Abstract
Acyl-CoA dehydrogenases (ACADs), which are key enzymes in fatty acid and amino acid catabolism, form a large, pan-taxonomic protein family with at least 13 distinct subfamilies. Yet most reported ACAD members have no subfamily assigned, and little is known about the taxonomic distribution and evolution of the subfamilies. In completely sequenced genomes from approximately 210 species (eukaryotes, bacteria and archaea), we detect ACAD subfamilies by rigorous ortholog identification combining sequence similarity search with phylogeny. We then construct taxonomic subfamily-distribution profiles and build phylogenetic trees with orthologous proteins. Subfamily profiles provide unparalleled insight into the organisms’ energy sources based on genome sequence alone and further predict enzyme substrate specificity, thus generating explicit working hypotheses for targeted biochemical experimentation. Eukaryotic ACAD subfamilies are traditionally considered as mitochondrial proteins, but we found evidence that in fungi one subfamily is located in peroxisomes and participates in a distinct β-oxidation pathway. Finally, we discern horizontal transfer, duplication, loss and secondary acquisition of ACAD genes during evolution of this family. Through these unorthodox expansion strategies, the ACAD family is proficient in utilizing a large range of fatty acids and amino acids—strategies that could have shaped the evolutionary history of many other ancient protein families.
INTRODUCTION
From the last two decades of intensive research especially in mammals, acyl-CoA dehydrogenases (ACADs) are now known as a large and biologically important enzyme family. Genetic defects of the corresponding genes cause severe health problems in human, including hypoglycemia, neuromuscular pathology and even death (1). While ACAD proteins occur in all three domains of life, animals possess the largest number of distinct subfamilies. In human, for example, 11 different ACAD enzymes have been recognized (2–12). These proteins, which in eukaryotes are localized in mitochondria, share up to ∼50% amino acid identity among each other (Table 1) and catalyze similar biochemical reactions: the oxidation of diverse acyl-CoA compounds, produced during the degradation of fat and protein, to enoyl-CoA (Figure 1).
Table 1.
Pairwise sequence similarities between human ACAD subfamily membersa
| ACD11 | ACADS | ACADM | ACADL | ACADV | ACADV2 | ACDSB | GCDH | IVD | IBD | fadE | fadE12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACD10 | 46 | 30 | 28 | 25 | 26 | 26 | 27 | 25 | 24 | 24 | 29 | 25 |
| ACD11 | 29 | 26 | 26 | 23 | 26 | 26 | 23 | 22 | 27 | 31 | 25 | |
| ACADS | 36 | 31 | 35 | 36 | 38 | 30 | 36 | 35 | 26 | 27 | ||
| ACADM | 32 | 34 | 34 | 37 | 27 | 34 | 33 | 24 | 25 | |||
| ACADL | 28 | 30 | 33 | 27 | 34 | 32 | 20 | 27 | ||||
| ACADV | 45 | 34 | 30 | 32 | 30 | 24 | 24 | |||||
| ACADV2 | 38 | 29 | 34 | 33 | 25 | 22 | ||||||
| ACDSB | 28 | 33 | 35 | 25 | 24 | |||||||
| GCDH | 28 | 27 | 34 | 24 | ||||||||
| IVD | 32 | 24 | 23 | |||||||||
| IBD | 21 | 22 | ||||||||||
| fadE | 24 |
aAll subfamily members are from human, except for fadE and fadE12, which are prokaryotic subfamilies. Percentage of identical residues in aligned region by BLAST. Sequences are obtained from SwissProt. Sequence IDs are listed in Table 2.
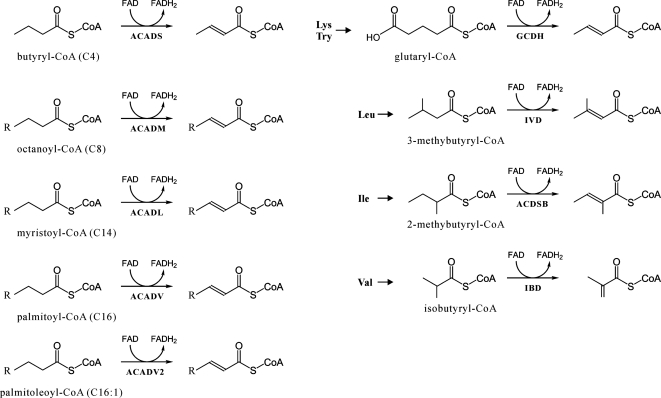
Figure 1.
Optimal substrates of ACAD subfamilies. C4, etc., length of the acyl-CoA chain. C16:1, unsaturated fatty acid with one double bond. Subfamilies in the left part of the figure are involved in fatty acid degradation. Those in the right part are involved in amino acid degradation. ‘R’ represents straight alkyl chain.
ACAD subfamilies are distinguished by the metabolic pathways in which they participate, and by their substrate specificity (Figure 1, Table 2). Five subfamilies participate in β-oxidation of fatty acids, with optimal activity for acyl-CoA substrates of particular chain length, short (ACADS), medium (ACADM), long (ACADL), or very long (ACADV and ACADV2) (11–15). Four other subfamilies are implicated in amino acid degradation. After removal of the amino groups from isoleucine, leucine, lysine/trytophan and valine, the remaining branched acyl-CoA is dehydrogenated by short/branched chain acyl-CoA dehydrogenase (ACDSB), isovaleryl-CoA dehydrogenase (IVD), glutaryl-CoA dehydrogenase (GCDH) and isobutyryl-CoA dehydrogenase (IBD), respectively (3,5–7). The most recently identified subfamilies, ACD10 and ACD11, are of yet unknown function (8,9). Two additional subfamilies have been reported in bacteria: fadE degrades a broad range of substrates from short to long chain acyl-CoAs (16,17), while fadE12 prefers medium-chain length molecules (18). The reaction mechanism and 3D structure of ACAD enzymes have been reviewed by others (19,20).
Table 2.
