Abstract
Large population sizes, rapid growth and 3.8 billion years of evolution firmly establish microorganisms as a major source of the planet's biological and genetic diversity. However, up to 99% of the microorganisms in a given environment cannot be cultured. Culture-independent methods that directly access the genetic potential of an environmental sample can unveil new proteins with diverse functions, but the sequencing of random DNA can generate enormous amounts of extraneous data. Integrons are recombination systems that accumulate open reading frames (gene cassettes), many of which code for functional proteins with enormous adaptive potential. Some integrons harbor hundreds of gene cassettes and evidence suggests that the gene cassette pool may be limitless in size. Accessing this genetic pool has been hampered since sequence-based techniques, such as hybridization or PCR, often recover only partial genes or a small subset of those present in the sample. Here, a three-plasmid genetic strategy for the sequence-independent recovery of gene cassettes from genomic libraries is described and its use by retrieving functional gene cassettes from the chromosomal integron of Vibrio vulnificus ATCC 27562 is demonstrated. By manipulating the natural activity of integrons, we can gain access to the caches of functional genes amassed by these structures.
INTRODUCTION
Integrons are natural gene cloning and expression systems that incorporate circularized open reading frames, called gene cassettes, and convert them to functional genes (1–3). The most notable integron gene cassettes code for antibiotic resistance determinants (4,5) and the stockpiling of these cassettes can produce what are often referred to as resistance integrons (RIs), which contain several antibiotic resistance genes in tandem. Five classes of RIs have been defined based on the homology of the integrase genes (6–12) and each is associated with a mobile DNA element. The paradigm for integron function is the class 1 integron (13). The integron platform consists of an integrase gene and an adjacent primary recombination site (attI). The integrase is a member of the tyrosine recombinase superfamily (14) and catalyses recombination between its cognate attI site and a secondary target called an attC site. The attC site is normally found associated with a single open reading frame (ORF), and the ORF–attC structure is termed a gene cassette (15). Integron gene cassettes are compact structures. They typically consist of the ORF, the attC site and little non-coding sequence (16). Consequently, most gene cassettes are promoterless. However, recombination between the attI and attC sites leads to insertion of the gene cassette downstream of a resident promoter, PC, within the integron that drives expression of the encoded product. The strongest variant of PC constitutes a perfect Escherichia coli consensus promoter and is six times more efficient than the de-repressed PTac promoter (17). The integron platforms are sedentary, but the common association of integrons with mobile DNA elements such as transposons and conjugative plasmids facilitates the transit of their amassed genes across phylogenetic boundaries, and augments the impact of integrons on bacterial evolution (2,18,19). The proficiency of this partnership is confirmed by the marked differences in codon usage among cassettes within the same RI, indicating that the captured genes are of diverse origins (20–23).
The ancestors of RIs are the chromosomal integrons (CIs) (24,25). The first was discovered in the Vibrio cholerae genome (26). Located on the smaller of the two circular chromosomes, this element spanned 126 kb and gathered at 179 cassettes into a single structure (24,27), dwarfing previously identified RIs. CIs have since been identified throughout the proteobacterial phylum through both systematic searches (25,28–30) and large-scale microbial genome sequencing efforts (27,31–33). CIs can span hundreds of kilobases and harbour hundreds of gene cassettes. The CIs of the Vibrionaceae are among the largest identified to date, with those from the clinical V. vulnificus strains CMCP6 and YJ016 stretching 153 kb and 138 kb long, and containing 219 and 188 gene cassettes, respectively (34).
Just five Vibrio CIs contain a gene cassette reservoir that is equivalent in size to a small genome (35,36). Furthermore, PCR on environmental DNA samples with degenerate primers targeting the conserved regions of attC sites (37) has led to the recovery of novel integrase genes and hundreds of diverse gene cassettes (38–40). Importantly, accumulation analysis of the gene cassettes showed no signs of saturation (39), supporting the notion that the integron gene cassette pool is likely immense, and that the process of gene cassette genesis is constant and efficient. Although, the vast majority of the thousands of gene cassettes identified in CIs and environmental samples thus far have no counterparts in the database, or the sole homologues are unassigned ORFs, several have been shown to code for proteins that adopt characteristic three-dimensional structures and assemblies indicative of biological activity (41). The gene cassettes for which an activity has been experimentally demonstrated encoded proteins related to simple adaptive functions, including –metabolism (25,42), virulence (43,44) and antibiotic resistance (45–47). Others appear to be of viral, bacterial or eukaryotic origin, indicating their recruitment from all kingdoms of life (38–40,48). Thus, integron gene cassettes are an important source of new enzymes and novel proteins.
Difficulties in sampling or recognizing integron gene cassettes have limited our capacity to exploit this genetic resource. Although useful, a PCR-based approach for the sequence-independent retrieval of functional gene cassettes from environmental samples is limited by the diversity of the attC sites, most of which are unique in sequence and length (25,49). This will undoubtedly preclude the detection of many of the gene cassettes that may be present in a given sample. CI cassettes have been demonstrated to be substrates for recombination by the class 1 integrase of RIs (26,33,45,50,51). Here, the ability of the class 1 integron-integrase, IntI1, to recognize diverse attC target sites is exploited to develop an integron-based genetic selection system to recover functional gene cassettes from genomic libraries.
MATERIALS AND METHODS
Strains, plasmids and media
The E. coli strains, DH10B and EPI300, were from Invitrogen and EpiCentre, respectively. Vibrio vulnificus 75.4T (ATCC 27562), Vibrio cholerae N16961, Xanthomonas campestris pv. campestris 100069T (ATCC 33913) and Shewanella oneidensis 106686 (ATCC 700550; MR-1) were obtained from the Pasteur Institute (Paris, France). All strains were grown in LB media (Sigma). The class 1 integron-integrase gene, int1, was amplified with Platinum Pfx polymerase (Invitrogen) using primers intI1BspHI (5′-ggaattctcatgaaaaccgccactgcgcc) and intI1XbaI (5′-ctagtctagactacctctcactagtgaggg) and cloned into the NcoI and XbaI sites of pTRC99A. R388 and its In3 integron have been previously described (5,52,53) and the pCC1FOS fosmid vector was from EpiCentre. Plasmid DNA purifications were performed using a Sigma Plasmid Miniprep and Midiprep kits. Antibiotics were used at the following concentration when indicated: ampicillin (Ap), 100 µg/ml; rifampicin (Rf), 100 µg/ml; chloramphenicol (Cm), 10 µg/ml; kanamycin (Km), 25 µg/ml; tetracycline (Tc), 15 µg/ml; IPTG 1 mM; l-arabinose (L-ara), 0.02%.
Overall strategy of the three-plasmid integron selection system
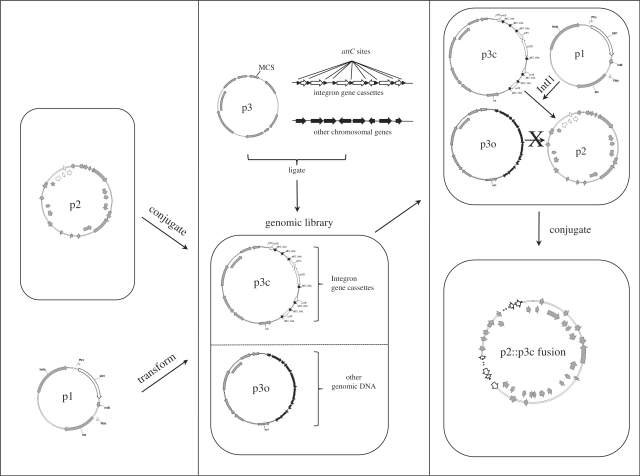
The overall strategy for the selection system is depicted in Figure 1. The system consists of three plasmids: p1 expresses the class 1 integron-integrase gene, intI1; p2 is a self-transmissible recipient plasmid that contains a class 1 integron; p3 is the substrate plasmid into which genomic DNA is cloned and some of the genomic DNA will contain integron gene cassette arrays. The genomic library is constructed in a specialized E. coli host strain (βDH10B) that allows for easy antibiotic-independent counter-selection by the simple omission of a specific nutrient from the media. p2 is transferred by conjugation to the E. coli strain containing the library. The transconjugants are pooled and transformed with p1 to complete assembly of the system. Integrase-mediated recombination between the attI1 site of p2 and target attC sites on p3 plasmids harbouring integron gene cassettes fuses these plasmids together, and the fusions are selected following conjugation to a second E. coli strain and plating on the appropriate selective media that lacks the specific nutrient.
Figure 1.
