Abstract
Analyses of suppressor mutations have been extremely valuable in understanding gene function. However, techniques for mapping suppressor mutations are not available for most bacterial species. Here, we used high-throughput sequencing technology to identify spontaneously arising suppressor mutations that enabled disruption of rpoE (which encodes σE) in Vibrio cholerae, the agent of cholera. The alternative sigma factor σE, which is activated by envelope stress, promotes expression of factors that help preserve and/or restore cell envelope integrity. In Escherichia coli, rpoE is an essential gene that can only be disrupted in the presence of additional suppressor mutations. Among a panel of independent V. cholerae rpoE mutants, more than 75% contain suppressor mutations that reduce production of OmpU, V. cholerae’s principal outer membrane porin. OmpU appears to be a key determinant of V. cholerae’s requirement for and production of σE. Such dependence upon a single factor contrasts markedly with regulation of σE in E. coli, in which numerous factors contribute to its activation and none is dominant. We also identified a suppressor mutation that differs from all previously described suppressors in that it elevates, rather than reduces, σE’s activity. Finally, analyses of a panel of rpoE mutants shed light on the mechanisms by which suppressor mutations may arise in V. cholerae.
INTRODUCTION
The alternative sigma factor σE (also known as RpoE) is one of several regulators that enable bacteria to respond to perturbation of the cell envelope (1). In unstressed growing cells, σE activity is relatively low, as it is tethered to the bacterial inner membrane by the antisigma factor RseA, and hence cannot interact with RNA polymerase to direct transcription from σE-dependent promoters (2,3). However, if misfolded outer membrane proteins (OMPs) accumulate within the periplasm, they trigger the degradation of RseA and release of σE, thereby allowing increased expression of the σE regulon, which includes genes encoding periplasmic proteases, foldases and chaperones that aid in periplasmic folding, as well as rpoE itself (4–6). Activation of σE also induces transcription of several small non-coding RNAs (sRNAs) that repress production of OMPs (7). Heightened expression of these factors is thought to aid in elimination of the inducing stimulus and restoration of envelope homeostasis. σE activity is also induced as cells enter stationary phase. In Escherichia coli, this response has been shown to be dependent upon the alarmone ppGpp, which typically accumulates when nutrients are limited (8,9).
σE is classically thought of as regulating a stress response; however, it has been found to be essential for viability in E. coli and Yersinia enterocolitica, suggesting that it plays a more fundamental role in bacterial physiology as well (10,11). σE was not initially recognized to be essential in E. coli, both because suppressor mutations can mask this phenotype and because the growth (or lack thereof) of E. coli rpoE mutants is highly dependent upon the selective criteria employed. However, comparisons of independently derived E. coli rpoE mutants suggested that these strains might contain suppressor mutations, and several suppressors have been identified to date (12,13). These include a loss of function mutation in ydcQ, which encodes a putative DNA-binding protein, and multicopy plasmids containing ptsN or yhbW. Little is known about the mechanisms by which these suppressors function; however, it has been observed that all the suppressors reduce basal activity of other envelope stress response pathways as well. Somewhat surprisingly, suppressors that counteract E. coli’s requirement for the proteases RseP and DegS, which are needed for degradation of RseA and thus for activation of σE in response to periplasmic stresses, did not also function as suppressors for rpoE mutations. For example, when production of OmpA and OmpC is reduced, either via deletion of both genes or via overexpression of RseX, an sRNA that negatively regulates them, degS and rseP are no longer essential; however, rpoE is still required (14). This may reflect the fact that several pathways have been shown to activate σE in E. coli.
Studies of rpoE in V. cholerae, the etiologic agent of cholera, have not directly addressed whether rpoE is essential or whether rpoE mutants contain suppressor mutations. As with early studies of rpoE in E. coli, the ability to generate rpoE mutants may have led to the assumption that the gene is not required, without consideration of the possibility of suppressors. In V. cholerae, rpoE has been shown to be required for intestinal colonization in an animal model of disease and for resistance to an antimicrobial peptide, and likely members of the regulon have been identified (15–17). Furthermore, we have found that expression of σE is reduced in the absence of OmpU, an abundant outer membrane protein in V. cholerae whose expression is controlled by the key virulence regulator ToxR (18). However, many aspects of the biology of rpoE mutants remain to be explored, and phenotypic comparisons of independently derived mutants have not been reported.
Identification of suppressor mutations can be hampered by a lack of scorable/selectable phenotypes and/or of tools for genetic mapping. In V. cholerae, there are currently no robust genetic tools for mapping spontaneous suppressor mutations. However, the recent development of massively parallel sequencing technology provides a potential path around these experimental barriers. This technology enables generation of high coverage genomic sequence data at a cost that permits analysis of multiple samples, and hence allows for direct identification of differences between the DNA sequences of related strains. The approach is ideally suited for unbiased identification of genetic changes, particularly if multiple changes with interdependent effects are present within a single strain [e.g. as observed in (19)], since such effects might be missed in phenotypic assays of individual changes.
We generated and characterized a set of V. cholerae rpoE mutants and found that they displayed a range of phenotypes, consistent with the possibility that they might contain distinct suppressor mutations. Using high-throughput sequencing, we identified several differences between wt and rpoE strains that were subsequently confirmed to be suppressor mutations. Remarkably, the majority of strains that lacked σE contained additional mutations that reduced expression of V. cholerae’s major OMP, OmpU. An ompU-independent suppression pathway was also identified, which, unlike other rpoE suppressors, elevated the basal activity of this envelope stress response pathway. Overall, our data suggest that a key role for σE is to respond to an endogenously generated stimulus transmitted from OmpU. They also shed light on the mechanisms used by V. cholerae to maintain appropriate OMP expression, and upon the mechanisms by which suppressor mutations may arise in this organism. Finally, our results illustrate the utility of high-throughput sequencing technology in carrying out a genomics-based approach to suppressor genetics.
MATERIALS AND METHODS
Bacterial strains and culture conditions
All V. cholerae strains generated for this study are derivatives of the sequenced clinical isolate N16961 and are resistant to streptomycin. For analysis of rpoE production by V. cholerae lacking various OMPs, transposon insertion mutants derived from C6706 were used (20). A related mutant with an insertion in vca0199 was also used, both with the KnR transposon still present (BD2034) and with the majority of the transposon excised by flippase (BD2041). Transposons were also excised from a subset of the C6706 OMP mutants, namely those in which the transposon ‘scar’ remaining after excision contained a stop codon to prevent production of the full length gene product. Removal of the transposon, which contains lacZ, was necessary prior to monitoring rpoE::lacZ activity. C6706 ΔtoxRS was obtained from JJ Mekalanos. Escherichia coli strains DH5αλpir and SM10λpir were used for cloning and conjugation of suicide vectors, respectively. Bacteria were grown in LB at 37°C unless otherwise noted, using antibiotics at the following concentrations: streptomycin, 200 μg/ml; carbenicillin, 50 μg/ml; kanamycin, 50 μg/ml. Negative selection against strains containing sacB was performed on plates containing 10% sucrose at 30°C. LB plates containing 0.25% SDS were used to assess the sensitivity of rpoE mutants to detergents.
