG 430 2.0 arrays (mouse) and the HG U133 Plus 2.0 arrays (human) many genes are represented by multiple probe sets (Supplementary Figure S5). Due to quality differences between probe sets and experimental noise, half-life measurements of different probe sets for a single gene often result in dramatically different results. We solved this problem by calculating a probe set quality score (PQS) for each probe set based on the difference between the sum of measurements for newly transcribed and pre-existing RNA and total RNA levels for each probe set (see ‘Materials and Methods’ section and Supplementary Data). To evaluate the performance of this procedure, we determined probe set quality scores independently for each replicate of total, newly transcribed and pre-existing RNA of human B-cells and murine fibroblasts and identified the optimal probe set for each gene for the corresponding replicate. In our study, the same probe sets were independently identified as optimal for the corresponding genes in all three replicates significantly more often than expected by chance (binomial test, FDR corrected P < 0.05). In addition, when considering only the optimal probe set for each gene in each replicate, we observed decreased variations between replicates and increased reproducibility of results compared to the standard averaging approach (Supplementary Figure S5). This indicates that our approach can identify individual, incorrect measurements but also distinct quality differences between probe sets in an experimental setting.
Conservation of RNA half-life
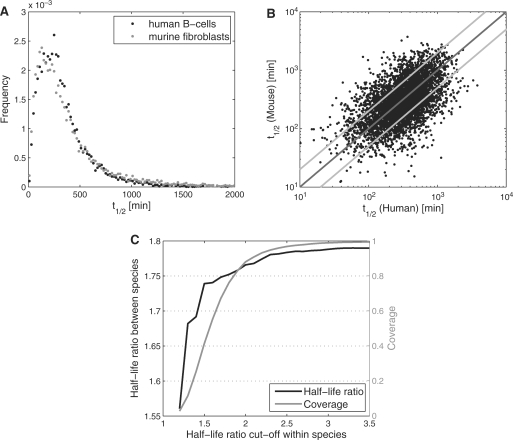
For both murine fibroblasts and human B-cells, we determined RNA half-lives for more than 8000 genes based on newly transcribed/total RNA ratios using only the optimal probe set for each gene (Supplementary Tables S1 and S2). Median RNA half-life t1/2m was determined at 315 min (95% confidence interval: 240–382 min) in human B-cells and 274 min in murine fibroblasts (225–323 min) (see ‘Materials and Methods’ section and Supplementary Data). Although median half-lives differ between the two cell lines, this difference is not significant (t-test for unequal variance, P ∼0.38). Furthermore, the distribution of half-lives is similar (Figure 2A) and follows approximately a log-normal distribution in both species. Using orthology tables between human and mouse genes from the MGD (27), transcript half-lives were compared for 4825 genes with RNA half-lives for both species (see Figure 2B). The average half-life ratio (hlr, see ‘Materials and Methods’ section) between human and mouse was 1.8 and ∼67% of genes were within the two-fold range. Although variation between species was significantly larger than between different replicates for the same species and experiment (hlr = 1.2–1.3), it was significantly lower than the variation between different replicates after 1 h Act-D treatment in murine fibroblasts (hlr = 1.97). In addition, differences in transcript half-life between human and mouse were reduced by filtering genes based on the variation between different replicates for the same species. The lower the half-life ratios between different replicates in one species, the lower were the half-life ratios between the two species (Figure 2C). Furthermore, for only 18 genes (∼0.37%) the deviation between species was significant (t-test for unequal variance, FDR corrected P < 0.01). A list of these genes is provided in Supplementary Table S3.
Figure 2.
(A) Distribution of transcript half-lives for human B-cells and murine fibroblasts. (B) Comparison of half-lives between human B-cells and murine fibroblasts for about 5000 genes based on orthology assignments from the MGD database (27). (C) Average half-life ratios between species were calculated only for genes for which half-life ratios among different replicates for the same species were below a specific cut-off (x-axis). For each cut-off, average half-life ratios between species (black) and coverage (grey), i.e. fraction of genes selected by the cut-off, are shown. Half-life ratios decreased significantly when more selective cut-offs were chosen.