Seed sequences used for BLAST searches
| Protein name | Molecular function | Seed from | Sequence IDa | Evidence |
|---|---|---|---|---|
| ACADV | Oxidation of very long chain fatty acid | Homo sapiens | P49748 | Experiment |
| ACADV2 | Homo sapiens | Q9H845 | Experiment | |
| ACADL | Oxidation of long chain fatty acid | Homo sapiens | P28330 | |
| Monosiga brevicollisa | gi|167537125 | BLAST and phylogeny | ||
| ACADM | Oxidation of medium chain fatty acid | Homo sapiens | P11310 | |
| Batrachochytrium dendrobatidisa | BDEG_05327 | BLAST and phylogeny | ||
| Monosiga brevicollisa | gi|167534479 | |||
| ACADS | Oxidation of short chain fatty acid | Homo sapiens | P16219 | |
| Monosiga brevicollisa | gi|167515960 | BLAST and phylogeny | ||
| ACDSB | Oxidation of isoleucine | Homo sapiens | P45954 | Experiment |
| Batrachochytrium dendrobatidisa | BDEG_04739 | BLAST and phylogeny | ||
| IVD | Oxidation of leucine | Homo sapiens | P26440 | Experiment |
| Batrachochytrium dendrobatidisa | BDEG_02262 | BLAST and phylogeny | ||
| Monosiga brevicollisa | gi|167524148 | |||
| GCDH | Oxidation of lysine and tryptophan | Homo sapiens | Q92947 | Experiment |
| Batrachochytrium dendrobatidisa | BDEG_05264 | BLAST and phylogeny | ||
| Monosiga brevicollisa | gi|167524186 | |||
| IBD | Oxidation of valine | Homo sapiens | Q9UKU7 | Experiment |
| Batrachochytrium dendrobatidisa | BDEG_03936 | BLAST and phylogeny | ||
| Monosiga brevicollisa | gi|167524677 | |||
| ACD10 | Function unknown | Homo sapiens | Q6JQN1 | cDNA |
| ACD11 | Function unknown | Homo sapiens | Q709F0 | cDNA |
| fadE | Oxidation of fatty acids of different chain length | Escherichia coli | Q47146 | Experiment |
| fadE12 | Oxidation of medium chain fatty acid | Mycobacterium tuberculosis | P71539 | Experiment |
aSeeds added in the second round of BLAST search.
In eukaryotes, β-oxidation involving ACAD enzymes takes place in mitochondria. Eukaryotes also possess peroxisomal β-oxidation catalyzed by acyl-CoA oxidase (ACOX) instead of ACAD proteins. The two families resemble each other in several aspects. ACOX proteins share remote yet significant sequence similarity with ACAD proteins, and also catalyze the conversion of acyl-CoA to enoyl-CoA. But unlike the ACAD family, ACOX proteins occur predominantly in eukaryotes, are located exclusively in peroxisomes and function by a distinct enzymatic mechanism: ACOX proteins are re-oxidized by molecular oxygen, generating H2O2 (20); ACAD enzymes, in contrast, having only low reactivity with molecular oxygen, are re-oxidized by electron-transferring flavoproteins, which in turn pass the electrons to the respiratory chain, generating H2O. Insight into the origin of the ACOX family will critically depend on a better understanding of the ACAD family, which is the focus of the study reported here.
Our current knowledge about ACAD proteins is limited to a few model organisms. There has been no comprehensive survey of ACAD enzymes, except for genome-wide in silico screens in fungi without subfamily identification (21,22). Further, it is unclear whether the 11 subfamilies recognized in human are conserved throughout animals or even beyond. One reason for these shortcomings is that in public data repositories, sequences are generally annotated indistinctively as ‘acyl-CoA dehydrogenase’. This is because in BLAST searches, remote ACAD homologs often match members from different subfamilies with similar scores. For example, a protein from Janthinobacterium (gi|152980951) shares identities of 28% with ACADS from Mycobacterium, 27% with ACDSB from rat and 27% with ACADV2 from human. Evidently, such a lack of distinction by similarity scores has hampered research on subfamily distribution, diversity and evolution.
As a large number of complete genome sequences from prokaryotes and eukaryotes have become available, large-scale subfamily classification and phylogenetic analysis of the ACAD family are now tractable. Our first step in this investigation was assignment of ACAD proteins to defined subfamilies. The most direct way to do so is via sequence similarity search as employed in previous protein family studies (23,24). But as illustrated above, it is difficult to distinguish members of different ACAD subfamilies by sequence similarity alone. Another widely used approach employs sequence profiles, e.g. PFAM domains (25,26) or hidden Markov models (HMMs) generated from subfamilies (27,28). But for ACAD enzymes, the number of confirmed sequences in each subfamily is not large enough to make reliable profiles. Here we identify ACAD subfamily members by rigorous ortholog detection via phylogenetic analysis, an approach successfully employed in certain genome annotation and comparison studies (29,30). Our procedure involves reiterative phylogenetic tree construction combined with a two-round BLAST search. Then, based on comprehensive subfamily assignment, we ascertain the taxonomic distribution of ACAD proteins and make inferences of their molecular function and, more generally, the energy sources of a given organism. We also attempt the inference of a global ACAD family tree, which, however, proves by far more difficult than anticipated. Still, for eukaryotic ACAD genes, we have been able to discern several recurring evolutionary patterns that we present in the last section of this article.
MATERIALS AND METHODS
Data collection
We collected the genome-deduced protein sequences of completed or coding regions-completed genome projects from 212 species, mostly taken from NCBI (http://www.ncbi.nlm.nih.gov/Genomes/), Broad Institute of MIT and Harvard (http://www.broad.mit.edu) and DOE Joint Genome Institute (http://www.jgi.doe.gov/). This dataset is composed of 91 bacteria, 29 archaea and 92 eukaryotes. To increase taxonomic coverage in phylogenetic analyses, we included proteins of 32 eukaryotes whose genome sequence is only partially completed, as well as Expressed Sequence Tag (EST) clusters from six jakobids that were generated by us in the context of the Canadian collaborative Protist EST project and retrieved from the Taxonomically Broad eukaryote EST DataBase (TBestDB) (31). Jakobids are a group of heterotrophic flagellates that are believed to diverge close to the eukaryotic origin (32–35). Since no genome sequences are available from jakobids, we included EST data of six jakobid species to obtain a more comprehensive view of the evolution of ACAD enzymes. A detailed list of species names and data sources is compiled in Table S1. Sequences of enzymatically characterized ACAD proteins were retrieved from SwissProt (Table 2) and used as seeds to search for ACAD subfamilies in collected genomes. In addition, we included sequences of ACOX in the BLAST seed (listed in Table S2) to exclude potential mix-up of ACAD and ACOX homologs.