Strategy of the three-plasmid integron selection system. The system consists of three plasmids: p1 expresses the class 1 integron-integrase gene, intI1 (white arrow); p2 is a self-transmissible recipient plasmid that contains a class 1 integron (white arrows); p3 is the substrate plasmid into which genomic DNA is cloned. A genomic library is constructed in p3 (middle panel), some of which will contain integron gene cassettes (p3c; white arrows, orfs; black arrowheads, attC sites). The rest will contain other chromosomal DNA (p3o; black arrows, orfs). A specialized E. coli strain that allows for easy antibiotic-independent counter-selection by the simple omission of a specific nutrient from the media is used as the host strain. p2 is transferred by conjugation (top left panel) to the E. coli strain containing the library. The transconjugants are pooled and transformed with p1 (bottom left panel) to complete assembly of the system (top right panel). Integrase-mediated recombination between the attI1 site of p2 and target attC sites on p3c fuses these plasmids together, while p3o plasmids are not recombined. The fusions are selected following conjugation to a second E. coli strain and plating on the appropriate selective media that lacks the specific nutrient (bottom right panel).
Constructing the βDH10B host strain
In E. coli strain β2155, the dapA gene is disrupted by a construct containing an erythromycin (erm) cassette and the pir gene (54). The disrupted gene was amplified by PCR with primers DAP1 (5′-atgttcacgggaagtattgtc) and DAP2 (5′-cagcaaaccggcatgcttaa) and purified. E. coli DH10B was transformed with pKOBEGA, which carries the λ red (redαβγ) recombination system under the control of the arabinose-inducible PBAD promoter (55). This system can be used to generate gene knockouts in E. coli by allelic exchange. DH10B/pKOBEGA cells were made electrocompetent following arabinose induction and were transformed with the linear DAP1/DAP2 PCR product to create strain βDH10B. Transformants were selected on LB Erm plates containing diaminopimelic acid (DAP). Disruption of the dapA gene was conformed by PCR and the strains inability to grow on plates lacking DAP.
Fosmid library construction
Genomic DNA was prepared using the DNAzol genomic DNA isolation reagent (Invitrogen) as per the manufacturer's instructions. The DNA was then sheared by passage through a Hamilton syringe, end-repaired and ligated into Eco72I-digested pCC1FOS using the CopyControl Fosmid Library Production Kit (EpiCentre). The ligation was then transformed into the βDH10B strain and colonies were selected on LB DAP Cm plates. Alternatively, sheared DNA was end-repaired and separated on a 1% low melting point PFGE gel. Gel slices containing fragments of 40 kb were excised and digested with gelase (InVitrogen) to recover the DNA. The DNA was concentrated by ethanol precipitation when necessary. The 40-kb DNA fragments were then ligated into Eco72I-digested pCC1FOS. MaxPlax lambda packaging extracts (Epicentre) were used to package the library and transduce βDH10B cells. Transductants were selected on LB DAP Cm plates at 37°C. The colonies were grown overnight at 37°C. To determine how many clones were required in order to obtain a given genome sequence with 99% probability, I used the equation N = ln (1 − P)/ln (1 – I/GS), where N = number of clones, P = probability, I = insert size and GS = genome size (56). Six hundred clones of a 40-kb insert genomic library were assembled. Screening on X-gal indicated that >90% of the clones were recombinant.
Integrase-mediated recombination of fosmid clones containing gene cassettes
The R388ISceKT plasmid was conjugated from E. coli DH5α cells to the βDH10B library. βDH10B transconjugants were selected on LB DAP Cm Km plates at 37°C. These cells were then pooled and transformed with pTRC99A::intI1 and transformants were selected on LB DAP Cm Km Ap plates at 37°C. The specific recovery of fosmid clones containing CI gene cassettes from the library was monitored as follows. The cells were pooled and incubated overnight in LB liquid media containing DAP Cm Km Ap and IPTG at 37°C with shaking. The next day, recombinant plasmids were isolated by conjugation to E. coli EPI300 cells and selection of transconjugants on LB Cm plates that lacked DAP. Higher yield of recombinant plasmids for PFGE, sequencing and activity screening was obtained by growing the transconjugants in media containing the induction solution that increases expression of trfA in the EPI300 strain.
Sequencing and protein analysis
Fusion plasmids were used in PCR reactions with primers intI1 and VVR1, VCR1, XCR1 or SPR1-3. The intI1 primer anneals within intI1 and VVR1, VCR1 and XCR1 hybridize to the signature attC sites of V. vulnificus, V. cholerae and X. campestris, respectively (25,28). SPR1, SPR2 and SPR3 hybridize to three of the attC sites from the CI of S. oneidensis (27,52). PCR was conducted as follows: 94°C, 30 s; 60°C, 30 s; 72°C, 2 min, for 30 cycles with Pfu polymerase. Neither primer alone yielded a PCR product, nor did PCR with intI1 and the respective second primer on uninduced cultures. The amplified fragments were sequenced directly with the intI1 primer to confirm integration at the attI site. Sequencing was done at The Centre for Applied Genomics (TCAG, Hospital for Sick Children, Toronto). Homology searches were conducted using BLAST analysis (57), and protein parameters and domain predictions were performed with ProtParam (58) and the PROSITE research tool from the Expert Protein analysis system (59). Protein structure predictions were made using Phyre (60). The 35-kb CI region of V. vulnificus ATCC 27562 was sequenced at The Joint Genome Institute (JGI, University of California) and was deposited at GenBank (accession no. GQ292873).
RESULTS
The three-plasmid integron selection system
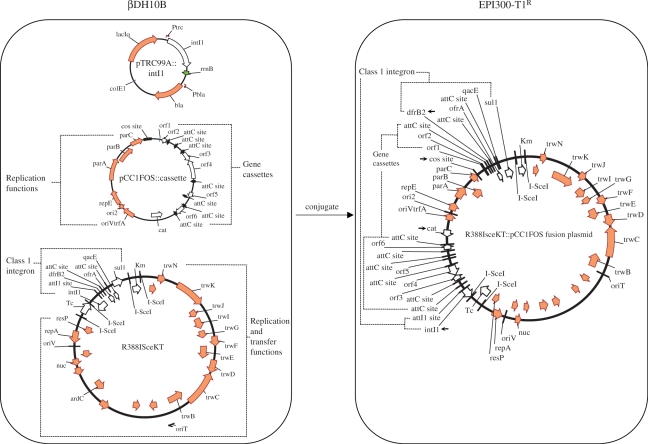
Based on the strategy outlined in Figure 1, a three-plasmid retrieval system was developed for the sequence-independent recovery of complete, functional open reading frames from CIs (Figure 2). The first plasmid, pTRC99A::intI1, expresses the integron-integrase gene, intI1, from the IPTG-inducible PTRC promoter (61). The second, R388ISceKT, is the recipient plasmid. It is a Km- and Tc-resistant derivative of the 34-kb self-transmissible plasmid R388, which carries an attI1 site within its class 1 integron, In3 (52). In3 contains two genes: dfrB2, which confers trimethoprim resistance, and orfA, of unknown function (62). However, dfrB2 is the first gene cassette of In3. Recombination of an attC site with the attI1 site would displace the promoterless dfrB2 from the Pc promoter, potentially leading to trimethoprim sensitivity due to a lack of dfrB2 expression. Furthermore, a defined media that lacks any thymine derivatives must be used when selecting with trimethoprim. The addition of the aph gene permits selection in complex media and avoids the potential problem of clone loss that can occur during growth of genomic libraries in defined media. The plasmid also includes a Tc resistance marker and I-SceI sites, which aid in the isolation of the insert. The third, pCC1FOS, is the substrate plasmid into which genomic DNA is cloned. pCC1FOS is an 8-kb fosmid that carries an F-factor origin of replication (ori2) and partitioning functions that maintain the fosmid in single copy. This circumvents the problem of under-representation of clones carrying genes that may be toxic when present in multiple copies. A second TrfA-dependent origin of replication, oriV, is also present on the fosmid. Genomic fragments of any size can be cloned into the unique Eco72 I (blunt end) site within lacZα and recombinants are selected by blue/white screening. If size selection is desired, the fosmid also contains cos sites for packaging into phage particles in vitro, ensuring that the library is comprised only of plasmids containing inserts of ∼40 kb under these conditions. The host strain for maintaining libraries is a derivative of the E. coli strain DH10B that has a disruption in dapA, a component of the diaminopimelic acid (DAP) biosynthetic pathway. DAP is incorporated into the bacterial cell wall peptidoglycan. E. coli mutants altered in the DAP pathway require exogenous DAP for growth. Use of this strain (βDH10B) as a donor in conjugation experiments allows for absolute counter-selection in an antibiotic-independent manner even in complex media such as LB, making it easy to isolate transconjugants. βDH10B has a high transformation efficiency for both large and small clones, readily accepts large DNA for the construction of large-insert genomic libraries, and is endA1 and recA1 to ensure high yields of DNA and greater stability of large cloned inserts.
Figure 2.