Association of transcript half-life and gene function
Previous studies of RNA half-lives based on inhibition of transcription have shown that mRNAs of transcriptional genes are preferentially short-lived while transcripts involved in the metabolism of the cell are quite stable (5,8,31). Based on the RNA half-lives determined in this study, we performed a comprehensive analysis of GO terms (for detailed results see Supplementary Table S4 and S5) to associate functional categories with differences in half-life distribution (see ‘Materials and Methods’ section). In both species, short-lived transcripts were characteristic for genes involved in the regulation of transcription (P < 10−16) and signal transduction (P < 0.007) (Figure 3A and Supplementary Figure S6). Signal transduction has so far only been associated with fast transcript decay in Arabidopsis (5). Interestingly, short half-life was specific only for regulators of transcription and signal transduction as a comparison of the half-life distribution for regulator genes neither involved in transcription nor signal transduction against the distribution for all genes showed no significant difference. Furthermore, previous observations of fast transcript decay for apoptosis and cell cycle transcripts (8) could not be confirmed as no significant difference in the half-life distribution was observed (Supplementary Figure S6).
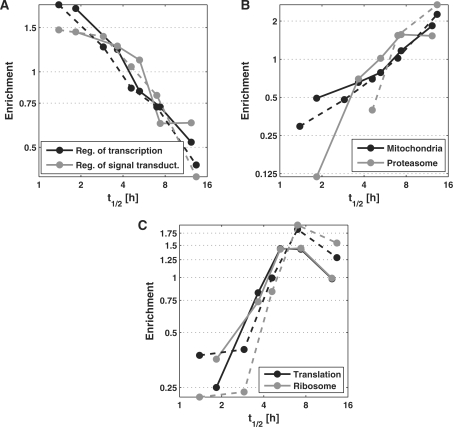
Figure 3.
To illustrate the correlation between gene function and transcript half-life, we determined the enrichment of specific functional category (y-axis) within different intervals of transcript half-life (x-axis) for human B-cells (solid lines) and mouse fibroblasts (dashed lines). For this purpose, the range of transcript half-lives in human B-cells and mouse fibroblasts, respectively, was divided into five intervals each containing approximately the same number of genes. Enrichment for a specific functional category in each interval was then calculated as the ratio of the frequency for this functional category within this interval divided by the overall frequency for this category. Results are shown for (A) transcripts involved in regulation of transcription (black) and signal transduction (grey), (B) mitochondrial (black) and proteasomal transcripts (grey) and (C) translational (black) and ribosomal (grey) transcripts.
Significant enrichment for very long RNA half-lives was found for genes involved in cellular respiration and energy metabolism as well as translation and protein decay by the proteasome. Enzymes and protein complexes involved in all parts of energy metabolism consistently had half-lives of more than 5 h both in human and mouse (Supplementary Figure S6 and Table S6). Interestingly, our method revealed a behavior of translational genes not previously described. While the frequency of transcription and signal transduction regulators steadily decreased with increasing RNA half-life (Figure 3A) and the frequency of genes involved in metabolic processes like energy metabolism and the proteasome steadily increased (Figure 3B), translational genes, encoding e.g. for ribosomal subunits or translation initiation factors, clustered in the medium-to-long half-life range and showed a lower frequency on either side (Figure 3C).
Regulation of biological processes by transcript half-life
Since protein family members generally have similar functions, we investigated whether this translates to similar transcript half-lives. Indeed, we found that similarity of transcript half-lives was significantly increased in protein families (P < 10−4). However, we also identified a large number of protein families with substantial variations in transcript half-life. 111 of 738 protein families (15%) with at least two members for which we obtained half-lives in both human and mouse contained family members with both fast- and slow-decaying transcripts (see ‘Materials and Methods’ section). A detailed list of these families containing both the genes with the shortest and longest transcript half-life conserved between mice and men is provided in Supplementary Table S7. The median size of these families [22] was significantly larger than for families without such large differences in transcript half-lives (5, Wilcoxon rank sum test P < 10−16). The most likely explanation for this finding is that these larger families simply contain more diverse family members with a wider range of functions. Indeed, we found that both similarity of molecular functions and biological processes was significantly lower than for the other families (Wilcoxon rank sum test P < 0.05, Supplementary Figure S7). Surprisingly however, we observed that the proteins with the largest difference in half-life did not necessarily show the largest differences in their molecular functions or biological processes. For 44 (39.6%) of the 111 families, functional similarity was actually higher when comparing proteins with short-lived transcripts to proteins with long-lived transcripts than when the other proteins in the family were compared. This indicates that these proteins have a similar function but differential types of regulation. A good example for this finding is the hexokinase gene family. While hexokinase I (HK-I) transcripts decayed slowly (t1/2 ∼ 9 h) as do those of most other genes involved in cellular respiration, transcripts of hexokinase II (HK-II) decayed fast (t1/2 ∼ 1–3.6 h) in both human and mouse. Phosphorylation of glucose by hexokinases is the first step of the glycolysis and hexokinase I is considered a ‘housekeeping gene’ whose mRNA levels remain stable despite alterations in glucose or insulin levels (32) or feeding conditions (33). Contrary to that, expression of HK-II is induced by a variety of stimuli (32,34–36) thereby accelerating hexose catabolism and regulating blood sugar levels. Fast changes in the expression of HK-II are supported by the fast turnover of HK-II mRNA. In contrast, slow decay of HK-I transcripts prevents any rapid changes in gene expression. Thus, transcript half-lives for these enzymes are specifically adjusted according to their functional role. We noted the same phenomenon for the family of cytosine triphosphate (CTP) synthases. Here, CTP synthase 1 (CTPS) has a short transcript half-life (t1/2 ∼ 3 h) in both mice and men while CTPS 2 has a long transcript half-life (t1/2 > 11 h). Although CTP synthases are essential enzymes (37), little is known about functional differences and transcriptional regulation. From our results we predict a similar regulatory pattern as for the two hexokinases. One isoform (CTPS) can be rapidly and transiently induced by transcriptional regulation, e.g. during cell cycle, while RNA concentrations of the other isoform (CTPS2) are stable and provide basal enzyme activity levels.