Subfamily assignment
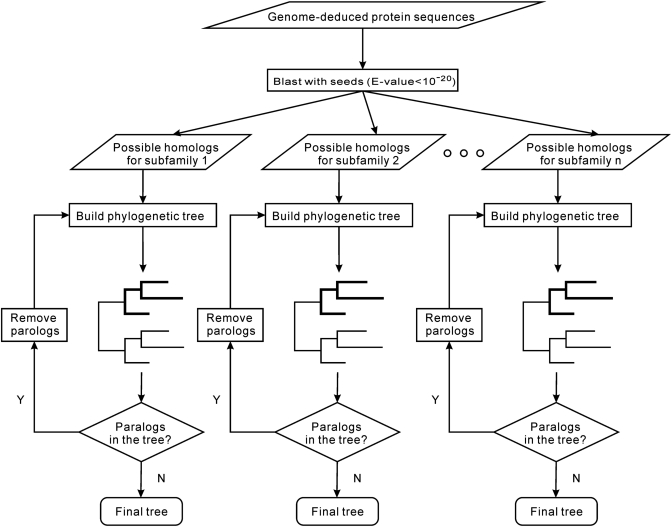
Orthologs of subfamilies were identified by a two-round procedure combining BLAST search with phylogenetic inference. Each round included BLAST searches and data selection followed by phylogenetic analysis. The difference between round one and two was the set of seed sequences used for BLAST searches. In round one, the genome-deduced protein sequences from each species were compared by BLAST with known ACAD proteins (seeds listed in Table 2), at a threshold of e = 1 × 10−20. In total, 2258 sequences matched at least one seed under this condition. From each species, we selected up to three top matches for each ACAD subfamily and preliminarily annotated the corresponding proteins as potential homologs of the corresponding subfamily. As certain query sequences matched multiple different subfamilies, we analyzed these a second time by applying the following rule: if a given sequence matched multiple subfamilies and the e-values of the matches differed by more than 10-fold, then the sequence was assigned to the subfamily with the lowest e-value. This case applied to 1572 sequences. Otherwise, if e-values of the multiple matches differed less than 10-fold, all preliminary subfamily assignments were retained and the final annotation was based on the subsequent phylogenetic analysis. This category included 341 sequences. We built a maximum likelihood phylogenetic tree for each protein subfamily (see procedure below) using all sequences assigned to this subfamily. From these trees, we selected slowly evolving and unambiguous orthologs of the initial mammalian seed sequences, i.e. proteins from Monosiga, the closest unicellular relative of animals (36), and Batrachochytrium, a member of the earliest divergence in fungi (36). These, combined with the first set of seeds, formed the second set of seeds used for round two of BLAST searches (Table 2). The same screening procedure was applied as in round one, followed by construction of a phylogenetic tree for each subfamily. Inspection of the trees showed that certain species possessed multiple members of the same subfamily. These extra copies were removed from the dataset to save computational cost during subsequent analyses (especially bootstrap, see below), yielding a non-redundant data set of 861 sequences. In order to detect paralogs, phylogenetic trees were built again for each subfamily, this time with the non-redundant data set. Paralogs were removed from the subfamily until the gene trees were reconciled with the species tree (Figure 2). The sequences removed in this step (32 in total) are considered as ACAD proteins of unknown subfamily (Table S3). A special procedure was applied to ACD10 and ACD11. Numerous potential homologs of ACD10 displayed similar BLAST e-values to ACD11 and vice versa, and subfamily assignment as described above was possible only for a few members. The remaining proteins were grouped into a provisional subfamily termed as ACD10/11 and further analyzed by a special procedure as described in the ‘Results and Discussion’ section.
Figure 2.
Flow chart of the procedure for assigning ACAD subfamilies. Blast searches combined with reconciliation of gene and species trees were used to identify orthologs (see ‘Materials and Methods’ section). Y, yes; N, no.
Phylogenetic inference
Multiple protein sequence alignments were constructed with MUSCLE (37), and alignment logos were created by WebLogo (38). For phylogenetic analysis, ambiguously aligned and highly divergent regions of the alignment were eliminated using Gblocks (39). Maximum likelihood trees were constructed using RAxML (40) with the WAG+Γ model and four discrete γ-rate categories. The statistical support of branches was evaluated by 100 bootstrap replicates.
Protein domain search
To locate functional domains in ACD10 and ACD11 homologs, we used InterProtScan (41). Protein sequences were searched against PROSITE patterns, PROSITE profiles, PRINTS, PFAM, PRODOM, SMART, TIGRFAMs, PIR SuperFamily and SUPERFAMILY.
Taxonomic distribution profiling
After the subfamily assignment of ACAD proteins, we compiled the presence/absence of subfamilies in the genome-derived proteomes of the species included in this study. This information was mapped on NCBI's taxon tree (http://www.ncbi.nlm.nih.gov/sites/entrez?db=taxonomy).
Targeting peptide prediction
We predicted subcellular location of subfamilies based on recognition of targeting peptide from four predictors: TargetP (42), Predotar (43), Protein Prowler (44) and MitoProt (45). Annotation-based predictors, such as PA-SUB (46), were excluded from the analysis to preclude ‘prejudicial’ association, because ACAD enzymes are traditionally annotated as mitochondrial proteins in public databases. All results were obtained from online servers, except for MitoProt, which was installed and run locally. For most proteins, the predictors gave contradictory results. Therefore, we integrated these predictions via YimLOC, a tool employing machine learning (MTP-DT predictor) (47) that is significantly more accurate than any of the individual predictors. To detect the peroxisomal targeting signal, we used the web service PTS1 predictor (http://mendel.imp.ac.at/mendeljsp/sat/pts1/PTS1predictor.jsp) (48).
RESULTS AND DISCUSSION
Subfamily assignment of previously unclassified ACAD proteins
To identify ACAD subfamilies in the genome-derived proteomes of 250 species (species names are listed in Table S1 and S3), we initially searched for homologs of well-characterized subfamily members by BLAST. This approach failed to distinguish subfamilies in many instances, especially when query and target sequences were from taxonomically distant species. As illustrated by the example of Janthinobacterium (see ‘Introduction’ section), remote homologs often match several subfamilies with similar scores, and the top hit may not correspond to the protein's true affiliation. Therefore, we developed a procedure that combines BLAST searches with phylogenetic analysis, as described in the ‘Materials and Methods’ section. By this procedure, a total of 702 sequences from 177 species were unambiguously assigned to one of the ACAD subfamilies (Table S1), with the exception of ACD10 and ACD11. As all non-animal proteins that match ACD10 also match ACD11 with similar score and vice versa, these proteins were classified provisionally as ACD10/11 and further analyzed with a distinct approach (see subsequently). Only 32 (4%) proteins, mostly from prokaryotes and protists, remained unassigned (Table S3). These proteins do not form a new subfamily, because sequence similarity was observed only between proteins from the same genus and not across larger phylogenetic distances. Notably, 60 out of the 250 investigated species, predominantly bacteria, appear to completely lack ACAD genes (Table S3). The subfamily distribution across taxa is analyzed in more detail further below.