The three-plasmid integron selection system. An example of the βDH10B host strain, which requires exogenous DAP for growth, carrying a pCC1FOS library clone containing gene cassettes is shown. The same strain also carries the pTRC99A::intI1 and R388ISceKT plasmids. IPTG-induced expression from pTRC99A::intI1 leads to integrase-mediated recombination between pCC1FOS plasmids containing attC sites and the attI1 site of the class 1 integron on the conjugative R388 plasmid. The recombinant plasmid is specifically selected by conjugation to the EPI300-T1R recipient and plating on media containing chloramphenicol but lacking DAP. The original class 1 integron of R388 is disrupted by the fusion. Small black arrows denote primers that can be used to sequence the gene cassettes of the recovered recombinant plasmids.
The expression of intI1 catalyzed recombination between the attI1 site of In3 on R388ISceKT and pCC1FOS fosmids from the library that contained inserts with at least one functional attC site, resulting in fusion of the two plasmids. Since the mobilizable R388ISceKT plasmid was KmR, and the CmR pCC1FOS fosmid did not contain an origin of conjugative transfer, the recombinant plasmids were easily isolated by conjugation to a recipient E. coli strain (EPI300-T1R) and selection on LB Km Cm plates that lacked DAP. EPI300-T1R cells are endA1 and recA1, and carry a copy-up allele of the trfA gene integrated into the chromosome under control of an inducible promoter. Induced expression of trfA allows for replication from oriV of pCC1FOS. The resulting 40–80-fold increase in the fosmid copy number provided for better fosmid yields for sequencing of the gene cassettes and aided activity-based screening of gene cassettes expressed from PC of In3. If desired, the integrated fosmid of the recombinant plasmid could be recovered by digestion with I-SceI, self-ligation, transforming EPI300 cells and selecting clones on LB Cm plates. The fosmid harbouring the chromosomal fragment can replicate via the oriV of pCC1FOS.
Recovery of functional genes from a genomic library with the integron-based recombination system
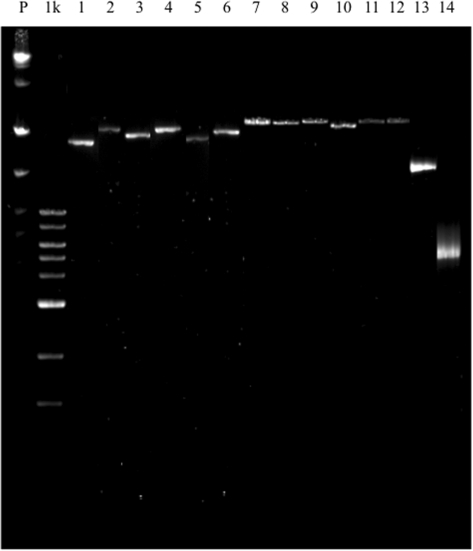
The 153-kb CI of V. vulnificus strain CMCP6 represents 3% of its genome (34). Genomic DNA from strain CMCP6 was ligated into pCC1FOS and the library was either directly transformed into βDH10B or packaged into phage and transduced into the host strain. R388IsceKT and pTRC99A::intI1 were then moved to these strains to complete assembly of the three-plasmid system. The expression of intI1 was induced with IPTG and recombinant plasmids were retrieved by conjugation to EPI300-T1R recipient cells. A representative gel of several random recombinant plasmids from the transformed and transduced libraries is shown in Figure 3. The wider size range for recombinant plasmids obtained with the transformed library relative to the more uniform plasmid size obtained from the transduced library was reflective of the size-selection imposed by packaging of the ligation reaction into phage particles prior to transduction. Restriction digests of randomly chosen recombinant plasmids suggested that they were indeed the products of recombination between the pCC1FOS library and R388 (data not shown). Specific recombination between the attC site of the substrate plasmid (pCC1FOS library) and the attI1 site of recipient plasmid (R388ISceKT) was verified by PCR and sequencing.
Figure 3.
Pulse-field gel of the R388-pCC1FOS recombinant plasmids obtained from the transformed and transduced cosmid libraries. Plasmids were isolated and separated by PFGE. P, PFGE marker; 1 k, 1-kb maker; lanes 1–6, plasmids chosen at random from the transformed substrate library; lanes 7–12, plasmids chosen at random from the transduced substrate library; lane 13, R388; lane 14, the empty pCC1FOS cosmid.
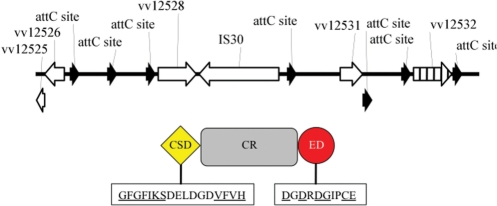
One of the sequenced recombinant clones from the transformed pCC1FOS library contained a 5383-bp insert with 7 attC sites and corresponded to genes vv12525-vv12533 of strain CMCP6 (Figure 4). One of the partial gene cassettes contained two small overlapping orfs (vv12525 and vv12526) that coded for a toxin-antitoxin system that we previously characterized from V. vulnificus strain 27562 (63). Toxin/Antitoxin (TA) systems encode a stable toxin and a specific but unstable antitoxin (64–66). They enhance plasmid maintenance by preventing the proliferation of plasmid-free progeny because the continued de novo synthesis of the short-lived antitoxin is necessary to ensure cell survival. These toxins have proven useful in various molecular biology applications. The TA toxin CcdB (control of cell death), which poisons bacterial DNA gyrase causing degradation of the host chromosome and cell death (67–69), is a key component of the pSW2644T allelic replacement vector, where expression of ccdB is used as a counter-selection marker (70,71). Invitrogen's Zero Background™ cloning methodology exploits the same toxin to eliminate high background in blunt-end cloning strategies. When an insert is ligated into the vector, the ccdB gene is disrupted, enabling only recombinant colonies to grow.
Figure 4.
Schematic of the insert from a R388-pCC1FOS recombinant plasmid obtained from the transformed cosmid library of V. vulnificus strain CMCP6. (Top) The 5383-bp insert contained seven attC sites (black arrows) and corresponded to genes vv12525-vv12533 of the CI of strain CMCP6. One of the partial gene cassettes contained two small overlapping orfs (vv12525 and vv12526) that coded for a toxin–anti-toxin system that we previously characterized from V. vulnificus strain 27562 (28,63). Another cassette, vv12532 (striped), coded for a putative cold shock protein (CSP). (Bottom) The domain structure of VV12532. Cold shock domains (CSDs) contain 11 residues (underlined) that are important for CSP function. Excalibur domains (ED) bind extracellular calcium and contain the conserved DxDxDGxxCE motif (underlined). CR, central region.
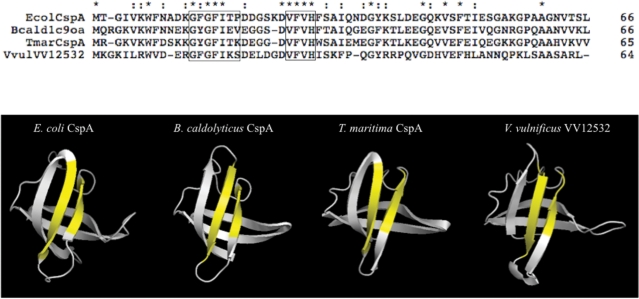
Another cassette (vv12532) coded for a protein with an Excalibur domain and a putative cold shock domain (CSD) (Figure 4). Excalibur domains bind extracellular calcium and contain the conserved DxDxDGxxCE motif, which is strikingly similar to the Ca2+-binding loop of calmodulin-like EF-hand domains (72). CSD are commonly found in cold shock proteins (CSP) (73). CSP typically acts as transcription anti-terminators to prevent the formation of secondary structures in RNA molecules, thus facilitating translation at low temperature. Eleven residues that are important for CSP function are proposed to form two RNA-binding motifs, RNP-1 and RNP-2, on the surface of the CSP (74–79). An alignment between VV12532 and the major CSP of E. coli, Thermotoga maritima and Bacillus caldolyticus suggested that the RNP-1 and RNP-2 motifs were present in VV12532 (Figure 5). The VV12532 CSD was predicted to adopt an oligomer binding (OB)-fold similar to that of the E. coli, T. maritime and B. caldolyticus CSP, and the surface location of the RNP-1 and RNP-2 motifs was also conserved (Figure 5).
Figure 5.
Comparison of the CSD of VV12532 to the major CSP of E. coli, Thermotoga maritima and Bacillus caldolyticus. (Top) Alignment between the CSD of VV12532 and the major CSP of the indicated species. Protein lengths are indicated on the right. Asterisks and colons mark identical and similar amino acids, respectively. The 11 residues that form the two RNA-binding motifs, RNP-1 and RNP-2, and that are important for CSP function are boxed. Bottom, the known structures of the major CSP of E. coli, T. maritima and B. caldolyticus are shown. The two RNA-binding motifs, RNP-1 and RNP-2 (yellow regions), are on the surface of the CSP. A proposed structure for the CSD of VV12532 is also shown.