Short- and long-lived transcripts were also observed for the BCL-2 family which contains both pro- and anti-apoptotic proteins (38) (Table 1). Interestingly, long RNA half-lives were only observed for pro-apoptotic family members—although not for all of them. This provides additional evidence that an arrest in transcription following severe stress conditions could lead to a selective decline of the short-lived anti-apoptotic genes but not the pro-apoptotic ones thereby promoting apoptosis (21). Interestingly, the pro-apoptotic BCL-2 family members BAX and BAK, which share a common domain structure and are generally assumed to substitute for each other (39) stand out with respect to their different transcript half-lives (>12 h versus 2–3 h, respectively) in both mice and men. There is evidence that BAX and BAK play non-redundant roles and are regulated in different ways (40–43). Our results support this non-redundant role indicating that it is facilitated by the differences in transcript turnover. This implies that transcriptional regulation is important for BAK-mediated regulation of apoptosis while BAX activity may be preferentially regulated by post-transcriptional means.
Table 1.
RNA half-lives for genes in the BCL-2 family containing both anti- and pro-apoptotic genes
| Gene | Half-life (h) (human) | Half-life (h) (mouse) |
|---|---|---|
| Anti-apoptotic | ||
| BCL2 | 3.80 | 3.74 |
| BCL2L1 | 1.96 | 1.11 |
| BCL2L2 | NA | 1.75 |
| BCL2A1 | 3.72 | NA |
| MCL1 | 1.07 | 0.70 |
| Pro-apoptotic | ||
| BAX | 38.77 | 12.14 |
| BAK1 | 2.13 | 3.28 |
| BOK | 4.64 | NA |
| BID | 10.02 | 4.21 |
| BCL2L11 | 3.99 | 0.58 |
| BAD | 11.46 | NA |
| HRK | 5.14 | NA |
| BBC3 | 1.47 | NA |
| BIK | 4.66 | NA |
| Uncategorized | ||
| BCL2L13 | 4.09 | 2.22 |
Classification into anti- and pro-apoptotic genes was taken from Youle and Strasser (38). Half-lives longer than the median half-life of ∼5 h are shown in boldface. NA means that transcript half-life for this gene could not be determined in the corresponding species due to low expression.
These examples show that analysis of transcript half-life can reveal different types of regulation of proteins which otherwise appear to have very similar function.
Regulation of protein complexes by transcript half-life
For yeast, it has been previously reported that decay rates of transcripts encoding subunits of the same protein complex are similar (31). To investigate this for human and mouse, we analyzed transcript half-lives for the 1434 non-redundant but partially overlapping human and mouse protein complexes taken from the CORUM database (29) (see ‘Materials and Methods’ section). In both human B-cells and murine fibroblasts, transcripts with long half-lives were significantly over-represented among transcripts encoding subunits of these protein complexes (P < 0.0003). As expected, this was particularly prominent for subunits of the large protein complexes involved in energy and protein metabolism (see Supplementary Table S6). In addition, we found that transcript half-lives for subunits of the same protein complex were significantly more similar to each other compared to random expectation (hlr = 1.8–2.0, P < 10−4) even if we accounted for the overall high transcript half-life in complexes by randomizing complex memberships instead of half-lives.