Distinction of ACD10 and ACD11
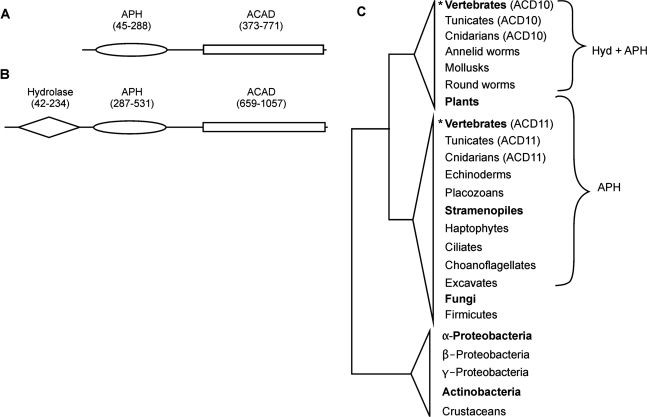
The two ACAD subfamilies of unknown function, ACD10 and ACD11, have only recently been discovered in human and a few other mammals (8,9). Our subfamily assignment procedure clearly distinguishes ACD10 and ACD11 in animals, but fails to do so for other taxa. In the tree built with all identified ACD10 and ACD11 sequences and the provisional class ACD10/11 (127 proteins from 110 taxa in total; Figure 3C and Figure S1A), only vertebrate ACD10 and ACD11 form well supported, distinct and coherent clades. Sequences from other taxa cannot be placed with confidence.
Figure 3.
Distinction of ACD10 and ACD11. Protein domains of human ACD11 and ACD10 (Q709F0, A) and ACD10 (Q6JQN1, B). InterProt domain IDs are as follows: hydrolase domain, IPR005834; APH domain, IPR002575. The ACAD domain is composed of three parts: ACAD N-terminal domain, IPR013786; ACAD central domain, IPR006091; ACAD C-terminal domain, IPR013764. (C) Domain content of ACD10, ACD11 and provisional ACD10/11 homologs mapped onto the phylogenetic tree. Taxa representing more than three species are shown in bold. Clades with bootstrap support value >90 are labeled with asterisk. Taxa that appear twice in the tree are distinguished by the labels ‘ACD10’ and ‘ACD11’. In animals, ACD11 includes (in addition to the common ACAD domains) an APH domain, and ACD10 possesses an APH plus a hydrolase (Hyd) domain. Exceptions are gi|115941654 of the echinoderm Strongylocentrotus purpuratus, which is more similar to ACD10 but lack the hydrolase domain, and jgi|Dappu1|346313 of the crustacean Daphnia pulex, which shares equal sequence similarity with ACD10 and ACD11 and lacks both extra domains. Homologs of other eukaryotes, which have an APH domain, but no hydrolase domain, are classified as ACD11. Sequences lacking both domains are all homologs of fungi, the green algae Volvox carteri and Ostreococcus lucimarinus and the stramenopiles Aureococcus anophagefferens and Phytophthora ramorum. Bacterial homologs also lack both domains. Those lacking both domains are classified as ACD11n, see text.
Further distinction of ACD10 and ACD11 comes from protein domain analysis. Mammalian ACD10 and ACD11 proteins are conspicuously longer than those from other ACAD subfamilies. Domain search with InterProScan shows that the human ACD11 proteins carry in their N-terminal region an aminoglycoside phosphotransferase (APH) domain. In bacteria, this domain is involved in antibiotic resistance (49) (Figure 3A), but its role in eukaryotes is unknown. ACD10 has in addition to APH an N-terminal hydrolase domain (Figure 3B). Both domains are absent from other ACAD families. While screening non-metazoan ACD10/11 for these domains, we did not detect a single protein including the hydrolase domain; the majority of these sequences carry APH and some lack both domains (Figure 3C). Phylogenetic trees of fungal and animal homologs place animal ACD10 and ACD11 into two monophyletic clades to the exclusion of fungal proteins, suggesting that ACD11 is an ancestral eukaryotic gene from which ACD10 has arisen in the animal lineage by gene duplication and subsequent addition of the hydrolase domain (Figure S1B). Therefore, we classify non-metazoan homologs carrying APH as ACD11 and those without either domain as ACD11n.
Are eukaryotic ACD10, ACD11 and ACD11n indeed mitochondrial proteins as traditionally assumed for the entire ACAD family? A proteomics study of rat peroxisomal proteins (50) reports the peptides whose sequence match the mouse homolog of ACD11 according to our subfamily classification (gi|28280023). In addition, an enzymatic study of peroxisomal β-oxidation in Magnaporthe grisea speculates that some of the fungus’ ACAD proteins are imported into peroxisomes to substitute for the ACOX enzyme, whose gene is missing from the genome (51). Here we predict by in silico methods the subcellular localization of all ACAD subfamilies present in Magnaporthe. Indeed, ACD11n is the only ACAD member that has the propensity to enter peroxisomes, pinpointing ACD11n as the hypothetical protein participating in peroxisomal β-oxidation. The same situation most likely applies to other fungi that lack ACOX in their genome, specifically Nectria haematococca, Hypocrea jecorina and Hypocrea virens (22). This hypothesis can be readily tested experimentally, e.g. by co-localization of tagged ACAD protein with subcellular structures. If true, the reaction mechanism of ACD11n must have undergone a fundamental adaptation to the peroxisomal environment.
Notably, peroxisomal localization is predicted for most eukaryotic members of ACD11n and the entire ACD11 subfamily (exceptions are listed in Table S4), while ACD10 displays features typical of mitochondrial proteins. The predicted subcellular localization should guide experimental approaches to elucidate these proteins’ molecular function and the specific roles of the APH and hydrolase domains.