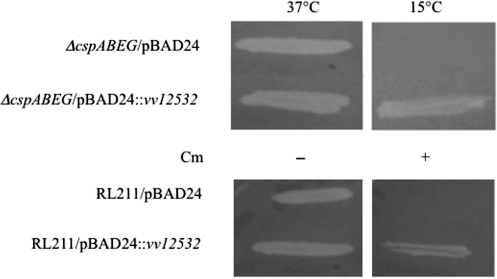
These observations suggested that VV12532 might function as a CSP. At least four separate csp genes must be deleted in E. coli to obtain a cold-sensitive phenotype due to overlapping activity of the CSP family of proteins (80). In E. coli, complementation by any one allele can revert the cold-sensitive phenotype of the quadruple deletion ΔcspABGE strain (81). The genomes of strain YJ016 and CMCP6 contain 4 and 5 CSP homologues, respectively. If a similar situation to that in E. coli occurs in V. vulnificus, targeted disruption of the putative vv12532 csp gene cassette alone would likely have little or no effect on the adaptation of V. vulnificus to cold temperature. Therefore, I opted to test the VV12532 CSP homologue by expressing it in the ΔcspABGE E. coli cold-sensitive mutant. The vv12532 gene was cloned into the arabinose-inducible pBAD24 expression vector (82) and transformed into the E. coli cspABEG quadruple mutant. The cold-sensitive phenotype of the quadruple-deletion strain was suppressed in cells that expressed vv12532 but not in cells that carried the empty vector (Figure 6), suggesting that VV12532 was a CSP. The protein was thus designated CspAvvu. To determine if CspAvvu acted as a transcription anti-terminator, pBAD24::cspAvvu was transformed into the E. coli strain RL211 (83,84). RL211 contains a cat gene that is preceded by the ρ-independent trpL terminator. This strain is sensitive to chloramphenicol but becomes chloramphenicol resistant when transcription termination at trpL is reduced and thus provides a measure of antitermination activity. RL211 cells carrying pBAD24::cspAvvu grew on plates containing chloramphenicol under inducing conditions, whereas the parental strain carrying pBAD24 did not (Figure 6). Hence, CspAvvu was a CSP that appeared to act as a transcription antiterminator.
Figure 6.
CspAvvu is a CSP that causes transcription antitermination. Top, the E. coli cspABEG quadruple mutant with pBAD24::vv12532 or the empty vector were streaked on plates containing L-ara and were incubated at the indicated temperatures. Bottom, E. coli RL211 cells containing a cat gene cassette positioned downstream of the trpL terminator were transformed with pBAD24::cspAvvu and streaked on plates containing 25 µg/ml chloramphenicol and arabinose. Plates were incubated overnight at 37°C.
There were only 27 bp between the end of the 5′ attC site and the start ATG of cspAvvu. A translation initiation region (TIR) was apparent but the sequence did not include a discernable promoter. Hence, I sought to directly screen for the activity of CspAvvu in vivo by relocating gene cassettes from the V. vulnificus CMCP6 CI to the attI1 site of the class 1 integon on R388ISceKT. This would place any incorporated CI cassettes downstream of the PC promoter to ensure their expression. R388ISceKT and pTRC99A::intI1 were introduced into V. vulnificus CMCP6 by conjugation. Following induction of intI1 gene expression, R388 was transferred to the E. coli ΔcspABGE cold-sensitive mutant and transconjugants were selected on LB plates containing Tc and Km at 15°C. Sequence analysis of several transconjugants revealed that a CI cassette, corresponding to cspAvvu, had been precisely inserted into In3.
Comparative genomic analysis of a 35-kb contig containing CI gene cassettes from V. vulnificus strain ATCC 27562
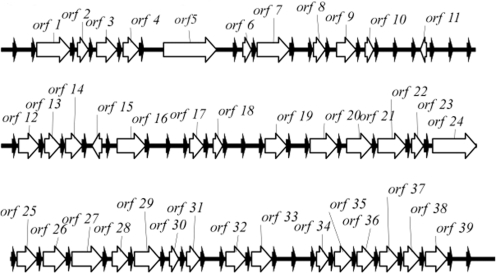
Genomic comparisons of the CI cassette contents of the two sequenced V. vulnificus strains CMCP6 and YJ016 revealed that they share only 30–39% of their gene cassettes (34,85). The genome of strain 27562 has not been sequenced but we previously demonstrated that it contained a CI (28). To recover fosmids carrying large inserts from the CI, genomic DNA from strain 27562 was ligated into pCC1FOS, the library was packaged into phage and then transduced into the βDH10B host strain. R388IsceKT and pTRC99A::intI1 were then moved to this strain to complete assembly of the system. Recombination was induced with the addition of IPTG and recombinant plasmids were isolated by conjugation to strain EPI300. The 35-kb insert of one of the recovered clones (p2190) was sequenced and annotated (Figure 7), and the attC sites and gene cassette boundaries were extracted with the XXR program we developed (28). Of the 55 complete and two partial gene cassettes that we identified, 39 (71%) contained a discernable orf (Table 1), 23 (59%) had homologues in the CIs of other members of the Vibrionaceae and only 16 (29%) had homologues in either the CMCP6 or YJ016 CIs. Furthermore, only about 12% of the CI cassettes from the V. vulnificus CMCP6, YJ016 and 27562 strains had homologues in the CI of V. cholerae N16961. These results support the notion that CIs contribute to the extensive heterogeneity of the Vibrionaceae and that the pool of gene cassettes available for exchange among bacteria is likely immense.
Figure 7.
Schematic of a 35-kb region of the V. vulnificus ATCC 27562 CI. The insert of p2190 was sequenced and annotated. Gene cassettes with discernable orfs (white arrows) are numbered. The attC sites are shown as black arrows.
Table 1.
Homologues of the orfs from the 27562 CI
| orf | Bacterial species | E-value | Function | Accession no. |
|---|---|---|---|---|
| 1 | NH | Hypothetical protein | ||
| 2 | NH | Hypothetical protein | ||
| 3 | Shewanella sp. MR7 | 8e−30 | preprotein translocase subunit SecA | YP_736195.1 |
| 4 | NH | Hypothetical protein | ||
| 5 | Legionella pneumophila | 3e−152 | Reverse transcriptase | YP_001251128.1 |
| 6 | V. vulnificus | 5e−44 | Hypothetical protein | NP_934729.1 |
| 7 | NH | Hypothetical protein | ||
| 8 | V. vulnificus | 4e−55 | Hypothetical protein | NP_761344.1 |
| 9 | NH | Hypothetical protein | ||
| 10 | Aeromonas salmonicida | 6e−31 | Hypothetical protein | YP_001143449.1 |
| 11 | Shewanella sp. MR4 | 2e−17 | Hypothetical protein | YP_734249.1 |
| 12 | S. lioihica PV4 | 7e−5 | Hypothetical protein | YP_001095272.1 |
| 13 | V. vulnificus | 2e−80 | Hypothetical protein | NP_934596.1 |
| 14 | Aliivibrio salmonicida | 4e−18 | Acetyltransferase | YP_002262874 |
| 15 | V. vulnificus | 1e−41 | Hypothetical protein | NP_761251.1 |
| 16 | V. fischeri | 3e−108 | Esterase/lipase | YP_002156256.1 |
| 17 | V. vulnificus | 5e−73 | Hypothetical protein | NP_761320.1 |
| 18 | S. lioihica PV4 | 9e−19 | Hypothetical protein | YP_001094978.1 |
| 19 | NH | Hypothetical protein | ||
| 20 | V. parahaemolyticus | 3e−151 | CBS domain protein | NP_798204.1 |
| 21 | V. vulnificus | 5e−131 | Hypothetical protein | NP_761333.1 |
| 22 | V. vulnificus | 2e−165 | Hypothetical protein | NP_761335.1 |
| 23 | V. parahaemolyticus | 1e−60 | Hypothetical protein | NP_798185.1 |
| 24 | Oenococcus oeni | 5e−59 | Permease of the major facilitator superfamily | YP_809940.1 |
| 25 | NH | Hypothetical protein | ||
| 26 | NH | Hypothetical protein | ||
| 27 | V. vulnificus | 2e−142 | Hypothetical protein | NP_761333.1 |
| 28 | A. hydrophila | 3e−10 | Hypothetical protein | |
| 29 | NH | Hypothetical protein | ||
| 30 | V. vulnificus | 5e−27 | Acetyltransferase | NP_761288.1 |
| 31 | Photobacterium profundum | 2e−17 | Hypothetical protein | YP_132162.1 |
| 32 | NH | Hypothetical protein | ||
| 33 | NH | Hypothetical protein | ||
| 34 | V. vulnificus | 4e−47 | Hypothetical protein | |
| 35 | NH | Hypothetical protein | ||
| 36 | V. vulnificus | 9e−71 | Lactoglutathione lyase | |
| 37 | V. vulnificus | 1e−86 | Hypothetical protein | NP_934598.1 |
| 38 | V. vulnificus | 7e−81 | 4-oxalocrotonate tautomerase | NP_761363.1 |
| 39 | V. vulnificus | 3e−109 | Adenylate kinase | NP_934722.1 |
Several of the CI cassettes showed homology to enzymes involved in energy metabolism and detoxification (Table 1). These included a 4-oxalocrotonate tautomerase that enables bacteria to use various aromatic hydrocarbons as their sole energy source (86), a carbohydrate transport protein of the major facilitator superfamily, an adenylate kinase that interconverts ADP to AMP and ATP for driving energy requiring processes and a lactoylglutathione lyase detoxification enzyme that converts toxic methylglyoxal (MG) to lactic acid (87).