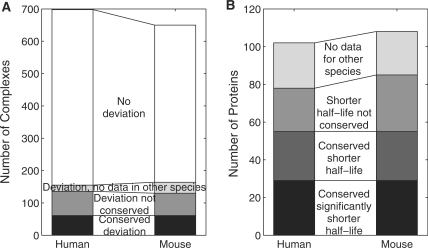
Interestingly, despite this relatively high similarity of RNA half-lives in complexes, individual subunits of some complexes deviated substantially in transcript half-life from the remaining subunits. In principle, a single protein may be involved in several different protein complexes which may be regulated in different ways and, thus, may be characterized by different median transcript half-lives. Therefore, for some subunits their deviation in transcript half-life to one complex might be explained by similarity to another. Nevertheless, even when excluding these proteins, we still identified 155 complexes in human and 164 in mouse (out of 698 and 650 complexes, respectively, for which we had half-lives for at least three subunits) which contained protein subunits with a considerably shorter half-life than the remaining subunits (Figure 4, see ‘Materials and Methods’ section for details). 61 (∼37%) of the complexes with deviating subunits were identified in both species which are significantly more than expected by chance (hypergeometric test, P < 10−5). Notably, we found that not only these complexes but also their deviating subunits were conserved. In total, we identified 102 and 108 proteins in human and mouse, respectively, which showed significantly shorter half-lives than the other members of the complexes they were part of (Figure 4). A complete list of these proteins and the complexes they are involved in is provided in Supplementary Tables S8 and S9. For 29 (28%) of these proteins identified in human B-cells the significantly shorter RNA half-life in the corresponding complex could be confirmed in murine fibroblasts (hypergeometric test, P < 10−9). For an additional 26 (25%) proteins, RNA half-life in the murine fibroblasts was shorter than the median half-life of the complex but the difference was not sufficiently pronounced.
Figure 4.
(A) Number of complexes containing at least one subunit with (i) significantly shorter transcript half-life than the median RNA half-life in the complex and (ii) no similarity in transcript half-life to any other complex it is contained in (black, grey and light grey) and number of complexes containing no such subunits (white). In the first case, we distinguished between complexes for which the deviation was conserved between species (black), for which it was not conserved (grey) or for which there were no data available in the other species (light grey). Only protein complexes were considered containing at least three subunits for which RNA half-lives could be determined in the respective species. (B) Number of proteins with significantly shorter transcript half-life than the remaining subunits in all protein complexes they are contained in. We distinguished between cases in which the significant shorter half-life was conserved (black), a shorter RNA half-life than the median RNA half-life for the complex was observed in the other species although this difference was not sufficiently pronounced (dark grey), the shorter half-life was not conserved (grey) and no data were available for the other species (light grey).
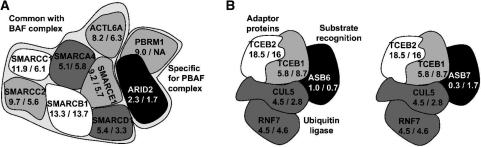
A short transcript half-life allows both fast up- and down-regulation of gene activity on the transcriptional level (see Supplementary Figure S8) (44). Our results suggest that transcriptional regulation of individual key subunits is an evolutionary conserved mechanism to regulate the activity of protein complexes despite overall long RNA half-lives. One example for this type of regulation is the PBAF (Polybromo- and BAF containing complex) chromatin remodeling complex. Here, only the ARID2 (AT rich interactive domain 2) protein is characterized by short transcript half-life (Figure 5A). This subunit has previously been shown to be essential for the stability of the PBAF complex (45). Due to its short transcript half-life, the activity of the PBAF complex can be efficiently regulated by transcriptional regulation of the ARID2 protein alone. Another example is the E3 ubiquitin ligase complex (Figure 5B) consisting of CUL5 (Cullin 5) and RNF7 (Ring finger protein 7) linked by the heterodimeric Elongin BC complex (TCEB1 and TCEB2) to an Ankyrin repeat and SOCS box (ASB) protein which serves as substrate-recognition component (46,47). Here, different ASB proteins are responsible for the recognition of different substrates. In our study, we found that two such ASB proteins, ASB6 and ASB7, showed significantly shorter transcript half-lives than the remaining subunits. As these proteins are responsible for substrate recognition, transcriptional regulation of only these subunits suffices to regulate the activity of the complex with regard to specific substrates. As all the other ligase subunits show long transcript half-lives, our results indicate that they are continuously available for binding with various ASB subunits targeting different substrates.
Figure 5.