Taxonomic distribution of ACAD subfamilies
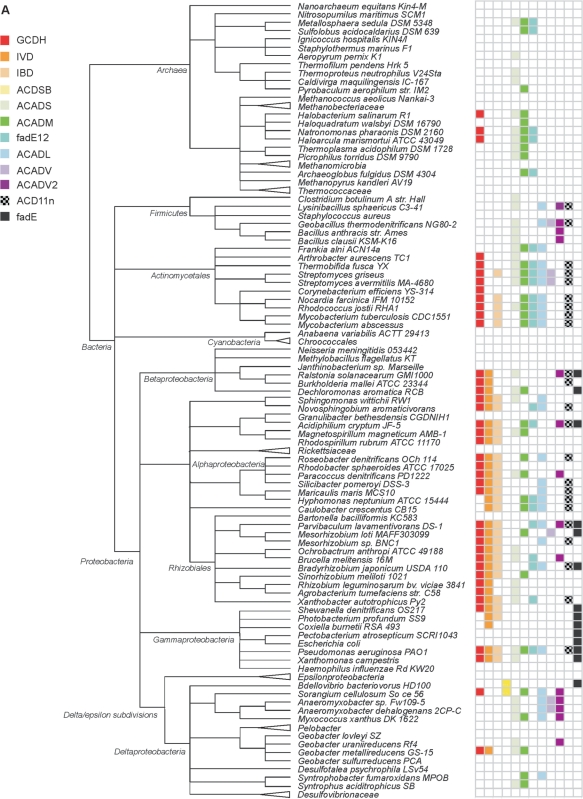
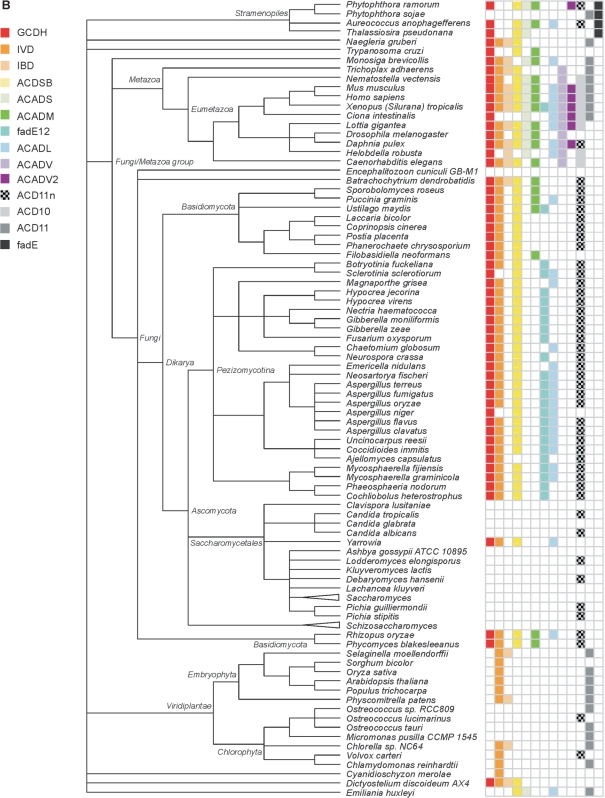
Based on the comprehensive annotation, we examined the presence/absence of ACAD subfamilies across the 250 species investigated here. The distribution profiles of subfamilies differ markedly (Figure 4). Large sets of ACAD subfamilies are typical for animals with 11 in vertebrates, as many as initially identified in mammals. In contrast, fungi possess on average only five subfamilies and these are involved in both mitochondrial β-oxidation and amino acid catabolism. A total lack of ACAD genes is observed in a few fungal lineages (Saccharomyces, Encephalitozoon and Schizosaccharomyces), all characterized by highly derived and reduced genomes. In Plantae, only two ACAD subfamilies, IVD and ACD11, are widely present. From the other eukaryotic lineages, there are not enough genome sequences available to infer specific profile features. Finally, Archaea have conspicuously small sets of ACAD subfamilies, and there is much variation among Bacteria.
Figure 4.
ACAD subfamily distribution mapped on the taxonomy hierarchy from NCBI. Only species whose genome has been completely sequenced are included in the figure. The sequence IDs are listed in ←Table S1. A triangle in front of a taxon name indicates that no ACAD subfamily was detected in the members of this taxon. (A) Subfamily distribution in prokaryotes; (B) subfamily distribution in eukaryotes.
Four subfamilies occur in all domains of life: ACADS (degrading short fatty acids), ACADM, fadE12 (both preferring medium-length fatty acids) and GCDH (involved in lysine and tryptophan catabolism). Subfamilies virtually restricted to a single domain are ACDSB, ACADV and ACADV2 in eukaryotes, and fadE in bacteria. Overall, ACAD subfamilies specialized in short-chain (straight and branched) acyl-CoAs are more broadly distributed than those preferring long-chain substrates.
We confronted the inferred ACAD subfamily profiles with experimental evidence. As mentioned earlier, animals possess 11 out of 13 ACAD subfamilies (Figure 4). Indeed, fatty acids and amino acids make up an important part of metazoan nutrition, requiring a host of specialized enzymes for degrading acyl-substrates of various length and steric structure. A comparably large repertoire of ACAD enzymes (the largest in bacteria) is present in the opportunistic human pathogen, Pseudomonas aeruginosa. This organism is notorious for its extraordinary metabolic versatility, capable of utilizing a wide range of organic compounds including fatty acids and amino acids as an energy source. The ACAD families in P. aeruginosa identified here explain the observed efficient use of fatty acids via β-oxidation (52,53). Only a single ACAD subfamily—ACADS—is found in Clostridium botulinum, a food-borne pathogen. Experimental studies confirm that C. botulinum cannot catabolize long-chain fatty acids. This explains the documented poor growth of this bacterium on ripened cheese (54). Finally, many intracellular parasites such as Rickettsiaceae lack all ACAD genes (Figure 4), reflecting their reliance on their host for nutrients. In sum, the above examples illustrate that the in silico-generated ACAD profiles are in strong agreement with, and explain well, the biochemical data. Therefore, the presence of ACAD subfamilies provides a window on an organism's biology based on genome sequence alone, when information on nutritional requirements and enzymatics are not available.
Multiple single-purpose enzymes versus single multi-purpose enzymes
Recent biochemical studies on Aspergillus nidulans revealed an unexpected substrate range of ACDSB, which in this organism catalyzes dehydrogenation of not only isobutyryl-CoA (derivative of isoleucine), but also 2-methyl-butyryl-CoA (derivative of valine) and short-chain acyl-CoA (55). In human, the latter two compounds are degraded by IBD and ACADS (Figure 1), two enzymes that are missing intriguingly in Aspergillus species and other fungi (Figure 4B).
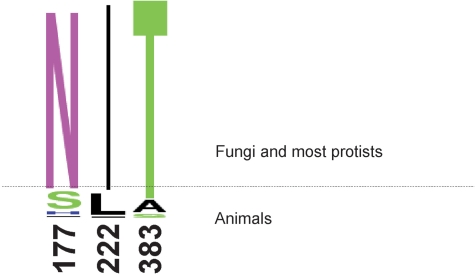
Insight into the molecular basis of such a broad substrate range comes from directed mutagenesis experiments of the human ACDSB protein (5). This study shows that the substitutions Ser177Asn, Leu222Ile and Ala383Thr lead to significantly higher turnover rates of hexanoyl-CoA and isobutyryl-CoA (the substrates optimally degraded by ACADS and IBD, respectively). This pinpoints Ser177, Leu222 and Ala383 as substrate specificity determinants of human ACDSB (in the following, the ‘specificity’ residues are indicated as NIT and SLA).
To find out the substrate range of ACDSB from other organisms where experimental data are lacking, we constructed a multiple sequence alignment and also superimposed the 3D structure of ACDSB, ACADS and IBD proteins (Figure S2). These alignments show that homologs from all primates and a few other mammals are of the human type (ACDSB-SLA), whereas all fungal enzymes are of the Aspergillus type (ACDSB-NIT, with a single minor exception, Figure 5). From this finding, together with the subfamily distribution pattern, we predict that all fungal ACDSB unite three functions in one enzyme, i.e. the functions of human ACDSB, ACADS and IBD. (For a hypothesis on the evolution of these three subfamilies, see Figure S3.)
Figure 5.