To demonstrate that the system could recover integron gene cassettes from different sources within the same sample, genomic DNA from V. cholerae, X. campestris pv campestris and S. oneidensis was isolated, mixed in equal quantities and cloned into pCC1FOS. While each of these species is known to harbour a CI (27,30,88), they are of varying size. The CIs of V. cholerae, X. campestris and S. oneidensis harbour 216, 23 and three gene cassettes, respectively (27,32,89), and represent 3, 0.3 and 0.1% of the genomes. The three-plasmid system was assembled and integrase-mediated recombination was induced. Recovered fusion plasmids were used in PCR reactions with primers intI1 and VVR1, VCR1, XCR1 or SPR1-3, and the products were sequenced. Products corresponding to integron gene cassettes from the CIs of each species were identified, suggesting that the system can indeed recover low abundance integron gene cassettes from different sources within the same sample.
Sequencing of integron gene cassette arrays with the integron-based recombination system
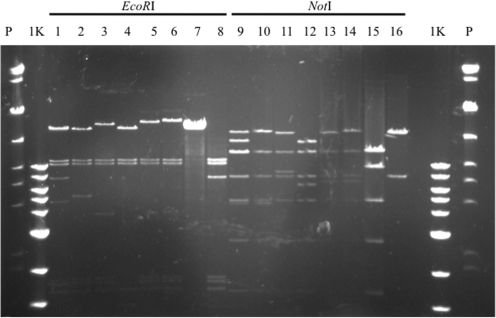
The system can also be applied to the sequencing of integron gene cassette arrays. The p2190 plasmid was used as the sole substrate plasmid in the integron-based recombination assay with R388ISceKT. Recombination was induced by the addition of IPTG and recombinant plasmids were isolated by conjugation. The recombinant plasmids from six randomly chosen transconjugants were digested with either EcoRI or NotI and separated by PFGE (Figure 8). Although each of the plasmids maintained the same overall size, each exhibited a different banding pattern, suggesting that multiple attC sites of p2190 could serve as substrates for recombination with the attI1 site on R388ISceKT. This can prove useful since a series of plasmids with differing restriction patterns can be sequenced using two sets of unique primers that anneal within the 5′ conserved region and the dfrb2 gene of the class 1 integron, and within the cat gene and cos site of pCC1FOS, to obtain the sequence of the gene cassette array (Figure 2).
Figure 8.
Pulse-field gel of R388-p2190 fusion plasmids. Fusion plasmids that were obtained by integrase-mediated recombination between the integron platform of R388 and the gene cassettes of p2190 were digested with NotI or EcoRI and separated by PFGE. Lanes are as follows: P, PFGE markers; 1 K, 1-Kb markers; 1 and 9, clone 1; 2 and 10, clone 2; 3 and 11, clone 3; 4 and 12, clone 4; 5 and 13, clone 5; 6 and 14, clone 6; 7 and 15, R388; 8 and 16, p2190.
DISCUSSION
Fundamentally, the integron platform is a simple structure. There is an integrase (intI) and an adjacent, cognate recombination site (attI). The substrates for this system are the compact gene cassettes, many of which code for functional proteins or proteins that can adopt a structure of known biological significance. Further interest in the nature of the gene cassette pool has emerged with the demonstration that metabolic (25,42), virulence (43,44) and antibiotic resistance (45–47) genes are structured as gene cassettes and that the gene cassette pool may be limitless in size (1,90). Accessing this genetic pool has been hampered since sequence-based techniques such as hybridization with probes based on known genes or PCR with primers directed at conserved sequence motifs, often lead to the recovery of only partial genes or a small subset of those present in the sample. A previous method for the sequence-independent recovery of integron gene cassettes from environmental samples relied on cassette detection by PCR using degenerate primers that targeted conserved sequences within the attC sites (37). Although useful, this approach is limited by the diversity of the attC sites, which will undoubtedly preclude the detection of many of the gene cassettes that may be present in a given sample. Here, a genetic system to extract fosmids harboring integron gene cassettes from a bulk genomic pool is described. The system is comprised of three plasmids. The first expresses the class 1 integron-integrase under the control of an IPTG-inducible promoter. The second is a self-transmissible recipient plasmid that carries an attI1 site for the integration of gene cassettes via recombination with their attC sites. The third is the donor plasmid that contains the genomic DNA library. IntI1-catalysed recombination between the attI site of recipient plasmid and target attC sites on the inserts of donor plasmids results in the fusion of the two plasmids. The fusion plasmid can then be isolated by conjugation into a recipient host and recovered for sequencing or activity screening of the gene cassettes.
The system has several advantages over existing methods for the detection of gene cassettes. Since integron-integrases are able to recognize diverse attC sites as targets for recombination (39,51,91,92), a recovery system based on the recombination activity of the integron integrase improves the likelihood of recovering gene cassettes from samples relative to detection by PCR or hybridization. Primers targeting the cos site, dfrB2, cat and intI1 genes can be used to sequence the insert of the fusion plasmid. The I-SceI sites flanking the Km and Tc resistance markers of the fusion plasmid (CmR, KmR, TcR) can be used to isolate clones lacking the R388 backbone (CmR KmS, TcS) by replica plating on media containing the appropriate antibiotics. Obtaining such clones may be desirable for sequencing purposes or for activity-based screening. Finally, the ability to directly implement activity-based screening is a significant advantage. The activity-based screening of gene banks allows for the recovery of novel enzymes without any prior knowledge of the sequence and relies only on the ingenuity of the screening method. Chitinases, amylases, DNAses, esterase/lipases, proteases, oxygenases, dehydrogenases and antimicrobial activities have been identified via activity screens of genomic libraries (93,94). Many of these enzymes are used as key active components in detergents, various food/feed processes, pulp and paper manufacturing, molecular biology applications and medical treatment protocols. Activity screening requires gene expression and proper folding of the resulting protein in a heterologous host. In E. coli, the minimal set of requirements for gene expression includes the presence of a promoter for transcription and a translation initiation region (TIR) comprised of a Shine–Dalgarno sequence, the initiation codon, and an adequate spacer between them. In such systems, three modes of gene expression can be anticipated: (i) independent gene expression with both the promoter and the TIR provided by the insert; (ii) expression as a transcriptional fusion with only the TIR located on the insert; and (iii) expression as a translational fusion depending on both the promoter and the TIR of the vector. Genes of the last category are virtually inaccessible by shotgun cloning because of the low frequency of functional constructs. Class 1 integrons naturally incorporate many of the attributes necessary for the proper expression of exogenous genes. The Pc promoter drives expression of promoterless gene cassettes integrated at the attI site. Often, a TIR can be recognized at the 5′ boundary of the gene cassette. Even cassettes that have no plausible TIR can be translated. A short open reading frame (ORF-11) that is separated by an octameric spacer from a consensus SD tetramer overlaps the attI site of class 1 integrons. Hanau-Berçot et al. (95) demonstrated that translation of the TIR-deficient aminoglycoside 6′-N-acetyltransferase (aac(6′)-Ib7) gene cassette was dependent on the translation of ORF-11 but not on the amino acid sequence of the corresponding peptide. These results indicated that ORF-11 and its TIR could substantially enhance expression at the translational level of TIR-deficient gene cassettes. Hence, the integron/gene cassette system can support all three modes of gene expression. Even potential translational attenuation signals, the attC sites themselves (96), are present. Using integron-based activity screening, we previously identified a novel cat gene in the CI of V. cholera (45) and here I demonstrate the activity of a CSP encoded by a V. vulnificus CI gene cassette.
Greater than 99% of the microorganisms present in many natural environments are unculturable. Hence their genetic wealth has eluded exploitation for medical, biotechnological or basic research purposes. Cultivation-independent approaches, in which DNA is directly isolated from environmental samples and used to construct complex genomic libraries, permits analysis of the genetically and metabolically rich microbial communities in their entirety (97). This obviates the need for cultivation and allows the direct cloning of the collective genomes of all organisms present in a habitat. The quest to identify functional genes in the vast amounts of genetic data contained in these metagenomic libraries presents a variety of challenges, not the least of which is filtering out the often large mounts of undesired DNA that codes for informational (transcription, translation and related processes) and conserved operational (housekeeping) genes (98,99). This can be overcome, in part, by focusing on genes that are structured as cassettes in integrons, since they tend to code for proteins of unknown function or with tremendous adaptive potential (1,100–104), including structural (105), metabolic (25,42,106), resistance (40,107), toxin (45,46,108) and virulence proteins (44,109). Thus, integron gene cassettes may be a tremendous source of new enzymes and novel proteins from diverse origins. By manipulating the natural activity of integrons, it should be possible to gain access to many of the functional gene cassettes contained within a metagenomic sample.