Strain and plasmid construction
Strains with insertion mutations in rpoE were generated using pBD1574, a derivative of the suicide vector pGP704 that contains 424 bp of an internal fragment of rpoE (21). Integration of this vector, via a single crossover event within the rpoE-derived sequence, results in gene disruption and confers carbenicillin resistance. Exconjugates were selected on plates containing streptomycin and carbenicillin. When primary exconjugates gave rise to colonies of varied sizes upon restreaking, which occurred for at least 90% of exconjugants, large colonies were selected for further analysis, in order to minimize analyses of mixed populations. Strains with insertion mutations in ompS and ompT were generated using pBD1664 and pBD1665, which contain internal gene fragments of 451 and 382 bp, respectively.
All allele replacements were generated using vectors derived from the suicide vector pCVD442, using standard allele exchange procedures (22,23). In brief, the targeting vector, which confers carbenicillin resistance, was transferred by conjugation to a V. cholerae recipient, where it integrated. Primary exconjugants (selected on plates containing streptomycin and carbenicillin) were then subjected to counterselection on sucrose plates to identify strains that no longer contained the plasmid backbone, and further screened via PCR to identify strains containing the mutation of interest. Strains containing deletions within rpoE were generated using pBD1578, which introduces a 342-bp deletion. Strains in which rpoE was replaced by an rpoE::aph allele were generated using pBD1999, in which rpoE is interrupted by insertion of an aph gene cassette from pUC71K. Mutants were selected on plates containing kanamycin as well as sucrose. (Primary selection on plates containing sucrose alone, followed by patching to plates containing kanamycin, led to similar conclusions.) Point mutations in vca0199, vc2413 and the ompU (vc0633) promoter and 5′UTR were introduced using plasmids pBD2038, pBD2036, pBD2049 and pBD2088, respectively. Deletions in ompU and ompT were introduced using the previously described vectors pKEK276 and pKEK309 (24). Deletion of vc0972 was performed using pBD1900.
All ompU::lacZ transcription reporter fusions are derivatives of pCB192N, which contains a promoterless lacZ cassette (H. Kimsey, manuscript in preparation). Each contains ompU-linked sequences extending from position –411 to +227 relative to the ompU transcriptional start site (25). The rpoE::lacZ transcription reporter fusion is a derivative of pCB192 (26) and contains sequences from –91 to +112 relative to the σE dependent transcriptional start site, as defined in (16).
Additional details regarding plasmid and strain construction are available upon request. The sequences of plasmids and the chromosomal loci they were used to alter were confirmed by Sanger sequencing.
Genome sequencing and analysis
Genome sequencing was performed by the Partners Healthcare Center for Genetics and Genomics using an Illumina Genome Analyzer. Sequence data was analyzed using MAQ (http://maq.sourceforge.net/index.shtml) (27) and Edena (http://www.genomic.ch/edena.php, (28)). MAQ was primarily used for identification of single nucleotide polymorphisms (SNPs), but also enabled detection of some single-nucleotide insertions and deletions in regions with extended mononucleotide tracts. The consensus sequence obtained for rpoE mutants was compared to a control sequence from the wt progenitor and to the published V. cholerae N16961 sequence, thereby enabling identification both of mutant-specific SNPs and SNPs distinguishing our isolate from the published genome (29). MAQ reports each candidate SNP with a consensus quality score that reflects the trustworthiness of the SNP prediction. Filtered SNPs with consensus quality scores lower than 50 were generally deemed unreliable.
De novo sequence assembly, using Edena, was performed on pooled data from all sequenced samples, using default parameters. Assembled contigs were then compared to the published N16961 genome using MUMmer to identify SNPs, insertions, and deletions (http://mummer.sourceforge.net; Supplementary Table 1). Comparable analyses using sequencing results from individual samples did not yield genome-wide results, as the depth of sequence coverage from individual samples was not always sufficient to permit contig assembly; however, a subset of differences could be identified.
The precise locations of sequences that differ between the published N16961 genome and individual rpoE mutants, which were confirmed by Sanger sequencing, are presented in Supplementary Table 2.
β-galactosidase assays
Reporter plasmids were transformed into strains made ΔlacZ using the plasmid pJL1. Assays were performed basically as described (30).
Western blots
Protein samples were generated from overnight cultures of V. cholerae unless otherwise indicated. Culture volumes were normalized based on OD600, then cells were pelleted and lysed in 1× NuPAGE LDS sample buffer (Invitrogen) containing 5% β-mercaptoethanol at 95°C. Samples were run on NuPAGE Bis–Tris gels (Invitrogen) and transferred to nitrocellulose membranes according to the manufacturer's instructions. Membranes were probed with polyclonal antisera to σE, OmpU, OmpT, OmpA, or with a commercially available anti 6-His antibody (Genetex). Horseradish peroxidase–conjugated secondary antibodies and Supersignal West Pico chemiluminescent substrate kit (Pierce) were used for detection of bound primary antibodies.
Northern blots
RNA was harvested from log phase cultures unless otherwise noted. RNA was extracted from cell pellets using Trizol (Invitrogen), then treated with DNase I (Qiagen). It was electrophoresed on glyoxal gels (Ambion), then transferred to Bright Star Plus nylon membranes (Ambion). RNA integrity was confirmed via assessment of EtBr-stained rRNA bands. Blots were hybridized to 32P-labeled in vitro transcribed probes in ULTRAhyb (Ambion) at 68°C and washed according to the manufacturer's instructions.
RESULTS
Isolation of V. cholerae rpoE mutants
We and others previously isolated rpoE mutants of V. cholerae; thus, it was clear that there was not an absolute barrier to obtaining rpoE mutants in this organism (16,17). However, some of our strategies for generating rpoE mutants had failed, while others seemed to have only a low frequency of success. Furthermore, we noted that the two published mutants did not share all phenotypes (e.g. EtOH sensitivity), suggesting that they might have acquired distinct suppressor mutations. Thus, although V. cholerae rpoE had not previously been reported to be essential, we undertook a systematic characterization of V. cholerae rpoE mutants and their potential suppressor mutations.
We first assessed the frequency of obtaining insertion mutants following conjugation of suicide vectors containing either an internal fragment of rpoE or a control gene (ompS or ompT), along with the selectable marker bla. Carbenicillin resistant (carbR) mutants of rpoE, ompS and ompT were obtained with approximately equal frequencies, suggesting that these rpoE mutants could arise in the absence of any suppressor mutations. However, we noticed that the initial selection for rpoE mutants routinely yielded smaller colonies than for the control genes, and that upon restreaking these colonies typically yielded a mixture of large and small colonies (Figure 1). Further characterization of the mutants revealed that they could be grouped into distinct classes. Mutants could be distinguished by their levels of OMP expression, their colony size and morphology, and their ability to grow on plates containing detergents. Together, these results suggested that our carbR rpoE insertion mutants frequently acquired at least one of several additional mutations that suppressed the small colony phenotype of an rpoE mutant and enabled more rapid growth.