Alignment of residues that affect substrate specificity of human ACDSB. The multiple protein alignment was generated using eukaryotic ACDSB homologs. The numbers refer to the residue position of the mature human ACDSB (i.e. not including the mitochondrial targeting peptide). A minor deviation from the NIT motif is found in Puccinia graminis, which has a ‘NIS’.
The above functional generalization has previously remained undetected by sequence similarity and phylogeny-based function prediction. But as exemplified here, the prediction of an enzyme's substrate range can be improved by integrating subfamily distribution profiles and function/structure data from ‘model’ enzymes. Such advanced function prediction provides valuable working hypotheses that can be tested by targeted biochemical experimentation.
Evolution of the ACAD family
In an attempt to unravel the origin and evolution of the ACAD protein family, we built maximum likelihood phylogenetic trees using proteins drawn from complete genome sequences and assigned to subfamilies as described above (for the proteins used, see Table S1). The global trees including all ACAD subfamilies have largely unsupported topologies likely due to three factors: the extensive sequence divergence, the relatively short length of ACAD proteins (100 or less residues after Gblocks processing, see the ‘Materials and Methods’ section) and the immense evolutionary time spans in question. Trees of individual ACAD subfamilies suffer to a lesser degree from this problem, but only a few trees (ACDSB and ACADV2) display supported species-tree topology (Figure S1C–M). Close inspection reveals events of horizontal gene transfers, not only within bacteria but also within eukaryotes and across domains, in addition to gene losses and multiple independent gene duplications. After carefully studying each subfamily tree, we are able to discern several intriguing evolutionary patterns in eukaryotes.
Early acquisition of ACAD enzymes in eukaryotes
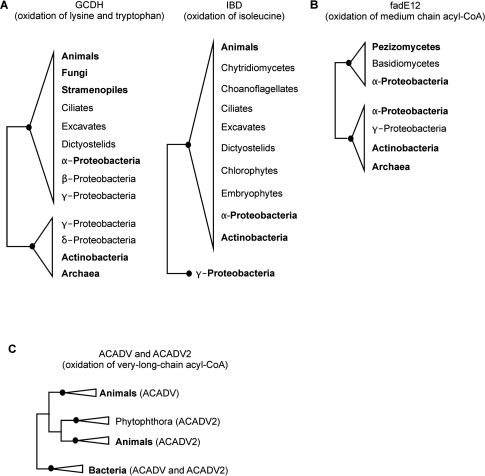
ACADS, GCDH, IVD and IBD occur in eukaryotes and bacteria (Figure 4). Subfamily trees unite eukaryotes mainly with α-Proteobacteria, to the exclusion of other prokaryotes. This trend is best supported by GCDH and IBD (Figure 6A). The tree topology, together with the taxonomic distribution and the predicted mitochondrial localization of these subfamilies (data not shown), suggests that eukaryotes acquired these genes from α-Proteobacteria via the endosymbiotic event leading to mitochondria. Yet, our current single-protein tree topologies have numerous branches with weak statistical support, and trees built with concatenated sequences of GCDH, IVD, IBD and ACADS do not provide more information either (Figure S1N). A rigorous test of this hypothesis would have to rule out horizontal transfer of the corresponding genes in bacteria, and improve tree robustness by substantially expanded taxon sampling.
Figure 6.
Schematic phylogenetic trees of ACAD subfamilies. The underlying explicit trees are provided in Figure S1. Branches with a bootstrap value ≥90 are labeled with filled dots. Taxa representing more than three species are shown in bold. (A) ACAD subfamilies with likely α-proteobacterial origin. In the trees of GCDH (left, Figure S1C) and IBD (right, Figure S1E), eukaryotic homologs group together with those from α-Proteobacteria. In the GCDH tree, eukaryotic and numerous α-proteobacterial taxa form a well-supported clade to the exclusion of Archaea plus Actinobacteria; both clusters include a few other Proteobacteria. The IBD tree unites eukaryotes and α-Proteobacteria to the exclusion of a few γ-Proteobacteria. (B) Secondary acquisition of α-proteobacterial homologs by certain fungal lineages. Some Basidiomycota and all investigated Ascomycota lack ACADM (Figure S1H). The ascomycete class Pezizomycotina possesses fadE12 (of same function as ACADM, Figure S1I) that associates strongly with α-Proteobacteria. Exceptions are Emericella nidulans, M. grisea and Chaetomium globosum, where fadE12 is absent. Ustilago maydis, the single basidiomycete possessing fadE12, likely acquired this gene from Pezizomycotina. (C) Gene duplication in animals and lateral transfer to other taxa. ACADV and ACADV2 are paralogs originating from a gene duplication prior to the divergence of animals (Figure S1O). The few bacterial ACADV homologs form a monophyletic clade, to the exclusion of animal proteins. ACADV2 from Phytophthora groups with animal homologs.
Loss of ACAD genes in fungi and recent recruitment from α-Proteobacteria
The two ACAD subfamilies fadE12 (typically prokaryotic) and ACADM (eukaryotic) have the same substrate specificity (Figure 4) (18). Pezizomycotina (including species such as Neurospora and Aspergillus) are an intriguing exception: they lack ACADM, but possess fadE12. Phylogenetic analysis of fadE12 proteins unites fungal and α-proteobacterial sequences with high support to the exclusion of other bacteria, strongly suggesting an α-proteobacterial origin of the Pezizomycotina genes (Figure 6B and Figure S1I; see legend of Figure 6 for exceptions). Based on the phylogeny and ACAD subfamily distribution, we propose that initially all fungi possessed ACADM, but this gene was later lost in the common ancestor of Ascomycota. After the ascomycete lineages had diverged, the predecessor of Pezizomycotina acquired the functionally equivalent fadE12 via horizontal gene transfer from α-Proteobacteria. A similar history involving loss and secondary acquisition of a bacterial ACAD gene by fungi apparently applies to ACADL (Figure S1J).
Duplication of ACAD genes in mammals and recent transfer to other lineages
As mentioned earlier, the two ACAD subfamilies degrading very long chain fatty acids, ACADV and ACADV2, are predominantly present in animals, with only a few exceptions in non-mammalian eukaryotes (i.e. Phytophthora species) and diverse bacteria (Figure 4). The phylogenetic tree including both subfamilies separates animal ACADV and ACADV2 into two well supported, distinct and coherent clades, indicating a gene duplication event prior to the divergence of animals (Figure 6C and Figure S1O). Homologs of Phytophthora affiliate (with moderate support) with animal ACADV2, and the same topology, now with 99% bootstrap support, are seen in the tree based on ACADV2 only (Figure S1L). The most parsimonious explanation is that the common ancestor of these oomycetes has acquired ACADV2 from animals, and transmitted it vertically to extant Phytophthora species.