FUNDING
The Natural Sciences and Engineering Research Council of Canada (293281) and the Canadian Institutes of Health Research (MOP-89794). Funding for open access charge: CIHR.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
I would like to thank Dr Jan-Fang Cheng (JGI) for sequencing of fosmid clones, and Erica Susky and Bella Kuchuk for technical assistance.
REFERENCES
- 1.Rowe-Magnus DA, Mazel D. Integrons: natural tools for bacterial genome evolution. Curr. Opin. Microbiol. 2001;4:565–569. doi: 10.1016/s1369-5274(00)00252-6. [DOI] [PubMed] [Google Scholar]
- 2.Mazel D. Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 2006;4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 3.Rowe-Magnus DA, Mazel D. Resistance gene capture. Curr. Opin. Microbiol. 1999;2:483–488. doi: 10.1016/s1369-5274(99)00004-1. [DOI] [PubMed] [Google Scholar]
- 4.Rowe-Magnus DA, Mazel D. The role of integrons in antibiotic resistance gene capture. Int. J. Med. Microbiol. 2002;292:115–125. doi: 10.1078/1438-4221-00197. [DOI] [PubMed] [Google Scholar]
- 5.Mazel D, Davies J. Antibiotic resistance in microbes. Cell Mol. Life Sci. 1999;56:742–754. doi: 10.1007/s000180050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez E, de la Cruz F. Transposon Tn21 encodes a RecA-independent site-specific integration system. Mol. Gen. Genet. 1988;211:320–325. doi: 10.1007/BF00330610. [DOI] [PubMed] [Google Scholar]
- 7.Martinez E, de la Cruz F. Genetic elements involved in Tn21 site-specific integration, a novel mechanism for the dissemination of antibiotic resistance genes. EMBO J. 1990;9:1275–1281. doi: 10.1002/j.1460-2075.1990.tb08236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansson K, Sundstrom L, Pelletier A, Roy PH. IntI2 integron integrase in Tn7. J. Bacteriol. 2002;184:1712–1721. doi: 10.1128/JB.184.6.1712-1721.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sundstrom L, Roy PH, Skold O. Site-specific insertion of three structural gene cassettes in transposon Tn7. J. Bacteriol. 1991;173:3025–3028. doi: 10.1128/jb.173.9.3025-3028.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arakawa Y, Murakami M, Suzuki K, Ito H, Wacharotayankun R, Ohsuka S, Kato N, Ohta M. A novel integron-like element carrying the metallo-beta-lactamase gene blaIMP. Antimicrob. Agents Chemother. 1995;39:1612–1615. doi: 10.1128/aac.39.7.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hochhut B, Lotfi Y, Mazel D, Faruque SM, Woodgate R, Waldor MK. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 2001;45:2991–3000. doi: 10.1128/AAC.45.11.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sorum H, Dommarsnes K, Sandersen K, Sundstrom L, Gullberg M, Solberg A. 2001. Genbank accession no. AJ277063. [Google Scholar]
- 13.Liebert CA, Hall RM, Summers AO. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 1999;63:507–522. doi: 10.1128/mmbr.63.3.507-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nunes-Duby SE, Kwon HJ, Tirumalai RS, Ellenberger T, Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall RM. Mobile gene cassettes and integrons: moving antibiotic resistance genes in gram-negative bacteria. Ciba Found. Symp. 1997;207:192–202. doi: 10.1002/9780470515358.ch12. [DOI] [PubMed] [Google Scholar]
- 16.Recchia GD, Hall RM. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 17.Levesque C, Brassard S, Lapointe J, Roy PH. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene. 1994;142:49–54. doi: 10.1016/0378-1119(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 18.Rowe-Magnus AD, Davies J, Mazel D. Impact of integrons and transposons on the evolution of resistance and virulence. Curr. Top. Microbiol. Immunol. 2002;264:167–188. [PubMed] [Google Scholar]
- 19.Rowe-Magnus DA, Davies J, Mazel D. In: Pathogenicity Islands and the Evolution of Pathogenic Microbes. Hacker J, Kaper J, editors. 264/2. 2002. pp. 167–188. [Google Scholar]
- 20.Davies JE. Origins, acquisition and dissemination of antibiotic resistance determinants. Ciba Found. Symp. 1997;207:15–27. discussion 27–35. [PubMed] [Google Scholar]
- 21.Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science. 1994;264:375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- 22.Peters ED, Leverstein-Van Hall MA, Box AT, Verhoef J, Fluit AC. Novel gene cassettes and integrons. Antimicrob. Agents Chemother. 2001;45:2961–2964. doi: 10.1128/AAC.45.10.2961-2964.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Recchia GD, Hall RM. Origins of the mobile gene cassettes found in integrons. Trends Microbiol. 1997;5:389–394. doi: 10.1016/S0966-842X(97)01123-2. [DOI] [PubMed] [Google Scholar]
- 24.Rowe-Magnus DA, Guerout A-M, Mazel D. Super-Integrons. Res. Microbiol. 1999;150:641–651. doi: 10.1016/s0923-2508(99)00127-8. [DOI] [PubMed] [Google Scholar]
- 25.Rowe-Magnus DA, Guerout AM, Ploncard P, Dychinco B, Davies J, Mazel D. The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc. Natl Acad. Sci. USA. 2001;98:652–657. doi: 10.1073/pnas.98.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazel D, Dychinco B, Webb VA, Davies J. A distinctive class of integron in the Vibrio cholerae genome. Science. 1998;280:605–608. doi: 10.1126/science.280.5363.605. [DOI] [PubMed] [Google Scholar]
- 27.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowe-Magnus DA, Guerout AM, Biskri L, Bouige P, Mazel D. Comparative analysis of superintegrons: engineering extensive genetic diversity in the vibrionaceae. Genome Res. 2003;13:428–442. doi: 10.1101/gr.617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaisvila R, Morgan RD, Posfai J, Raleigh EA. Discovery and distribution of super-integrons among Pseudomonads. Mol. Microbiol. 2001;42:587–601. doi: 10.1046/j.1365-2958.2001.02604.x. [DOI] [PubMed] [Google Scholar]
- 30.Gillings MR, Holley MP, Stokes HW, Holmes AJ. Integrons in Xanthomonas: a source of species genome diversity. Proc. Natl Acad. Sci. USA. 2005;102:4419–4424. doi: 10.1073/pnas.0406620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coleman N, Tetu S, Wilson N, Holmes A. An unusual integron in Treponema denticola. Microbiology. 2004;150:3524–3526. doi: 10.1099/mic.0.27569-0. [DOI] [PubMed] [Google Scholar]
- 32.Heidelberg JF, Paulsen IT, Nelson KE, Gaidos EJ, Nelson WC, Read TD, Eisen JA, Seshadri R, Ward N, Methe B, et al. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotech. 2002;20:1118–1123. doi: 10.1038/nbt749. [DOI] [PubMed] [Google Scholar]
- 33.Holmes AJ, Holley MP, Mahon A, Nield B, Gillings M, Stokes HW. Recombination activity of a distinctive integron-gene cassette system associated with Pseudomonas stutzeri populations in soil. J. Bacteriol. 2003;185:918–928. doi: 10.1128/JB.185.3.918-928.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen CY, Wu KM, Chang YC, Chang CH, Tsai HC, Liao TL, Liu YM, Chen HJ, Shen AB, Li JC, et al. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 2003;13:2577–2587. doi: 10.1101/gr.1295503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 36.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li BC, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stokes HW, Holmes AJ, Nield BS, Holley MP, Nevalainen KM, Mabbutt BC, Gillings MR. Gene cassette PCR: sequence-independent recovery of entire genes from environmental DNA. Appl. Environ. Microbiol. 2001;67:5240–5246. doi: 10.1128/AEM.67.11.5240-5246.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nield BS, Holmes AJ, Gillings MR, Recchia GD, Mabbutt BC, Nevalainen KM, Stokes HW. Recovery of new integron classes from environmental DNA. FEMS Microbiol. Lett. 2001;195:59–65. doi: 10.1111/j.1574-6968.2001.tb10498.x. [DOI] [PubMed] [Google Scholar]