RNA half-lives for the PBAF (A) and ubiquitin ligase (B) complexes. Half-lives (in hours) for human/mouse are indicated and mapped to grey scales ranging from black (short RNA half-lives) to white (long RNA half-lives). For PBRM1, no transcript half-life could be obtained for murine fibroblasts. For SMARCD1, its RNA half-life in murine fibroblasts was taken from 30 min labeling experiments (14). (A) The PBAF complex consists of several proteins in common with the BAF complex and two specific proteins (ARID2 and PBRM1) (45). ARID2, the only subunit with short transcript half-life in this complex, has been found to be essential for complex stability, potentially by recruiting PBRM1 (45). As most physical interactions in the complex are not characterized, the arrangement of the proteins in this figure does not necessarily represent the true complex structure. (B) Substrate-specificity of the ubiquitin ligase containing CUL5 and RNF7 is determined by binding to different ASB proteins (46,47). TCEB1 and TCEB2 are adaptor proteins which form a heterodimeric complex (Elongin BC) and additionally link the ligase subunits (CUL5 and RNF7) and the ASB protein. Short transcript half-life of the ASB6 and ASB7 subunits allows efficient regulation of complex activity with regard to specific substrates.
DISCUSSION
Regulation of biological processes occurs at the transcriptional, translational and post-translational level. Optimal control is only achieved by coordinated regulation at all levels. Yet, important information on functional characteristics on many biological processes can already be obtained by analyzing transcriptional regulation. While measurements of differential gene expression indicate which genes are regulated on the transcriptional level in a specific condition, we show in this study that analysis of RNA decay and turnover can provide insights on transcriptional regulation on a more general level.
RNA decay has been studied in a wide range of species: E. coli (3), yeast (31), Arabidopsis (4,5) and human (8). These studies were based on measurements of RNA decay after transcriptional arrest. In this article, we showed that these RNA half-lives, although quite accurate for short half-lives, are unreliable for medium to long half-lives. Contrary to that, measurements of RNA de novo transcription provide reliable and precise results on the whole range of RNA half-lives. Probe set quality scores determined for every probe set based on the combined analysis of newly transcribed, unlabeled pre-existing RNA and total cellular RNA further improved data quality.
One potential bias, which might affect half-life measurements based on newly transcribed RNA, is insufficient capture of short transcripts due to their low number of uracil residues. This would result in underestimation of newly transcribed/total RNA ratios and overestimation of corresponding half-lives. Such a bias was noted in a recent study by Miller et al. (30) which used 4-thiouracil (4tU) instead of 4-thiouridine (4sU) which we used in our study. By correlating uracil number of transcripts with newly transcribed/total RNA ratios we demonstrated that 4sU labeling for both human B-cells and murine fibroblasts resulted in sufficient 4sU incorporation to ensure efficient capture of transcript even with rather few (<100) uracil residues. Note that 4sU incorporation into nascent RNAs can be easily enhanced by increasing the applied 4sU concentration in the cell culture medium and, thus, transcript size bias can be experimentally controlled. In contrast, 4tU labeling requires the co-expression of uracil phosphoribosyltransferase (UPRT) of the protozoa Toxoplasma gondii (18). We found 4tU/UPRT based labeling to be strongly dependent on UPRT expression levels as well as the cell type under study but not on the concentration of 4tU (unpublished data). Therefore, labeling efficiency can not be significantly increased by simply adding more 4tU but transcript length bias needs to be controlled for by bioinformatic means (30).
Using our new approach, we determined precise RNA half-lives for more than 8000 genes in both human B-cells and mouse fibroblasts. By choosing two completely unrelated cell types, we focused on regulatory mechanisms not specific for only individual cell types. For about 5000 orthologous genes, we obtained RNA half-lives in both species and cell types. For the large majority of these orthologous genes, transcript half-lives are conserved across species and cell types. Only 18 out of the 4825 genes compared, i.e. only ∼0.37%, actually showed a significant difference in transcript half-life between the two species. Furthermore, variation between species was correlated to the variation observed in the individual experiments for each species. This suggests that to a large degree the observed variations between species were due to variations within the individual experiments and do not constitute important inter-species differences. This does not imply that RNA decay is a static process and that no significant differences in transcript half-life exist in between these two cell lines or species. Our results only show that for conserved genes expressed in both cell lines, transcript half-life is also conserved.