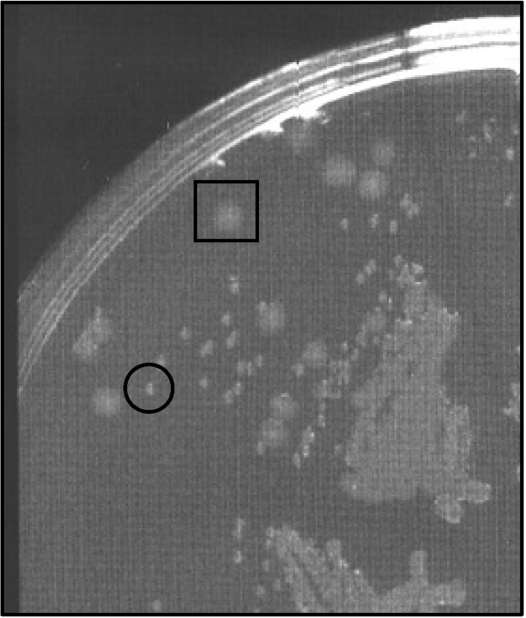
Figure 1.
An rpoE insertion mutant restreaked on plates containing Sm and Carb following the primary selection. The mutant is a derivative of wt V. cholerae N16961. Both large (boxed) and small (circled) colonies are detectable.
Derivatives of wt V. cholerae in which rpoE was deleted, using a targeting vector conferring carbR and sucrose sensitivity, were obtained much less readily than rpoE insertion mutants. Identification of deletion mutants required a biased screening of sucrose resistant colonies, namely preferential testing of small or differentially colored colonies. Mutants in which rpoE was replaced by rpoE::aph could not be obtained. Thus, rpoE appeared to be essential for V. cholerae growth under the selection conditions for rpoE::aph.
Identification of candidate suppressor mutations
To identify the putative suppressor mutations in V. cholerae rpoE mutants, we took advantage of newly developed massively parallel sequencing technology. Whole genome sequencing of DNA from a subset of insertion mutants and deletion mutants as well as from wt V. cholerae N16961, from which the mutants were derived, was performed on the Solexa Genome Analyzer. This approach yielded an average of 2.8 million 36-nt sequence reads per sample (range of 1.2–4.0 million). Sequences were subsequently mapped to the published V. cholerae N16961 genome sequence (29) using the freely available software MAQ [Mapping and Assembly with Qualities; http://maq.sourceforge.net/index.shtml (27)]. MAQ uses the Solexa nucleotide sequences and their associated quality scores (which reflect the confidence with which each nucleotide within a 36-mer was called by the Genome Analyzer) to predict SNPs, positions where the sequenced sample differs from the reference genome. MAQ also calculates its own consensus quality score for these potential SNPs. An average of 88% of the Solexa sequences could be mapped to the reference genome. Numerous potential SNPs were identified; however, most were identified as candidate SNPs in wt as well as rpoE mutant samples (and thus correspond to differences between our strain and the published sequence) or had relatively low consensus quality scores. Based on visual inspection of sequence alignments (using MAQ’s companion program, MAQview) as well as Sanger sequencing of some candidate loci, we determined that candidate SNPs with consensus quality scores of less than 50 were likely to be false. Removing these loci from the list of mutant-specific candidates left a relatively small number of candidates, suitable for confirmation using Sanger sequencing and/or elimination via visual inspection of aligned reads. Through this process we verified the presence of five candidate suppressor mutations, located in the vca0199 and vc2413-coding regions (one each, in strains BD1965 and BD1963 respectively) and in the intergenic region between vc0633 and vc0634 (three distinct mutations, in strains BD1962, BD1956 and BD1987). The effects of these mutations are described below.
Comparison of newly sequenced isolates to the published V. cholerae genome
Solexa reads were also used as inputs for the de novo assembly program Edena [Exact DE Novo Assembler (28)], which generated contigs that could be searched for differences relative to the published V. cholerae N16961 sequence using MUMmer. The advantage of this approach is that it performs gapped as well as ungapped alignments, and hence can be used to identify small insertions and deletions, which generally cannot be detected in our data set using MAQ. Disadvantages are that it does not utilize the Solexa quality scores or provide a means to identify high quality versus low quality candidate SNPs, and it requires a greater number of sequences to generate a consensus, often more than we obtained for a single sample. Edena confirmed the presence of most of the candidate suppressors identified using MAQ; however, its principal utility was in identification of differences between our starting strain and the reference strain, for which we pooled all our Solexa sequences to generate the Edena input. With this comprehensive set of sequences, contigs spanning 3.9 million bases, or >88% of the V. cholerae genome, could be assembled, and 145 differences between our strains and the reference strain could be identified (Supplementary Table 1). These included 40 SNPs, most of which had also been predicted by MAQ and 79 small insertions or deletions. A subset of these correspond to loci identified in a recent study as likely errors within the published genomic sequence (31). An approximate position for many of the insertions/deletions could also be obtained by performing BLAST analyses on sequences that could not be mapped by MAQ. This approach can be utilized even if sequence depth does not meet the stringent conditions required by Edena, and hence can be used to search for insertions and deletions in sequence from an individual sample as well as pooled samples. However, BLAST analyses did not identify any insertions or deletions in the rpoE mutants that were not also present in our wt strain.
Many rpoE mutants contain mutations upstream of ompU that reduce its expression
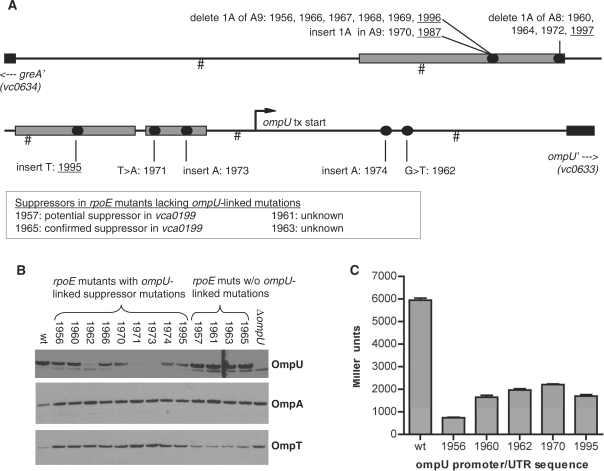
The intergenic sequence between vc0633 and vc0634, which was found to contain three independently derived candidate suppressor mutations, contains the promoters for both genes. Notably, vc0633 encodes OmpU, which we have previously shown is a determinant of σE levels (18). Given the frequency of mutations obtained in this region and our knowledge concerning the gene's role, we resequenced this region in all our rpoE mutants. Remarkably, these analyses revealed that 17 of 21 rpoE mutants contained mutations in this region (Figure 2A). Identical mutations were detected in multiple strains, almost all of which are known to have arisen independently. Most of the mutations are within confirmed binding sites for ToxR, which activates transcription of ompU; however, two of the mutations are not within the promoter, and instead lie within ompU’s 5′UTR (21,25,32). OmpU-coding sequences were also examined for many of the strains, but no mutations were detected. Western blots showed that all of the strains with ompU-linked mutations contained reduced amounts of OmpU, although the levels varied widely (Figure 2B). No mutations in this region were identified in the strains found to contain candidate suppressor mutations at loci other than ompU, and OmpU levels in rpoE strains lacking ompU mutations were equal to those in wt V. cholerae.