- 39.Collis CM, Recchia GD, Kim MJ, Stokes HW, Hall RM. Efficiency of recombination reactions catalyzed by class 1 integron integrase IntI1. J. Bacteriol. 2001;183:2535–2542. doi: 10.1128/JB.183.8.2535-2542.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nemergut DR, Martin AP, Schmidt SK. Integron diversity in heavy-metal-contaminated mine tailings and inferences about integron evolution. Appl. Environ. Microbiol. 2004;70:1160–1168. doi: 10.1128/AEM.70.2.1160-1168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson A, Wu PS, Harrop SJ, Schaeffer PM, Dosztanyi Z, Gillings MR, Holmes AJ, Helena Nevalainen KM, Stokes HW, Otting G, et al. Integron-associated mobile gene cassettes code for folded proteins: the structure of Bal32a, a new member of the adaptable alpha+beta barrel family. J. Mol. Biol. 2005;346:1229–1241. doi: 10.1016/j.jmb.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 42.Nield BS, Willows RD, Torda AE, Gillings MR, Holmes AJ, Nevalainen KM, Stokes HW, Mabbutt BC. New enzymes from environmental cassette arrays: functional attributes of a phosphotransferase and an RNA-methyltransferase. Protein Sci. 2004;13:1651–1659. doi: 10.1110/ps.04638704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogawa A, Takeda T. The gene encoding the heat-stable enterotoxin of Vibrio cholerae is flanked by 123-base pair direct repeats. Microbiol. Immunol. 1993;37:607–616. doi: 10.1111/j.1348-0421.1993.tb01683.x. [DOI] [PubMed] [Google Scholar]
- 44.Barker A, Clark CA, Manning PA. Identification of VCR, a repeated sequence associated with a locus encoding a hemagglutinin in Vibrio cholerae O1. J. Bacteriol. 1994;176:5450–5458. doi: 10.1128/jb.176.17.5450-5458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rowe-Magnus DA, Guerout AM, Mazel D. Bacterial resistance evolution by recruitment of super-integron gene cassettes. Mol. Microbiol. 2002;43:1657–1669. doi: 10.1046/j.1365-2958.2002.02861.x. [DOI] [PubMed] [Google Scholar]
- 46.Melano R, Petroni A, Garutti A, Saka HA, Mange L, Pasteran F, Rapoport M, Rossi A, Galas M. New carbenicillin-hydrolyzing beta-lactamase (CARB-7) from Vibrio cholerae non-O1, non-O139 strains encoded by the VCR region of the V. cholerae genome. Antimicrob. Agents Chemother. 2002;46:2162–2168. doi: 10.1128/AAC.46.7.2162-2168.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petroni A, Melano RG, Saka HA, Garutti A, Mange L, Pasteran F, Rapoport M, Miranda M, Faccone D, Rossi A, et al. CARB-9, a carbenicillinase encoded in the VCR region of Vibrio cholerae non-O1, non-O139 belongs to a family of cassette-encoded beta-lactamases. Antimicrob. Agents Chemother. 2004;48:4042–4046. doi: 10.1128/AAC.48.10.4042-4046.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tennstedt T, Szczepanowski R, Braun S, Pühler A, Schlüter A. Occurrence of integron-associated resistance gene cassettes located on antibiotic resistance plasmids isolated from a wastewater treatment plant. FEMS Microbiol. Ecol. 2003;45:239–252. doi: 10.1016/S0168-6496(03)00164-8. [DOI] [PubMed] [Google Scholar]
- 49.Stokes HW, O'Gorman DB, Recchia GD, Parsekhian M, Hall RM. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 1997;26:731–745. doi: 10.1046/j.1365-2958.1997.6091980.x. [DOI] [PubMed] [Google Scholar]
- 50.Drouin F, Melancon J, Roy PH. The IntI-like tyrosine recombinase of Shewanella oneidensis is active as an integron integrase. J. Bacteriol. 2002;184:1811–1815. doi: 10.1128/JB.184.6.1811-1815.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Biskri L, Bouvier M, Guerout AM, Boisnard S, Mazel D. Comparative study of class 1 integron and Vibrio cholerae superintegron integrase activities. J. Bacteriol. 2005;187:1740–1750. doi: 10.1128/JB.187.5.1740-1750.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Avila P, de la Cruz F. Physical and genetic map of the IncW plasmid R388. Plasmid. 1988;20:155–157. doi: 10.1016/0147-619x(88)90019-4. [DOI] [PubMed] [Google Scholar]
- 53.Hall RM, Brown HJ, Brookes DE, Stokes HW. Integrons found in different locations have identical 5′ ends but variable 3′ ends. J. Bacteriol. 1994;176:6286–6294. doi: 10.1128/jb.176.20.6286-6294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Demarre G, Guerout AM, Matsumoto-Mashimo C, Rowe-Magnus DA, Marliere P, Mazel D. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res. Microbiol. 2005;156:245–255. doi: 10.1016/j.resmic.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 55.Chaveroche MK, Ghigo JM, d'Enfert C. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 2000;28:E97. doi: 10.1093/nar/28.22.e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clarke L, Hitzeman R, Carbon J. Selection of specific clones from colony banks by screening with radioactive antibody. Methods Enzymol. 1979;68:436–442. doi: 10.1016/0076-6879(79)68033-3. [DOI] [PubMed] [Google Scholar]
- 57.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. In: The Proteomics Protocols Handbook, Humana Press. Walker JM, editor. NJ: Humana Press; 2005. pp. 571–607. [Google Scholar]
- 59.Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJA. The PROSITE database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 61.Amann E, Ochs B, Abel KJ. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 62.Huovinen P. Trimethoprim resistance. Antimicrob. Agents Chemother. 1987;31:1451–1456. doi: 10.1128/aac.31.10.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szekeres S, Dauti M, Wilde CDM, Rowe-Magnus DA. Chromosomal toxin-antitoxin loci can diminish large-scale genome reductions in the absence of selection. Mol. Microbiol. 2007;43:1657–1669. doi: 10.1111/j.1365-2958.2007.05613.x. [DOI] [PubMed] [Google Scholar]
- 64.Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2006;2:e135. doi: 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003;301:1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- 66.Engelberg-Kulka H, Glaser G. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 1999;53:43–70. doi: 10.1146/annurev.micro.53.1.43. [DOI] [PubMed] [Google Scholar]
- 67.Bernard P, Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J. Mol. Biol. 1992;226:735–745. doi: 10.1016/0022-2836(92)90629-x. [DOI] [PubMed] [Google Scholar]
- 68.Bernard P, Kezdy KE, Van Melderen L, Steyaert J, Wyns L, Pato ML, Higgins PN, Couturier M. The F plasmid CcdB protein induces efficient ATP-dependent DNA cleavage by gyrase. J. Mol. Biol. 1993;234:534–541. doi: 10.1006/jmbi.1993.1609. [DOI] [PubMed] [Google Scholar]
- 69.Critchlow SE, O'Dea MH, Howells AJ, Couturier M, Gellert M, Maxwell A. The interaction of the F plasmid killer protein, CcdB, with DNA gyrase: induction of DNA cleavage and blocking of transcription. J. Mol. Biol. 1997;273:826–839. doi: 10.1006/jmbi.1997.1357. [DOI] [PubMed] [Google Scholar]
- 70.Bernard P, Gabant P, Bahassi EM, Couturier M. Positive-selection vectors using the F plasmid ccdB killer gene. Gene. 1994;148:71–74. doi: 10.1016/0378-1119(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 71.Le Roux F, Binesse J, Saulnier D, Mazel D. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl. Environ. Microbiol. 2007;73:777–784. doi: 10.1128/AEM.02147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rigden DJ, Jedrzejas MJ, Galperin MY. An extracellular calcium-binding domain in bacteria with a distant relationship to EF-hands. FEMS Microbiol. Lett. 2003;221:103–110. doi: 10.1016/S0378-1097(03)00160-5. [DOI] [PubMed] [Google Scholar]
- 73.Ermolenko DN, Makhatadze GI. Bacterial cold-shock proteins. Cell Mol. Life Sci. 2002;59:1902–1913. doi: 10.1007/PL00012513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feng W, Tejero R, Zimmerman DE, Inouye M, Montelione GT. Solution NMR structure and backbone dynamics of the major cold-shock protein (CspA) from Escherichia coli: evidence for conformational dynamics in the single-stranded RNA-binding site. Biochemistry. 1998;37:10881–10896. doi: 10.1021/bi980269j. [DOI] [PubMed] [Google Scholar]