Conclusion
Our study of the large and biologically important ACAD protein family integrates three types of information, taxonomic distribution profiles, subfamily phylogenies as well as functional and structural data from model proteins. This allows analyses of broad scope leading to improved molecular function prediction of individual ACAD subfamilies, formulation of working hypotheses for targeted biochemical experimentation as well as to the discovery of a most ‘turbulent’ evolutionary history of the ACAD gene family. A study like this one relies critically on a rigorous method for identification of orthologs for each paralogous subfamily as we devised in this report.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institute for Advanced Research (CIfAR, salary support granted to G.B.). Y.-Q.S. is a Canadian Institute for Health Research (CIHR) Strategic Training Fellow in Bioinformatics, and B.F.L. holds a Canadian Research Chair. Funding for open access charge: Canadian Institute for Health Research, Institute of Genetics.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Henner Brinkmann and Dr Nicolas Lartillot (Université de Montréal) and Yu Liu (University of Toronto) for help in phylogenetic analyses. We also thank Emmet O’Brien (Université de Montréal) and the anonymous referees for improving the manuscript.
REFERENCES
- 1.Gregersen N, Bross P, Andresen BS. Genetic defects in fatty acid beta-oxidation and acyl-CoA dehydrogenases. Molecular pathogenesis and genotype-phenotype relationships. Eur. J. Biochem. 2004;271:470–482. doi: 10.1046/j.1432-1033.2003.03949.x. [DOI] [PubMed] [Google Scholar]
- 2.Ijlst L, Wanders RJ. A simple spectrophotometric assay for long-chain acyl-CoA dehydrogenase activity measurements in human skin fibroblasts. Ann. Clin. Biochem. 1993;30(Pt 3):293–297. doi: 10.1177/000456329303000311. [DOI] [PubMed] [Google Scholar]
- 3.Tiffany KA, Roberts DL, Wang M, Paschke R, Mohsen AW, Vockley J, Kim JJ. Structure of human isovaleryl-CoA dehydrogenase at 2.6 A resolution: structural basis for substrate specificity. Biochemistry. 1997;36:8455–8464. doi: 10.1021/bi970422u. [DOI] [PubMed] [Google Scholar]
- 4.Udvari S, Bross P, Andresen BS, Gregersen N, Engel PC. Biochemical characterisation of mutations of human medium-chain acyl-CoA dehydrogenase. Adv. Exp. Med. Biol. 1999;466:387–393. doi: 10.1007/0-306-46818-2_45. [DOI] [PubMed] [Google Scholar]
- 5.He M, Burghardt TP, Vockley J. A novel approach to the characterization of substrate specificity in short/branched chain Acyl-CoA dehydrogenase. J. Biol. Chem. 2003;278:37974–37986. doi: 10.1074/jbc.M306882200. [DOI] [PubMed] [Google Scholar]
- 6.Battaile KP, Nguyen TV, Vockley J, Kim JJ. Structures of isobutyryl-CoA dehydrogenase and enzyme-product complex: comparison with isovaleryl- and short-chain acyl-CoA dehydrogenases. J. Biol. Chem. 2004;279:16526–16534. doi: 10.1074/jbc.M400034200. [DOI] [PubMed] [Google Scholar]
- 7.Fu Z, Wang M, Paschke R, Rao KS, Frerman FE, Kim JJ. Crystal structures of human glutaryl-CoA dehydrogenase with and without an alternate substrate: structural bases of dehydrogenation and decarboxylation reactions. Biochemistry. 2004;43:9674–9684. doi: 10.1021/bi049290c. [DOI] [PubMed] [Google Scholar]
- 8.Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 9.Ye X, Ji C, Zhou C, Zeng L, Gu S, Ying K, Xie Y, Mao Y. Cloning and characterization of a human cDNA ACAD10 mapped to chromosome 12q24.1. Mol. Biol. Rep. 2004;31:191–195. doi: 10.1023/b:mole.0000043622.57408.6b. [DOI] [PubMed] [Google Scholar]
- 10.Saenger AK, Nguyen TV, Vockley J, Stankovich MT. Thermodynamic regulation of human short-chain acyl-CoA dehydrogenase by substrate and product binding. Biochemistry. 2005;44:16043–16053. doi: 10.1021/bi051048y. [DOI] [PubMed] [Google Scholar]
- 11.Merritt JL, 2nd, Matern D, Vockley J, Daniels J, Nguyen TV, Schowalter DB. In vitro characterization and in vivo expression of human very-long chain acyl-CoA dehydrogenase. Mol. Genet. Metab. 2006;88:351–358. doi: 10.1016/j.ymgme.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 12.Ensenauer R, He M, Willard JM, Goetzman ES, Corydon TJ, Vandahl BB, Mohsen AW, Isaya G, Vockley J. Human acyl-CoA dehydrogenase-9 plays a novel role in the mitochondrial ß-oxidation of unsaturated fatty acids. J. Biol. Chem. 2005;280:32309–32316. doi: 10.1074/jbc.M504460200. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda Y, Okamura-Ikeda K, Tanaka K. Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from rat liver mitochondria. Isolation of the holo- and apoenzymes and conversion of the apoenzyme to the holoenzyme. J. Biol. Chem. 1985;260:1311–1325. [PubMed] [Google Scholar]
- 14.Izai K, Uchida Y, Orii T, Yamamoto S, Hashimoto T. Novel fatty acid beta-oxidation enzymes in rat liver mitochondria. I. Purification and properties of very-long-chain acyl-coenzyme A dehydrogenase. J. Biol. Chem. 1992;267:1027–1033. [PubMed] [Google Scholar]
- 15.Roe CR, Ding J. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill Book Co.; 2001. [Google Scholar]
- 16.Pauli G, Overath P. ato Operon: a highly inducible system for acetoacetate and butyrate degradation in Escherichia coli. Eur. J. Biochem. 1972;29:553–562. doi: 10.1111/j.1432-1033.1972.tb02021.x. [DOI] [PubMed] [Google Scholar]
- 17.Iram SH, Cronan JE. The beta-oxidation systems of Escherichia coli and Salmonella enterica are not functionally equivalent. J. Bacteriol. 2006;188:599–608. doi: 10.1128/JB.188.2.599-608.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahadevan U, Padmanaban G. Cloning and expression of an acyl-CoA dehydrogenase from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 1998;244:893–897. doi: 10.1006/bbrc.1998.8354. [DOI] [PubMed] [Google Scholar]
- 19.Ghisla S, Thorpe C. Acyl-CoA dehydrogenases. A mechanistic overview. Eur. J. Biochem. 2004;271:494–508. doi: 10.1046/j.1432-1033.2003.03946.x. [DOI] [PubMed] [Google Scholar]
- 20.Kim JJ, Miura R. Acyl-CoA dehydrogenases and acyl-CoA oxidases. Structural basis for mechanistic similarities and differences. Eur. J. Biochem. 2004;271:483–493. doi: 10.1046/j.1432-1033.2003.03948.x. [DOI] [PubMed] [Google Scholar]