Figure 2.
Locations of rpoE suppressor mutations and their effects on OMP levels and promoter activity. (A) Schematic depiction of the locations of suppressor mutations in rpoE mutants. The ompU promoter region is drawn to scale; hash marks denote 100-bp intervals. The 5′ ends of the coding sequences of flanking genes are shown as black boxes; the ompU-coding sequence corresponds to that shown in (25) rather than the published genome sequence. Binding sites for ToxR are shown as grey rectangles as defined in (32). Circles represent the sites of suppressor mutations, and numbers above (e.g. 1956) correspond to the strain numbers of corresponding rpoE mutants. rpoE deletion mutants are underlined; all other strains are insertion mutants. In most cases, mutants were independently derived, despite detection of identical suppressors in multiple strains. Only the pairs 1966/1967 and 1968/1969 could each potentially consist of siblings. The precise positions (relative to the published N16961 genome) of all SNPs linked to suppression of rpoE mutations are listed in Supplementary Table 2. (B) Western blots showing expression of OmpU, OmpA and OmpT levels in whole cell lysates from wt, ΔompU, and a non-redundant subset of rpoE mutants. (C) β-galactosidase activity from ompU::lacZ transcription reporter fusions generated from wt V. cholerae and the indicated rpoE mutants. Activities shown are averages +SEM from three independent cultures and were generated from overnight cultures; log phase cultures yielded similar relative activities.
To confirm that the ompU-linked mutations are responsible for reduced OmpU levels, we generated ompU::lacZ transcription reporter fusions containing wt and mutant promoter/UTR sequences, and assessed their activity in wt V. cholerae. All of the mutant promoter fusions yielded at least 2-fold less activity, as did a reporter fusion containing a mutation within the ompU 5′UTR (Figure 2C), suggesting that the mutations do impair ompU expression, in most cases presumably due to reduced ToxR binding. However, the 5′UTR mutation (identified in strain BD1962) seems likely to influence processes other than, or in addition to, transcription, (e.g. mRNA translation or stability), as its effect on OmpU mRNA (data not shown) and protein levels (Figure 2B) was more dramatic than that of promoter mutations that yielded similar reporter activity. Additional northern and β-galactosidase assays suggest that the UTR and promoter mutations do not alter transcription of vca0634 (data not shown), although the intergenic region presumably contains its promoter as well.
OmpU-linked and vca0199 mutations reduce V. cholerae's requirement for σE through distinct processes
To determine whether the candidate rpoE suppressor mutations did in fact facilitate the growth of rpoE mutants, we regenerated candidate suppressor mutations in a wt strain, and assessed whether rpoE::aph mutants could be obtained in these new strains. Following chromosomal integration of the targeting vector, candidate rpoE::aph mutants were selected on plates containing sucrose and kanamycin, and then patched to plates containing carbenicillin to eliminate false positives from the sucrose selection. Using this selection protocol, all sucrose and kanamycin-resistant colonies isolated from the wt strain were also resistant to carbenicillin, i.e. no rpoE mutants were obtained (Table 1). In contrast, rpoE mutants were readily obtained in strains containing mutations within the ompU promoter or 5′ UTR, indicating that a reduction in OmpU levels obviates V. cholerae's need for σE under these selection conditions. A comparable result was observed using strains with deletions of ompU or toxRS (Table 1). Similarly, we found that rpoE::aph mutants were generated at high frequency in the presence of the vca0199 candidate suppressor mutation. However, no mutants were obtained in a strain containing the vc2413 candidate suppressor mutation, suggesting that the mutation does not function as a suppressor. The consequences of the vc2413 mutation were not pursued further.
Table 1.
Frequency of obtaining rpoE::aph mutants from sucrose/kanamycin selection
| Parental strain | Mutation | rpoE mutanta | Percentage rpoE::aph (n) |
|---|---|---|---|
| N16961 | NA | – | 0 (85) |
| BD2046 | vc2413 | BD1963 | 0 (49) |
| BD2048 | vca0199 (pt. mut.) | BD1965 | 91 (102) |
| BD2064 | ompU 5′UTR | BD1962 | 83 (31) |
| BD2096 | ompU promoter | BD1956 | 95 (68) |
| MKW156 | ΔompU | – | 96 (46) |
| JM3-3 | ΔompT | – | 0 (33) |
| BD1902 | Δvc0972 | – | 0 (45) |
| BD2139 | Δvca0199 | – | 45 (65) |
| C6706 | NA | – | 0 (48) |
| BD2041b | vca0199, vca0200 | 0 (52) | |
| BD2093b | ΔtoxRS | – | 94 (54) |
NA, not applicable; n, number of colonies tested for carbenicillin resistance.
aStrain in which candidate suppressor mutation was initially identified, if applicable.
bDerived from C6706; all other strains derived from N16961.
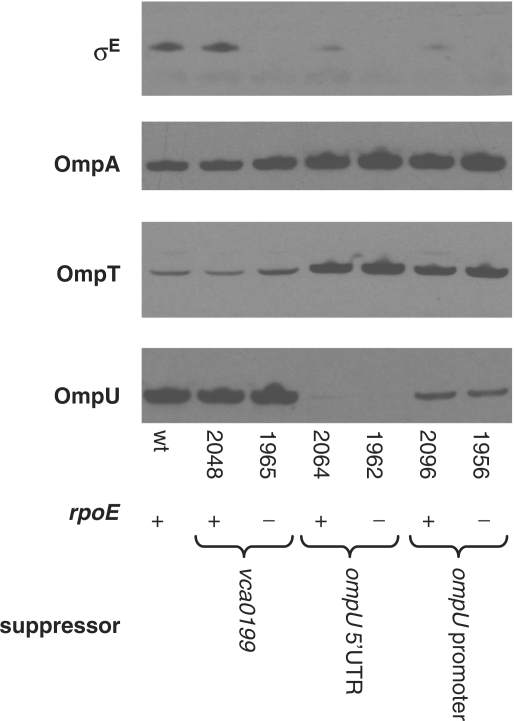
Suppressor mutations presumably reduce the cell's need for σE; consequently, it might be expected that production and activation of σE would be reduced in strains containing suppressor mutations. Our previous studies of OmpU and σE suggested that OmpU is a key determinant of basal levels of rpoE expression, as strains lacking OmpU contained markedly lower level of σE (18). Mutations that downregulate OmpU production (e.g. promoter and 5′ UTR mutations) also reduced the abundance of σE (Figure 3). In addition, such mutations cause a decrease in σE activity, as indicated by reduced β-galactosidase from an rpoE::lacZ transcription reporter fusion whose activity is dependent upon σE (Figure 4A and data not shown). In contrast, the point mutation in vca0199 that suppresses rpoE essentiality did not appear to influence production of σE, and increased σE activity several fold, suggesting that it influences survival via a different mechanism (Figures 3 and 4A). Additional attributes of vca0199 are discussed further below.