Fast transcript decay allows rapid alterations of steady-state RNA concentrations due to transcriptional changes. At the same time, these changes can also be rapidly reversed. Thus, a short transcript half-life is important for efficient regulation at transcriptional level. Assuming that protein levels and transcript levels are correlated, protein levels of these genes can be efficiently regulated by alteration in transcription rates alone. In contrast, up- or down-regulation of stable transcripts takes a very long time to result in altered total RNA levels which, once established, also persist much longer. Consistent with previous reports, we confirmed a shift towards short half-lives for genes involved in the regulation of transcription and observed this also for regulators of signal transduction. However, a similar shift for genes involved in the regulation of cell cycle or apoptosis as proposed earlier (8) or regulating genes in general (apart from transcriptional and signal transduction regulators) could not be confirmed. Thus, a short transcript half-life is not characteristic for regulators as such but only for regulators of transcription and signal transduction. The most stable transcripts were found for genes encoding for energy metabolism and protein translation and degradation. Interestingly, we observed that RNA half-lives of genes involved in translation cluster in the medium- to long-lived range and decrease in frequency on either side. This indicates that a greater degree of transcriptional control may be required for constituents of the translational machinery than for transcripts coding for proteins involved in protein degradation and energy metabolism. As measurements of RNA decay rates based on transcriptional arrest are inherently imprecise for medium- to long-lived transcripts, it is not surprising that this has been missed by previous studies.
So far, the biological processes and sequence features determining RNA decay are only poorly understood. Previous studies have suggested that certain RNA motifs in untranslated regions (8) or miRNA binding and the presence of introns (5) may play a role. Our approach now allows the analysis of RNA sequence features and motifs which determine fast and slow but also intermediate fast RNA decay. Therefore, the method and data we provide in this study will be valuable for more systematic studies on the mechanisms governing RNA decay.
We identified many biological processes in which closely related members of the same protein family with overlapping function differ significantly in RNA half-life. Here, differences in transcript half-life likely correspond to differences in regulation and, accordingly, functional roles of the corresponding genes. This is best exemplified by hexokinase I and II as well as the pro-apoptotic proteins BAX and BAK. These examples show that transcript half-lives are fine-tuned to support the regulation of cooperative but non-redundant roles of closely related family members. Based on these results, we predict similar regulatory patterns and provide a database for a large number of functionally less characterized genes and processes.
Most proteins function by interacting with other proteins in protein complexes. In this study, we confirmed previous observations in yeast that transcript half-lives for subunits of protein complexes are very similar (31). Furthermore, decay of transcripts for these subunits was found to be generally slow. This implies that most protein complexes are pre-dominantly regulated at the post-translational level. Nevertheless, for more than 150 complexes with overall long transcript half-lives in both human and mouse we identified individual key subunits with a short transcript half-life which deviate significantly from the remaining subunits in all complexes they are part of. For almost a third of these proteins, we found this pattern to be conserved across species. The probability of finding the same complexes and subunits in both species by chance is negligibly small. Therefore, we propose a generalized mechanism employed by cells to facilitate regulation of protein complexes in an efficient and targeted way. For complexes depending on the availability of specific essential components, regulation of complex activity is accomplished by regulating the abundance of only one or few of these key subunits. Thereby, complex activity can be regulated both faster—as most subunits are available and may have already assembled—and more energy efficient than by regulating all complex members. With the two examples of the PBAF complex and the E3 ubiquitin ligase complex, we demonstrated how transcriptional regulation of individual key subunits which are e.g. critical for either complex formation and stability (for the PBAF complex) or specificity (for the E3 ubiquitin ligase complex) can support efficient regulation of complex activity at the transcriptional level. A similar observation was made by de Lichtenberg et al. (48) for protein complexes of the yeast cell cycle. As most of these complexes contained both periodically and constitutively expressed subunits, they suggested a ‘just-in-time assembly’ (instead of ‘just-in-time synthesis’) in which the timing of the complex assembly is regulated by transcriptional regulation of only some subunits. Our results indicate that this may not be specific for the cell cycle but a general mechanism by which the function of large protein complex is regulated.
Based on this concept, we predict altogether about 100 key regulatory subunits in more than 150 complexes for both human and mouse. This list can be further extended by 85 and 67 proteins in human and mouse, respectively, which show a significantly shorter RNA half-life in at least one complex but not all complexes they are contained in. In these cases, their short transcript half-life may be explained by the fact that they are part of a complex for which all subunits have to be regulated strongly on the transcriptional level and, accordingly, have short transcript half-lives. Although these subunits were not included in the predictions for key regulatory subunits, they probably also have an important regulative function within the other complexes they are part of. Further studies on the regulatory subunits we predict in this study are required for a better understanding of the regulation of the involved protein complexes and the biological processes they govern.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Federal Ministry of Education and Research (BMBF NGFNplus 01GS0801 to L.D, U.K, C.C.F, R.Z.); the Friedrich-Baur Stiftung (to L.D.). Funding for open access charge: Ludwig-Maximilians-Universität München and BMBF.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Bernd Rädle for his excellent technical assistance.