- 75.Newkirk K, Feng W, Jiang W, Tejero R, Emerson SD, Inouye M, Montelione GT. Solution NMR structure of the major cold shock protein (CspA) from Escherichia coli: identification of a binding epitope for DNA. Proc. Natl Acad. Sci. USA. 1994;91:5114–5118. doi: 10.1073/pnas.91.11.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reid KL, Rodriguez HM, Hillier BJ, Gregoret LM. Stability and folding properties of a model beta-sheet protein, Escherichia coli CspA. Protein Sci. 1998;7:470–479. doi: 10.1002/pro.5560070228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schindelin H, Jiang W, Inouye M, Heinemann U. Crystal structure of CspA, the major cold shock protein of Escherichia coli. Proc. Natl Acad. Sci. USA. 1994;91:5119–5123. doi: 10.1073/pnas.91.11.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mueller U, Perl D, Schmid FX, Heinemann U. Thermal stability and atomic-resolution crystal structure of the Bacillus caldolyticus cold shock protein. J. Mol. Biol. 2000;297:975–988. doi: 10.1006/jmbi.2000.3602. [DOI] [PubMed] [Google Scholar]
- 79.Kremer W, Schuler B, Harrieder S, Geyer M, Gronwald W, Welker C, Jaenicke R, Kalbitzer HR. Solution NMR structure of the cold-shock protein from the hyperthermophilic bacterium Thermotoga maritima. Eur. J. Biochem. 2001;268:2527–2539. doi: 10.1046/j.1432-1327.2001.02127.x. [DOI] [PubMed] [Google Scholar]
- 80.Xia B, Ke H, Inouye M. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol. Microbiol. 2001;40:179–188. doi: 10.1046/j.1365-2958.2001.02372.x. [DOI] [PubMed] [Google Scholar]
- 81.Phadtare S, Inouye M, Severinov K. The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. J. Biol. Chem. 2002;277:7239–7245. doi: 10.1074/jbc.M111496200. [DOI] [PubMed] [Google Scholar]
- 82.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Landick R, Stewart J, Lee DN. Amino acid changes in conserved regions of the beta-subunit of Escherichia coli RNA polymerase alter transcription pausing and termination. Genes Dev. 1990;4:1623–1636. doi: 10.1101/gad.4.9.1623. [DOI] [PubMed] [Google Scholar]
- 84.Bae W, Xia B, Inouye M, Severinov K. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl Acad. Sci. USA. 2000;97:7784–7789. doi: 10.1073/pnas.97.14.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boucher Y, Nesbo CL, Joss MJ, Robinson A, Mabbutt BC, Gillings MR, Doolittle WF, Stokes HW. Recovery and evolutionary analysis of complete integron gene cassette arrays from Vibrio. BMC Evol. Biol. 2006;6:3. doi: 10.1186/1471-2148-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Whitman CP. The 4-oxalocrotonate tautomerase family of enzymes: how nature makes new enzymes using a beta-alpha-beta structural motif. Arch. Biochem. Biophys. 2002;402:1–13. doi: 10.1016/S0003-9861(02)00052-8. [DOI] [PubMed] [Google Scholar]
- 87.Ferguson GP, Totemeyer S, MacLean MJ, Booth IR. Methylglyoxal production in bacteria: suicide or survival? Arch. Microbiol. 1998;170:209–218. doi: 10.1007/s002030050635. [DOI] [PubMed] [Google Scholar]
- 88.Romine MF, Carlson TS, Norbeck AD, McCue LA, Lipton MS. Identification of mobile elements and pseudogenes in the Shewanella oneidensis MR-1 genome. Appl. Environ. Microbiol. 2008;74:3257–3265. doi: 10.1128/AEM.02720-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.da Silva AC, Ferro JA, Reinach FC, Farah CS, Furlan LR, Quaggio RB, Monteiro-Vitorello CB, Van Sluys MA, Almeida NF, Alves LM, et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature. 2002;417:459–463. doi: 10.1038/417459a. [DOI] [PubMed] [Google Scholar]
- 90.Stokes HW, Holmes AJ, Nield BS, Holley MP, Nevalainen KM, Mabbutt BC, Gillings MR. Gene cassette PCR: sequence-independent recovery of entire genes from environmental DNA. Appl. Environ. Microbiol. 2001;67:5240–5246. doi: 10.1128/AEM.67.11.5240-5246.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Collis CM, Grammaticopoulos G, Briton J, Stokes HW, Hall RM. Site-specific insertion of gene cassettes into integrons. Mol. Microbiol. 1993;9:41–52. doi: 10.1111/j.1365-2958.1993.tb01667.x. [DOI] [PubMed] [Google Scholar]
- 92.Collis CM, Hall RM. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J. Bacteriol. 1992;174:1574–1585. doi: 10.1128/jb.174.5.1574-1585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hardeman F, Sjoling S. Metagenomic approach for the isolation of a novel low-temperature-active lipase from uncultured bacteria of marine sediment. FEMS Microbiol. Ecol. 2007;59:524–534. doi: 10.1111/j.1574-6941.2006.00206.x. [DOI] [PubMed] [Google Scholar]
- 94.Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA, MacNeil IA, Minor C, et al. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 2000;66:2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hanau-Berçot B, Podglajen I, Casin I, Collatz E. An intrinsic control element for translational initiation in class 1 integrons. Mol. Microbiol. 2002;44:119–130. doi: 10.1046/j.1365-2958.2002.02843.x. [DOI] [PubMed] [Google Scholar]
- 96.Jacquier H, Zaoui C, Sanson-le Pors MJ, Mazel D, Berçot B. Translation regulation of integrons gene cassette expression by the attC sites. Mol. Microbiol. 1995;72:1475–1486. doi: 10.1111/j.1365-2958.2009.06736.x. [DOI] [PubMed] [Google Scholar]
- 97.Schmeisser C, Stockigt C, Raasch C, Wingender J, Timmis KN, Wenderoth DF, Flemming HC, Liesegang H, Schmitz RA, Jaeger KE, et al. Metagenome survey of biofilms in drinking-water networks. Appl. Environ. Microbiol. 2003;69:7298–7309. doi: 10.1128/AEM.69.12.7298-7309.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Charlebois RL, Doolittle WF. Computing prokaryotic gene ubiquity: rescuing the core from extinction. Genome Res. 2004;14:2469–2477. doi: 10.1101/gr.3024704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fitz-Gibbon ST, House CH. Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res. 1999;27:4218–4222. doi: 10.1093/nar/27.21.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Koenig JE, Boucher Y, Charlebois RL, Nesbo C, Zhaxybayeva O, Bapteste E, Spencer M, Joss MJ, Stokes HW, Doolittle WF. Integron-associated gene cassettes in Halifax Harbour: assessment of a mobile gene pool in marine sediments. Environ. Microbiol. 2008;10:1024–1038. doi: 10.1111/j.1462-2920.2007.01524.x. [DOI] [PubMed] [Google Scholar]
- 101.Michael CA. Mobile gene cassettes: a fundamental resource for bacterial evolution. Am. Nat. 2004;164:1–12. doi: 10.1086/421733. [DOI] [PubMed] [Google Scholar]
- 102.Rowe-Magnus DA, Zouine M, Mazel D. In: The Biology of Vibrios. Thompson FL, Austin B, Swings J, editors. Washington: American Society fro Microbiology; 2006. [Google Scholar]
- 103.Robinson A, Guilfoyle AP, Sureshan V, Howell M, Harrop SJ, Boucher Y, Stokes HW, Curmi PM, Mabbutt BC. Structural genomics of the bacterial mobile metagenome: an overview. Methods Mol. Biol. 2008;426:589–595. doi: 10.1007/978-1-60327-058-8_39. [DOI] [PubMed] [Google Scholar]
- 104.Robinson A, Wu PS, Harrop SJ, Schaeffer PM, Dosztanyi Z, Gillings MR, Holmes AJ, Nevalainen KM, Stokes HW, Otting G, et al. Integron-associated mobile gene cassettes code for folded proteins: the structure of Bal32a, a new member of the adaptable alpha+beta barrel family. J. Mol. Biol. 2005;346:1229–1241. doi: 10.1016/j.jmb.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 105.Barker A, Manning PA. VlpA of Vibrio cholerae O1: the first bacterial member of the alpha 2-microglobulin lipocalin superfamily. Microbiology. 1997;143:1805–1813. doi: 10.1099/00221287-143-6-1805. [DOI] [PubMed] [Google Scholar]
- 106.Koenig JE, Sharp C, Dlutek M, Curtis B, Joss M, Boucher Y, Doolittle WF. Integron gene cassettes and degradation of compounds associated with industrial waste: the case of the Sydney tar ponds. PLoS ONE. 2009;4:e5276. doi: 10.1371/journal.pone.0005276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sobecky PA, Coombs JM. Horizontal gene transfer in metal and radionuclide contaminated soils. Methods Mol. Biol. 2009;532:455–472. doi: 10.1007/978-1-60327-853-9_26. [DOI] [PubMed] [Google Scholar]
- 108.Fonseca EL, Dos Santos Freitas F, Vieira VV, Vicente AC. New qnr gene cassettes associated with superintegron repeats in Vibrio cholerae O1. Emerging Infect. Dis. 2008;14:1129–1131. doi: 10.3201/eid1407.080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Franzon VL, Barker A, Manning PA. Nucleotide sequence encoding the mannose-fucose-resistant hemagglutinin of Vibrio cholerae O1 and construction of a mutant. Infect. Immun. 1993;61:3032–3037. doi: 10.1128/iai.61.7.3032-3037.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]