Figure 3.
σE and OMPs produced by wt V. cholerae and strains containing rpoE and/or suppressor mutations. Western blots were probed with polyclonal antisera to σE, OmpA, OmpT and OmpU. Each rpoE mutant (strains BD1965, BD1962 and BD1956) is paired with a strain containing the suppressor mutation (e.g. in vca0199 or ompU-linked sequences) identified within the rpoE mutant. Extracts electrophoresed on NuPAGE gels were generated from cells cultured overnight in LB. Culture volumes used for extracts were adjusted based on culture OD600 to ensure equal loading.
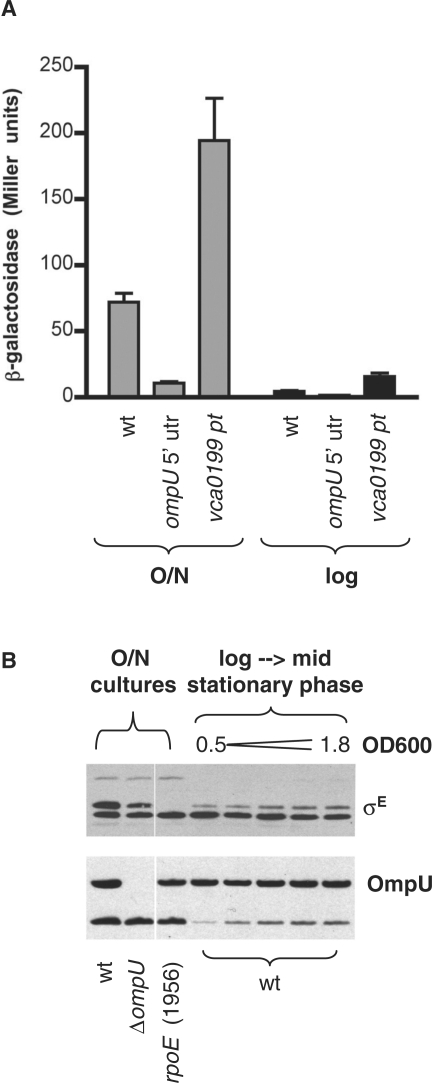
Figure 4.
Levels and activity of σE and OmpU produced by V. cholerae at various growth phases. (A) β-galactosidase activity produced from an rpoE-P2::lacZ transcription reporter fusion was assessed in a lacZ derivative of wt V. cholerae (wt) and in strains containing suppressor mutations in ompU (BD2064 lacZ; ompU 5′ UTR) and vca0199 (BD2048 lacZ; vca0199 pt). Activity was assessed in overnight (O/N) and log phase (O.D.600 ∼ 0.5–0.65; log) cultures. The reporter yielded no activity in a strain lacking rpoE (data not shown). Data shown is the mean and SEM for a representative experiment. (B) Western blots of OmpU and σE produced by wt V. cholerae and the indicated mutants during log and stationary phase growth.
Activation of σE is growth phase dependent
Although the abundance of OmpU is fairly consistent throughout all phases of V. cholerae growth, accumulation and activation of σE is largely limited to late stationary phase (Figure 4). Cells contain low amounts of σE throughout log phase, and σE activity in overnight cultures is at least 10-fold higher than in log phase cultures (Figure 4). Similar growth phase-dependent activation in E. coli has been attributed to production of ppGpp during stationary phase (8), and ppGpp may account for this activation in V. cholerae as well. It should be noted, however, that activation by OmpU is not limited to stationary phase; diminished ompU expression resulted in reduced activity (relative to the wt strain) in all phases of growth (Figure 4A).
ompU mutations modulate OmpT levels
All of the V. cholerae rpoE mutants contained higher levels of OmpA and OmpT than were detected in wt V. cholerae (Figure 2B). However, unlike the case for OmpA, the degree to which OmpT abundance increased was not uniform among these mutants. OmpT levels were only slightly increased in rpoE mutants that lacked ompU-linked suppressor mutations, while they were markedly higher in rpoE mutants that also contained ompU-linked mutations (Figures 2B and 3). These data suggest that σE–mediated control of OmpT accumulation [potentially due to σE–dependent expression of a regulatory sRNA, as seen for V. cholerae OmpA and several E. coli OMPs, (33,34)] is relatively minor. Furthermore, they suggest that ompU may regulate accumulation of OmpT in a σE–independent fashion. Notably, even relatively slight reductions in OmpU had a more significant effect on OmpT accumulation than did disruption of rpoE in a strain with normal OmpU production (Figure 3, compare strains BD2096 and BD1965).
OMPs other than OmpU are not major determinants of σE abundance and activity in V. cholerae
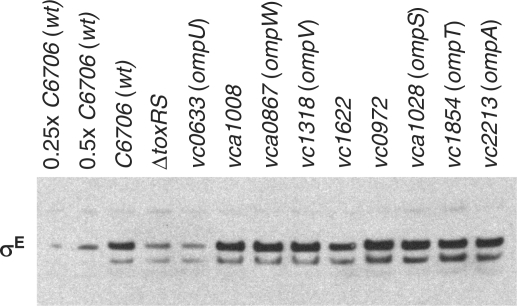
Western blot and rpoE::lacZ transcription reporter analyses of a panel of V. cholerae OMP mutants suggest that OmpU plays a privileged role with respect to σE levels in V. cholerae. Strains with mutations in a variety of OMP-encoding genes were obtained from a comprehensive transposon insertion library of V. cholerae mutants (20) and monitored for production of σE. Disruption of ompU or of toxRS, key activators of ompU expression, consistently reduced the abundance of σE 2- to 4-fold. In contrast, we found that disruption of genes encoding OMPs other than OmpU generally did not influence σE abundance, although disruption of ompA and vc1622 did slightly reduce σE levels in a subset of experiments (Figure 5 and data not shown). The cause of inconsistent results with these two mutants is not known. We also monitored the activity of a σE-dependent rpoE::lacZ transcription reporter fusion in a subset of the OMP mutants (those in which excision of the lacZ-encoding transposon resulted in production of a truncated, rather than full length, OMP). Among this set—which included strains with mutations in ompA, vc1622, ompU and four additional OMP-encoding genes—the activity of the transcription reporter was reduced by more than 2× only in the ompU mutant (data not shown). Together, these data suggest that, at least for the growth conditions assayed, OmpU is the principal OMP underlying σE induction (Figure 5). Deletion of several OMP-encoding genes (ompT, vc0972) also did not reduce V. cholerae's requirement for σE; rpoE::aph mutants could not be obtained in strains lacking these genes (Table 1).