REFERENCES
- 1.Ross J. mRNA stability in mammalian cells. Microbiol. Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson AF, Lundgren M, Eriksson S, Rosenlund M, Bernander R, Nilsson P. Global analysis of mRNA stability in the archaeon Sulfolobus. Genome Biol. 2006;7:R99. doi: 10.1186/gb-2006-7-10-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S, Cohen SN. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl Acad. Sci. USA. 2002;99:9697–9702. doi: 10.1073/pnas.112318199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutierrez RA, Ewing RM, Cherry JM, Green PJ. Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc. Natl Acad. Sci. USA. 2002;99:11513–11518. doi: 10.1073/pnas.152204099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narsai R, Howell KA, Millar AH, O'Toole N, Small I, Whelan J. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell. 2007;19:3418–3436. doi: 10.1105/tpc.107.055046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raghavan A, Ogilvie RL, Reilly C, Abelson ML, Raghavan S, Vasdewani J, Krathwohl M, Bohjanen PR. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 2002;30:5529–5538. doi: 10.1093/nar/gkf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selinger DW, Saxena RM, Cheung KJ, Church GM, Rosenow C. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 2003;13:216–223. doi: 10.1101/gr.912603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang E, Nimwegen Ev, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JJ. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 10.Blattner C, Kannouche P, Litfin M, Bender K, Rahmsdorf HJ, Angulo JF, Herrlich P. UV-Induced stabilization of c-fos and other short-lived mRNAs. Mol. Cell Biol. 2000;20:3616–3625. doi: 10.1128/mcb.20.10.3616-3625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorospe M, Wang X, Holbrook NJ. p53-dependent elevation of p21Waf1 expression by UV light is mediated through mRNA stabilization and involves a vanadate-sensitive regulatory system. Mol. Cell Biol. 1998;18:1400–1407. doi: 10.1128/mcb.18.3.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melvin WT, Milne HB, Slater AA, Allen HJ, Keir HM. Incorporation of 6-thioguanosine and 4-thiouridine into RNA. Application to isolation of newly synthesised RNA by affinity chromatography. Eur. J. Biochem. 1978;92:373–379. doi: 10.1111/j.1432-1033.1978.tb12756.x. [DOI] [PubMed] [Google Scholar]
- 14.Dölken L, Ruzsics Z, Rädle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, et al. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA. 2008;14:1959–1972. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenzelmann M, Maertens S, Hergenhahn M, Kueffer S, Hotz-Wagenblatt A, Li L, Wang S, Ittrich C, Lemberger T, Arribas R, et al. Microarray analysis of newly synthesized RNA in cells and animals. Proc. Natl Acad. Sci USA. 2007;104:6164–6169. doi: 10.1073/pnas.0610439104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ussuf KK, Anikumar G, Nair PM. Newly synthesised mRNA as a probe for identification of wound responsive genes from potatoes. Indian J. Biochem. Biophys. 1995;32:78–83. [PubMed] [Google Scholar]
- 17.Woodford TA, Schlegel R, Pardee AB. Selective isolation of newly synthesized mammalian mRNA after in vivo labeling with 4-thiouridine or 6-thioguanosine. Anal. Biochem. 1988;171:166–172. doi: 10.1016/0003-2697(88)90138-8. [DOI] [PubMed] [Google Scholar]
- 18.Cleary MD, Meiering CD, Jan E, Guymon R, Boothroyd JC. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat. Biotechnol. 2005;23:232–237. doi: 10.1038/nbt1061. [DOI] [PubMed] [Google Scholar]
- 19.R Development Core Team. R Foundation for Statistical Computing. Austria: Vienna; 2007. R: a language and environment for statistical computing. [Google Scholar]
- 20.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam LT, Pickeral OK, Peng AC, Rosenwald A, Hurt EM, Giltnane JM, Averett LM, Zhao H, Davis RE, Sathyamoorthy M, et al. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-10-research0041. RESEARCH0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001;29:1165–1188. [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Statist. Soc. B (Methodological) 1995;57:289–300. [Google Scholar]