- 21.Cornell MJ, Alam I, Soanes DM, Wong HM, Hedeler C, Paton NW, Rattray M, Hubbard SJ, Talbot NJ, Oliver SG. Comparative genome analysis across a kingdom of eukaryotic organisms: specialization and diversification in the fungi. Genome Res. 2007;17:1809–1822. doi: 10.1101/gr.6531807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen YQ, Burger G. Plasticity of a key metabolic pathway in fungi. Funct. Integr. Genomics. 2009;9:145–151. doi: 10.1007/s10142-008-0095-6. [DOI] [PubMed] [Google Scholar]
- 23.Kondrashov FA, Koonin EV, Morgunov IG, Finogenova TV, Kondrashova MN. Evolution of glyoxylate cycle enzymes in Metazoa: evidence of multiple horizontal transfer events and pseudogene formation. Biol. Direct. 2006;1:31. doi: 10.1186/1745-6150-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pereto J, Lopez-Garcia P, Moreira D. Phylogenetic analysis of eukaryotic thiolases suggests multiple proteobacterial origins. J. Mol. Evol. 2005;61:65–74. doi: 10.1007/s00239-004-0280-8. [DOI] [PubMed] [Google Scholar]
- 25.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown DP, Krishnamurthy N, Sjolander K. Automated protein subfamily identification and classification. PLoS Comput. Biol. 2007;3:e160. doi: 10.1371/journal.pcbi.0030160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hannenhalli SS, Russell RB. Analysis and prediction of functional sub-types from protein sequence alignments. J. Mol. Biol. 2000;303:61–76. doi: 10.1006/jmbi.2000.4036. [DOI] [PubMed] [Google Scholar]
- 28.Weston J, Leslie C, Ie E, Zhou D, Elisseeff A, Noble WS. Semi-supervised protein classification using cluster kernels. Bioinformatics. 2005;21:3241–3247. doi: 10.1093/bioinformatics/bti497. [DOI] [PubMed] [Google Scholar]
- 29.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 31.O’Brien EA, Koski LB, Zhang Y, Yang L, Wang E, Gray MW, Burger G, Lang BF. TBestDB: a taxonomically broad database of expressed sequence tags (ESTs) Nucleic Acids Res. 2007;35:D445–D451. doi: 10.1093/nar/gkl770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray MW, Lang BF, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Brossard N, Delage E, Littlejohn TG, et al. Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acids Res. 1998;26:865–878. doi: 10.1093/nar/26.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 34.Lang BF, Burger G, O’Kelly CJ, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Gray MW. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 35.Palmer JD. Genome evolution. The mitochondrion that time forgot. Nature. 1997;387:454–455. doi: 10.1038/387454a0. [DOI] [PubMed] [Google Scholar]
- 36.Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Pearlman RE, Roger AJ, Gray MW. The tree of eukaryotes. Trends Ecol. Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 37.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 40.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 41.Mulder N, Apweiler R. InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol. Biol. 2007;396:59–70. doi: 10.1007/978-1-59745-515-2_5. [DOI] [PubMed] [Google Scholar]
- 42.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 43.Small I, Peeters N, Legeai F, Lurin C. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- 44.Boden M, Hawkins J. Prediction of subcellular localization using sequence-biased recurrent networks. Bioinformatics. 2005;21:2279–2286. doi: 10.1093/bioinformatics/bti372. [DOI] [PubMed] [Google Scholar]
- 45.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 46.Lu Z, Szafron D, Greiner R, Lu P, Wishart DS, Poulin B, Anvik J, Macdonell C, Eisner R. Predicting subcellular localization of proteins using machine-learned classifiers. Bioinformatics. 2004;20:547–556. doi: 10.1093/bioinformatics/btg447. [DOI] [PubMed] [Google Scholar]
- 47.Shen YQ, Burger G. ‘Unite and conquer’: enhanced prediction of protein subcellular localization by integrating multiple specialized tools. BMC Bioinformatics. 2007;8:420. doi: 10.1186/1471-2105-8-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neuberger G, Maurer-Stroh S, Eisenhaber B, Hartig A, Eisenhaber F. Prediction of peroxisomal targeting signal 1 containing proteins from amino acid sequence. J. Mol. Biol. 2003;328:581–592. doi: 10.1016/s0022-2836(03)00319-x. [DOI] [PubMed] [Google Scholar]
- 49.Nurizzo D, Shewry SC, Perlin MH, Brown SA, Dholakia JN, Fuchs RL, Deva T, Baker EN, Smith CA. The crystal structure of aminoglycoside-3′-phosphotransferase-IIa, an enzyme responsible for antibiotic resistance. J. Mol. Biol. 2003;327:491–506. doi: 10.1016/s0022-2836(03)00121-9. [DOI] [PubMed] [Google Scholar]
- 50.Kikuchi M, Hatano N, Yokota S, Shimozawa N, Imanaka T, Taniguchi H. Proteomic analysis of rat liver peroxisome: presence of peroxisome-specific isozyme of Lon protease. J. Biol. Chem. 2004;279:421–428. doi: 10.1074/jbc.M305623200. [DOI] [PubMed] [Google Scholar]
- 51.Wang ZY, Soanes DM, Kershaw MJ, Talbot NJ. Functional analysis of lipid metabolism in Magnaporthe grisea reveals a requirement for peroxisomal fatty acid beta-oxidation during appressorium-mediated plant infection. Mol. Plant Microbe Interact. 2007;20:475–491. doi: 10.1094/MPMI-20-5-0475. [DOI] [PubMed] [Google Scholar]
- 52.Kang Y, Nguyen DT, Son MS, Hoang TT. The Pseudomonas aeruginosa PsrA responds to long-chain fatty acid signals to regulate the fadBA5 beta-oxidation operon. Microbiology. 2008;154:1584–1598. doi: 10.1099/mic.0.2008/018135-0. [DOI] [PubMed] [Google Scholar]
- 53.Son MS, Matthews WJ, Jr, Kang Y, Nguyen DT, Hoang TT. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect. Immun. 2007;75:5313–5324. doi: 10.1128/IAI.01807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grecz N, Wagenaar RO, Dack GM. Relation of fatty acids to the inhibition of Clostridium botulinum in aged surface ripened cheese. Appl. Microbiol. 1959;7:228–234. doi: 10.1128/am.7.4.228-234.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maggio-Hall LA, Lyne P, Wolff JA, Keller NP. A single acyl-CoA dehydrogenase is required for catabolism of isoleucine, valine and short-chain fatty acids in Aspergillus nidulans. Fungal Genet. Biol. 2008;45:180–189. doi: 10.1016/j.fgb.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.