Figure 5.
Production of σE by V. cholerae C6706 and transposon insertion mutants derived from it. The western blot was probed with polyclonal antisera to σE. Extracts electrophoresed on NuPAGE gels were generated from cells cultured overnight in LB. Culture volumes used for extracts were adjusted based on culture OD600 to ensure equal loading. Disrupted genes encode known and putative outer membrane proteins and porins, plus the positive regulator (ToxRS) of OmpU production. The first two lanes (0.25 × C6706 and 0.5 × C6706) were loaded with reduced (1/4 and 1/2) volumes of the wt sample to enable approximate quantitation of signal intensities.
Not all vca0199 mutations suppress V. cholerae’s requirement for rpoE
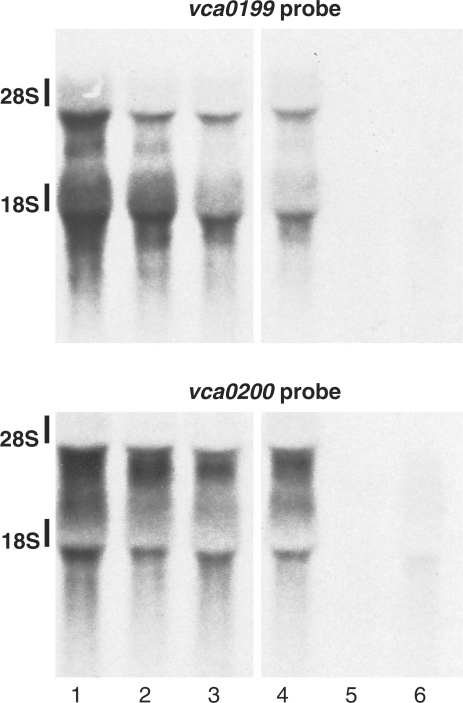
As noted above, a mutation within vca0199 appears to suppress V. cholerae’s requirement for σE by a different mechanism than does inhibition of OmpU production, since the vca0199 mutation increases, rather than diminishes, σE activity (Figure 4A). This effect was detectable both in log and stationary phase cultures. vca0199 does not appear to be a member of the σE regulon, based on Northern (data not shown) and previously published microarray analyses (17). Northern blotting also revealed that the suppressor mutation within vca0199 may cause a small reduction in its transcript and that of the nearby downstream gene, vca0200, although such a reduction was not evident in all experiments (Figure 6). vca0199 and vca0200 are probably cotranscribed, since an insertion mutation within vca0199 eliminates transcripts of both genes (Figure 6). We were unable to detect expression of chromosome-encoded epitope-tagged Vca0199 and Vca0200, suggesting that these proteins are probably produced at fairly low levels. However, cell fractionation studies performed using plasmid-encoded Vca0199 with a C-terminal His6 tag indicate that the product of vca0199 is probably a cytoplasmic protein (data not shown).
Figure 6.
Northern blots probed for transcripts of vca0199 and vca0200. Lanes 1–3 contain RNA from strains derived from N16961; lanes 4–6 contain RNA from strains derived from C6706. Lane 1, wt N16961; lane 2, BD2048 (vca0199 point mutation); lane 3, BD1965 (vca0199 point mutation and rpoE insertion); lane 4, wt C6706; lane 5, BD2034 (vca0199 insertion, KnR); lane 6, BD2041 (vca0199 insertion, KnS).
No function has yet been described for vca0199 or vca0200, and their putative protein products have strong homology only to hypothetical proteins. The closest homologs are in distantly related organisms, including Bacteriodes fragilis, Listeria innocua and Trichodesmium erythraeum; homologs are absent from vibrios other than V. cholerae and from most other gamma-proteobacteria as well, suggesting that these genes were probably acquired relatively recently via horizontal gene transfer. The vca0199 mutation detected by high-throughput sequencing is predicted to result in substitution of a Y for an S at amino-acid 594 of 638, a region with minimal conservation among vca0199 homologs. Interestingly, Sanger sequencing of vca0199 from other rpoE mutants lacking ompU-linked suppressors identified an additional vc0199 mutation, predicted to change P616L (in strain BD1957).
Since the effects on protein function of the vca0199 point mutations described above were not known, we assessed whether additional mutations in vca0199 could also suppress V. cholerae’s requirement for σE. Unexpectedly, these assays gave somewhat ambiguous results. An insertion mutation in vca0199, which eliminates expression of both vca0199 and vca0200, did not enable replacement of rpoE by rpoE::aph (Table 1; strain BD2041). In contrast, rpoE::aph mutants could be obtained when vca0199 contained an internal deletion, which should not alter expression of vca0200 (Table 1, strain BD2139). However, the rpoE mutants generated in the latter background grew very slowly; colonies were significantly smaller than those obtained in the presence of the vca0199 point mutation. Furthermore, mutants were obtained at a much lower frequency when vca0199 contained an internal deletion rather than the point mutation. Together, these data suggest that loss of Vca0199 function may facilitate growth of rpoE mutants, but only in the presence of Vca0200, and that the consequences of the point mutation are not simply a loss of protein function. Whether the SNP in vca0199 partially preserves wt protein function and/or results in a novel function remains to be determined.
DISCUSSION
We have assessed V. cholerae’s requirement for σE and found that it is essential for growth under some but not all selective conditions. Under permissive conditions for disruption of rpoE, mutants could readily be obtained but their initial growth was slow. More rapidly growing variants, which arose frequently from the initial isolates, were found to contain additional mutations at unlinked sites. Introduction of these unlinked mutations into wt V. cholerae enabled selection of rpoE mutants under otherwise non-permissive conditions, confirming that these mutations suppress V. cholerae’s need for σE. The majority of suppressor mutations reduced production of OmpU, a general diffusion porin that constitutes a significant portion of V. cholerae’s outer membrane protein (35,36). The basal level of σE declined in strains with ompU-linked mutations, presumably reflecting the diminished role for rpoE that rendered it non-essential. However, mutations within vca0199, which encodes a gene of unknown function, also were found to suppress V. cholerae’s need for rpoE. Unlike the ompU-linked suppressors and suppressors of rpoE essentiality identified in E. coli (13,37), suppressor mutations within vca0199 augmented, rather than reduced, σE activity, suggesting that they diminish the requirement for σE through a distinct mechanism, perhaps by influencing a process downstream of σE activation. High-throughput genomic sequencing was a crucial tool in the identification of suppressor mutations, particularly the latter class, which could not have been predicted based on previous knowledge of σE’s role and regulation in V. cholerae. The availability of this technology, which continues to improve, should greatly facilitate similar studies in other organisms.