- 26.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. The mouse genome database (MGD): new features facilitating a model system. Nucleic Acids Res. 2007;35:D630–D637. doi: 10.1093/nar/gkl940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlicker A, Domingues FS, Rahnenfuhrer J, Lengauer T. A new measure for functional similarity of gene products based on Gene Ontology. BMC Bioinformatics. 2006;7:302. doi: 10.1186/1471-2105-7-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruepp A, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, Stransky M, Waegele B, Schmidt T, Doudieu ON, Stumpflen V, et al. CORUM: the comprehensive resource of mammalian protein complexes. Nucleic Acids Res. 2008;36:D646–D650. doi: 10.1093/nar/gkm936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller MR, Robinson KJ, Cleary MD, Doe CQ. TU-tagging: cell type–specific RNA isolation from intact complex tissues. Nat. Methods. 2009;6:439–441. doi: 10.1038/nmeth.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. Proc. Natl Acad. Sci. USA. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Printz RL, Koch S, Potter LR, O'Doherty RM, Tiesinga JJ, Moritz S, Granner DK. Hexokinase II mRNA and gene structure, regulation by insulin, and evolution. J. Biol. Chem. 1993;268:5209–5219. [PubMed] [Google Scholar]
- 33.Soengas JL, Polakof S, Chen X, Sangiao-Alvarellos S, Moon TW. Glucokinase and hexokinase expression and activities in rainbow trout tissues: changes with food deprivation and refeeding. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006;291:R810–R821. doi: 10.1152/ajpregu.00115.2006. [DOI] [PubMed] [Google Scholar]
- 34.Osawa H, Printz RL, Whitesell RR, Granner DK. Regulation of hexokinase II gene transcription and glucose phosphorylation by catecholamines, cyclic AMP, and insulin. Diabetes. 1995;44:1426–1432. doi: 10.2337/diab.44.12.1426. [DOI] [PubMed] [Google Scholar]
- 35.Jones JP, Dohm GL. Regulation of glucose transporter GLUT-4 and hexokinase II gene transcription by insulin and epinephrine. Am. J. Physiol. 1997;273:E682–E687. doi: 10.1152/ajpendo.1997.273.4.E682. [DOI] [PubMed] [Google Scholar]
- 36.Riddle SR, Ahmad A, Ahmad S, Deeb SS, Malkki M, Schneider BK, Allen CB, White CW. Hypoxia induces hexokinase II gene expression in human lung cell line A549. Am. J. Physiol. Lung Cell Mol. Physiol. 2000;278:L407–L416. doi: 10.1152/ajplung.2000.278.2.L407. [DOI] [PubMed] [Google Scholar]
- 37.Stryer L. Biochemistry, 4th. New York, NY: W. H. Freeman & Company; 1995. [Google Scholar]
- 38.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 39.Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB, Korsmeyer SJ. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panaretakis T, Pokrovskaja K, Shoshan MC, Grander D. Activation of Bak, Bax, and BH3-only proteins in the apoptotic response to doxorubicin. J. Biol. Chem. 2002;277:44317–44326. doi: 10.1074/jbc.M205273200. [DOI] [PubMed] [Google Scholar]
- 41.Cartron PF, Juin P, Oliver L, Martin S, Meflah K, Vallette FM. Nonredundant role of Bax and Bak in Bid-mediated apoptosis. Mol. Cell Biol. 2003;23:4701–4712. doi: 10.1128/MCB.23.13.4701-4712.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klee M, Pimentel-Muinos FX. Bcl-X(L) specifically activates Bak to induce swelling and restructuring of the endoplasmic reticulum. J. Cell Biol. 2005;168:723–734. doi: 10.1083/jcb.200408169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samraj AK, Stroh C, Fischer U, Schulze-Osthoff K. The tyrosine kinase Lck is a positive regulator of the mitochondrial apoptosis pathway by controlling Bak expression. Oncogene. 2006;25:186–197. doi: 10.1038/sj.onc.1209034. [DOI] [PubMed] [Google Scholar]
- 44.Alon U. 2006. An Introduction to Systems Biology: Design Principles of Biological Circuits. Chapman and Hall/CRC, Boca Ration, FL. [Google Scholar]
- 45.Yan Z, Cui K, Murray DM, Ling C, Xue Y, Gerstein A, Parsons R, Zhao K, Wang W. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19:1662–1667. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kohroki J, Nishiyama T, Nakamura T, Masuho Y. ASB proteins interact with Cullin5 and Rbx2 to form E3 ubiquitin ligase complexes. FEBS Lett. 2005;579:6796–6802. doi: 10.1016/j.febslet.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 47.Heuze ML, Guibal FC, Banks CA, Conaway JW, Conaway RC, Cayre YE, Benecke A, Lutz PG. ASB2 is an Elongin BC-interacting protein that can assemble with Cullin 5 and Rbx1 to reconstitute an E3 ubiquitin ligase complex. J. Biol. Chem. 2005;280:5468–5474. doi: 10.1074/jbc.M413040200. [DOI] [PubMed] [Google Scholar]
- 48.de Lichtenberg U, Jensen LJ, Brunak S, Bork P. Dynamic complex formation during the yeast cell cycle. Science. 2005;307:724–727. doi: 10.1126/science.1105103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.