We previously observed that deletion of ompU reduced V. cholerae’s production of σE; however, the importance of this finding was not recognized at the time (18). Our new data highlight the central role played by OmpU as a regulator of σE; indeed, they suggest that one of σE’s most critical roles in V. cholerae may be to respond to this endogenously generated factor. Even relatively small reductions in OmpU levels have a dramatic effect on the organism's need for, and production of, this sigma factor. It appears a threshold level of OmpU must be present to generate a requirement for σE; simply reducing ompU promoter activity by ∼60% renders rpoE inessential, even though OmpU is still readily detectable. It is also notable that OmpU has a significant influence on activity both during log phase, when much OmpU must be synthesized, and during stationary phase, when OmpU transcript levels are quite low (data not shown). The latter observation raises the possibility that OmpU’s activating stimulus is transmitted from previously synthesized, rather than newly generated, protein. Factors may accumulate in the culture medium by late stationary phase that trigger membrane and OMP disruption and thereby reveal the activating motifs that are masked within properly folded proteins (4). Alternatively, exposure of activating domains within OmpU may be coupled to peptidoglycan remodeling during stationary phase, since OmpU has been reported to be associated with this polymer (36). It is also theoretically possible that the σE-activating stimulus is not due to OmpU misfolding but instead is a result of OmpU’s porin activity, which might enhance transmission of a signal from the culture media into the periplasmic compartment.
Our study reveals both significant similarities and differences between the role and regulation of σE in V. cholerae and E. coli. For both organisms, rpoE is absolutely essential under a subset of conditions tested, and no mutants can be derived from wt cells under these conditions (10). Under less stringent selection conditions, mutants can be isolated; however, they have been found to contain additional suppressor mutations. Such suppressor mutations also permit disruption of rpoE under the otherwise non-permissive selective conditions (14,38). Deletion of OMP-encoding genes reduces the activity of σE in both V. cholerae and E. coli. σE is also a negative regulator of OMP production in both organisms, although it should be noted that σE does not appear to influence V. cholerae’s production of its principal OMP, OmpU, since an rpoE mutant that lacks an ompU-linked suppressor produces wt amounts of this protein (Figure 3), and overexpression of σE also did not alter OmpU production (data not shown). A key difference between the species is that no single deletion or combination of deletions of OMP-encoding genes has yet rendered rpoE non-essential in E. coli (14), whereas the absence of a single V. cholerae OMP is sufficient to do so. Thus, it appears that the pathways for activation of σE in V. cholerae are more limited, and that monitoring and responding to OmpU (mis)folding is of paramount importance. Given that significant activation of σE is still detectable in stationary phase cultures of a strain with diminished OmpU, it is quite striking that rpoE is no longer essential in these cells.
Our work provides new evidence that bacteria have developed numerous safeguards to coordinate and balance expression of their OMPs. Excessive OMP accumulation was already known to trigger σE activity and the resulting transcription of sRNAs that downregulate OMP translation (33,38,39). In addition, it has been noted that OMP-encoding sequences are often adjacent to genes encoding negative regulators of other OMPs, suggesting that new OMP-encoding genes can more readily be acquired in conjunction with means for downregulating those that are already present (40). In this study, we observed that expression of OmpU has a significant negative effect on production of OmpT. It has long been recognized that the genes encoding these proteins respond in opposite fashions to the transcriptional regulator ToxR, and thus that their expression is coordinated (21,32,41); however, disruption of ompU has not previously been reported to augment OmpT production. It remains to be seen whether this interdependence of OmpU and OmpT production is mediated by OmpU’s porin function, by an sRNA that overlaps with or is coordinated with the ompU transcript, or by some alternate mechanism.
Finally, our identification of a panel of suppressor mutations provides clues about the mechanisms by which such suppressors may arise. Two aspects of these suppressor mutations are particularly striking. First, the same suppressor mutations were generated from distinct selection protocols (i.e. growth in the presence of antibiotics versus sucrose), suggesting that the process for suppressor formation is not determined solely by the selection used. Second, most suppressor mutations (11 of 21) were single nucleotide contractions of mononucleotide (A/T) repeats. In E. coli, such mutations are characteristic of the error-prone polymerase DinB, which is induced by specific stresses and is associated with temporary formation of a hypermutable state (42). Although mutation as a stress response has yet to be investigated in V. cholerae, our data are consistent with the possibility that formation of rpoE suppressor mutations may be facilitated by such a response. Transient emergence of a mutator phenotype may thus underlie the high frequency with which suppressors arise (e.g. Figure 1). Such a possibility could be explored by assessing whether factors often needed for formation of stress-linked mutations (e.g. DinB, RpoS and the SOS response) are also required for development of rpoE suppressor mutations. Additionally, blocking these processes might permit characterization of the true consequences of rpoE disruption, unmodulated by additional genomic changes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Howard Hughes Medical Institute; National Institutes of Health [AI-42347 to M.K.W.]. Funding for open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank H. Li, M. Chase, and J. Livny for helpful discussions regarding sequence analysis, A. C. Matin, S. Lynch, P. Nemani, and J. Kaper for providing antisera, and D. E. Cameron, J. J. Mekalanos and K. Klose for strains and plasmids.
REFERENCES
- 1.Ruiz N, Silhavy TJ. Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr. Opin. Microbiol. 2005;8:122–126. doi: 10.1016/j.mib.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Missiakas D, Mayer MP, Lemaire M, Georgopoulos C, Raina S. Modulation of the Escherichia coli σE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol. 1997;24:355–371. doi: 10.1046/j.1365-2958.1997.3601713.x. [DOI] [PubMed] [Google Scholar]
- 3.De Las Penas A, Connolly L, Gross CA. The σE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of sigmaE. Mol. Microbiol. 1997;24:373–385. doi: 10.1046/j.1365-2958.1997.3611718.x. [DOI] [PubMed] [Google Scholar]
- 4.Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 5.Dartigalongue C, Missiakas D, Raina S. Characterization of the Escherichia coli σE regulon. J. Biol. Chem. 2001;276:20866–20875. doi: 10.1074/jbc.M100464200. [DOI] [PubMed] [Google Scholar]
- 6.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 2006;4:e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valentin-Hansen P, Johansen J, Rasmussen AA. Small RNAs controlling outer membrane porins. Curr. Opin. Microbiol. 2007;10:152–155. doi: 10.1016/j.mib.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Costanzo A, Ades SE. Growth phase-dependent regulation of the extracytoplasmic stress factor, σE, by guanosine 3′,5′-bispyrophosphate (ppGpp) J. Bacteriol. 2006;188:4627–4634. doi: 10.1128/JB.01981-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costanzo A, Nicoloff H, Barchinger SE, Banta AB, Gourse RL, Ades SE. ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor σE in Escherichia coli by both direct and indirect mechanisms. Mol. Microbiol. 2008;67:619–632. doi: 10.1111/j.1365-2958.2007.06072.x. [DOI] [PubMed] [Google Scholar]
- 10.De Las Penas A, Connolly L, Gross CA. σE is an essential sigma factor in Escherichia coli. J. Bacteriol. 1997;179:6862–6864. doi: 10.1128/jb.179.21.6862-6864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heusipp G, Schmidt MA, Miller VL. Identification of rpoE and nadB as host responsive elements of Yersinia enterocolitica. FEMS Microbiol. Lett. 2003;226:291–298. doi: 10.1016/S0378-1097(03)00613-X. [DOI] [PubMed] [Google Scholar]
- 12.Button JE, Silhavy TJ, Ruiz N. A suppressor of cell death caused by the loss of sigmaE downregulates extracytoplasmic stress responses and outer membrane vesicle production in Escherichia coli. J. Bacteriol. 2007;189:1523–1530. doi: 10.1128/JB.01534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayden JD, Ades SE. The extracytoplasmic stress factor, σE, is required to maintain cell envelope integrity in Escherichia coli. PLoS ONE. 2008;3:e1573. doi: 10.1371/journal.pone.0001573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Douchin V, Bohn C, Bouloc P. Down-regulation of porins by a small RNA bypasses the essentiality of the regulated intramembrane proteolysis protease RseP in Escherichia coli. J. Biol. Chem. 2006;281:12253–12259. doi: 10.1074/jbc.M600819200. [DOI] [PubMed] [Google Scholar]
- 15.Mathur J, Waldor MK. The Vibrio cholerae ToxR-regulated porin OmpU confers resistance to antimicrobial peptides. Infect. Immun. 2004;72:3577–3583. doi: 10.1128/IAI.72.6.3577-3583.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kovacikova G, Skorupski K. The alternative sigma factorσE plays an important role in intestinal survival and virulence in Vibrio cholerae. Infect. Immun. 2002;70:5355–5362. doi: 10.1128/IAI.70.10.5355-5362.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding Y, Davis BM, Waldor MK. Hfq is essential for Vibrio cholerae virulence and downregulates σE expression. Mol. Microbiol. 2004;53:345–354. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]
- 18.Mathur J, Davis BM, Waldor MK. Antimicrobial peptides activate the Vibrio cholerae σE regulon through an OmpU-dependent signalling pathway. Mol. Microbiol. 2007;63:848–858. doi: 10.1111/j.1365-2958.2006.05544.x. [DOI] [PubMed] [Google Scholar]
- 19.Srivatsan A, Han Y, Peng J, Tehranchi AK, Gibbs R, Wang JD, Chen R. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 2008;4:e1000139. doi: 10.1371/journal.pgen.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cameron DE, Urbach JM, Mekalanos JJ. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc. Natl Acad. Sci. USA. 2008;105:8736–8741. doi: 10.1073/pnas.0803281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis BM, Waldor MK. RNase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA. Mol. Microbiol. 2007;65:373–385. doi: 10.1111/j.1365-2958.2007.05796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donnenberg MS, Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Provenzano D, Lauriano CM, Klose KE. Characterization of the role of the ToxR-modulated outer membrane porins OmpU and OmpT in Vibrio cholerae virulence. J. Bacteriol. 2001;183:3652–3662. doi: 10.1128/JB.183.12.3652-3662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sperandio V, Bailey C, Giron JA, DiRita VJ, Silveira WD, Vettore AL, Kaper JB. Cloning and characterization of the gene encoding the OmpU outer membrane protein of Vibrio cholerae. Infect. Immun. 1996;64:5406–5409. doi: 10.1128/iai.64.12.5406-5409.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider K, Beck CF. Promoter-probe vectors for the analysis of divergently arranged promoters. Gene. 1986;42:37–48. doi: 10.1016/0378-1119(86)90148-4. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernandez D, Francois P, Farinelli L, Osteras M, Schrenzel J. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res. 2008;18:802–809. doi: 10.1101/gr.072033.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller JH. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia Coli and Related Bacteria. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1992. [Google Scholar]
- 31.Feng L, Reeves PR, Lan R, Ren Y, Gao C, Zhou Z, Cheng J, Wang W, Wang J, Qian W, et al. A recalibrated molecular clock and independent origins for the cholera pandemic clones. PLoS ONE. 2008;3:e4053. doi: 10.1371/journal.pone.0004053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crawford JA, Kaper JB, DiRita VJ. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol. Microbiol. 1998;29:235–246. doi: 10.1046/j.1365-2958.1998.00925.x. [DOI] [PubMed] [Google Scholar]
- 33.Johansen J, Rasmussen AA, Overgaard M, Valentin-Hansen P. Conserved small non-coding RNAs that belong to the σE regulon: role in down-regulation of outer membrane proteins. J. Mol. Biol. 2006;364:1–8. doi: 10.1016/j.jmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Song T, Mika F, Lindmark B, Liu Z, Schild S, Bishop A, Zhu J, Camilli A, Johansson J, Vogel J, et al. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol. Microbiol. 2008;70:100–111. doi: 10.1111/j.1365-2958.2008.06392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simonet VC, Basle A, Klose KE, Delcour AH. The Vibrio cholerae porins OmpU and OmpT have distinct channel properties. J. Biol. Chem. 2003;278:17539–17545. doi: 10.1074/jbc.M301202200. [DOI] [PubMed] [Google Scholar]
- 36.Chakrabarti SR, Chaudhuri K, Sen K, Das J. Porins of Vibrio cholerae: purification and characterization of OmpU. J. Bacteriol. 1996;178:524–530. doi: 10.1128/jb.178.2.524-530.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Button JE, Silhavy TJ, Ruiz N. A suppressor of cell death caused by the loss of σE downregulates extracytoplasmic stress responses and outer membrane vesicle production in Escherichia coli. J. Bacteriol. 2007;189:1523–1530. doi: 10.1128/JB.01534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mecsas J, Rouviere PE, Erickson JW, Donohue TJ, Gross CA. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 1993;7:2618–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- 39.Figueroa-Bossi N, Lemire S, Maloriol D, Balbontin R, Casadesus J, Bossi L. Loss of Hfq activates the sigmaE-dependent envelope stress response in Salmonella enterica. Mol. Microbiol. 2006;62:838–852. doi: 10.1111/j.1365-2958.2006.05413.x. [DOI] [PubMed] [Google Scholar]
- 40.Guillier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev. 2006;20:2338–2348. doi: 10.1101/gad.1457506. [DOI] [PubMed] [Google Scholar]
- 41.Li CC, Crawford JA, DiRita VJ, Kaper JB. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in Vibrio cholerae. Mol. Microbiol. 2000;35:189–203. doi: 10.1046/j.1365-2958.2000.01699.x. [DOI] [PubMed] [Google Scholar]
- 42.Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.