Abstract
microRNA-155 (miR-155) has been implicated as a central regulator of the immune system, but its function during acute inflammatory responses is still poorly understood. Here we show that exposure of cultured macrophages and mice to lipopolysaccharide (LPS) leads to up-regulation of miR-155 and that the transcription factor c/ebp Beta is a direct target of miR-155. Interestingly, expression profiling of LPS-stimulated macrophages combined with overexpression and silencing of miR-155 in murine macrophages and human monocytic cells uncovered marked changes in the expression of granulocyte colony-stimulating factor (G-CSF), a central regulator of granulopoiesis during inflammatory responses. Consistent with these data, we show that silencing of miR-155 in LPS-treated mice by systemically administered LNA-antimiR results in derepression of the c/ebp Beta isoforms and down-regulation of G-CSF expression in mouse splenocytes. Finally, we report for the first time on miR-155 silencing in vivo in a mouse inflammation model, which underscores the potential of miR-155 antagonists in the development of novel therapeutics for treatment of chronic inflammatory diseases.
INTRODUCTION
MicroRNAs (miRNAs) are an abundant class of small endogenous noncoding RNAs that post-transcriptionally regulate gene expression by base-pairing to imperfect complementary target sites in the 3′ UTR of their target mRNAs, thereby mediating mRNA cleavage or translational repression (1). miRNAs have emerged as key regulators of diverse biological processes, including differentiation of hematopoeitic cells (2,3) and activation of the innate immune response (4,5). miR-155 is expressed in activated mature B and T lymphocytes (6,7) and in activated monocytes (8,9), while studies using miR-155 knockout mice have directly linked this miRNA to the functions of the immune system (6,7). In addition, miR-155 has been shown to regulate the production of cytokines, chemokines and transcription factors (6,7) and to be induced by endotoxins, such as bacterial lipopolysaccharide (LPS) (8–10). Recently, miR-155 was shown to direct the generation of immunoglobulin class-switched plasma cells (11) and to directly regulate the AID enzyme (activation-induced cytidine deaminase), which is responsible for the generation of functionally diverse antibody repertoires (12,13). Altogether, these observations strongly imply miR-155 as a central regulator of the immune system.
With the goal of gaining further insights into the biological function of miR-155 during acute inflammatory response, we have undertaken stimulation of cultured murine macrophage Raw264.7 and human THP-1 monocytic cells as well as treatment of mice by bacterial LPS. Previous studies have shown that the transcription factor CCAAT/enhancer binding protein Beta (c/ebp Beta) is induced in monocytes and macrophages by LPS (14) and that c/ebp Beta is involved in the regulation of proinflammatory cytokines as well as other genes associated with macrophage activation and the acute phase response (15,16). We report here that LPS treatment induces miR-155 expression in cultured mouse macrophages and in mouse splenocytes in vivo and that silencing of miR-155 leads to derepression of its direct target c/ebp Beta in vitro and in vivo. Additionally, we find that antagonism of miR-155 leads to down-regulation of G-CSF, a regulator of granulopoiesis, produced by activated macrophages during acute inflammatory responses. G-CSF belongs to the class of colony-stimulating factors (CSFs), which comprise granulocyte/macrophage CSF (GM-CSF), macrophage CSF (M-CSF) and granulocyte CSF (G-CSF). During progression of inflammatory diseases the CSFs have been suggested to constitute a pro-inflammatory CSF network, which at the site of inflammation involves CSF-mediated communication between stimulated macrophages and neighboring cell types (17). This positive-feedback loop model predicts that CSF blocking agents could be efficient anti-inflammatory drugs. Indeed, data from animal models indicate that depletion of CSFs has therapeutic value in inflammatory and autoimmune diseases. While early-phase clinical trials targeting GM-CSF and M-CSF have been initiated, no clinical trials on depletion of G-CSF have yet been reported (18).
Our data show for the first time that c/ebp Beta is regulated by miR-155 in vivo in mouse splenocytes during inflammatory responses. Moreover, we report that miR-155 mediates regulation of G-CSF expression, thereby underscoring the role of miR-155 in fine-tuning important regulatory networks during inflammation.
MATERIALS AND METHODS
Design and synthesis of LNA oligonucleotides
The LNA-antimiR oligonucleotides were synthesized as unconjugated and fully phosphorothiolated oligonucleotides. The perfectly matching LNA-antimiR oligonucleotide 5′-TcAcAATtaGmCAtTA-3′ was complementary to nucleotides 2–16 in the mature murine miR-155 sequence and 5′-TcAcGATtaGmCAtTA-3′ was complementary to nucleotides 2–16 in the mature human miR-155 sequence. The mismatch LNA control oligonucleotide was synthesized with the following sequence: 5′-TcAamCATtaGAmCtTA-3′ (uppercase: LNA; lowercase: DNA; mC denotes LNA methylcytosine).
Cell culture
Raw264.7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% FBS, 4 mM Glutamax I and 25 µg/ml Gentamicin (Invitrogen). Lipopolysaccharide (LPS) was purchased from Sigma and activation of Raw264.7 cells was induced by treating cells with 1, 10 or 100 ng/ml LPS for indicated time periods. THP-1 cells were grown in RPMI-1640 (Invitrogen) supplemented with 10% FBS, 4 mM Glutamax I and 25 µg/ml Gentamicin (Invitrogen). Raw264.7 and THP-1 cells were transfected with the Lipofectamine 2000 transfection reagent according to the manufacturer's protocol (Invitrogen) and the LNA-antimiR oligonucleotides were used at a final concentration of 5 nM unless otherwise stated. Human miR-155 precursor (pre-miR-155, Ambion) was cotransfected at a final concentration of 5 nM. HeLa cells were cultivated in Eagles MEM (Invitrogen) with 10% FBS, 2 mM Glutamax I, non-essential amino acids and 25 µg/ml Gentamicin (Invitrogen). HeLa cells were co-transfected with human pre-miR-155 (Ambion) at a final concentration of 50 nM and 0.1 µg luciferase reporter construct using Lipofectamine 2000. The transfections and luciferase activity measurements were carried out according to the manufacturer's instructions (Invitrogen Lipofectamine 2000/Promega Dual-luciferase kit). Relative luciferase activity levels were expressed as Renilla/firefly luciferase ratios.
Plasmids
The perfect match target sites for the human and the murine miR-155, respectively, were cloned downstream of the Renilla luciferase gene (XhoI/NotI sites) in the psiCHECK2 vector (Promega) using 5′ phosphorylated oligos: murine miR-155 forward 5′-tcgagcccctatcacaattagcattaagc-3′, and reverse 5′-ggccgcttaatgctaattgtgataggggc-3′; human miR-155 forward 5′-tcgagcccctatcacgattagcattaagc-3′ and reverse 5′-ggccgcttaatgctaatcgtgatagggg-3′. The 3′ UTR of human c/ebp Beta was cloned downstream of the Renilla luciferase gene (XhoI/NotI sites) in the psiCHECK2 vector. PCR primers used for amplification of the human c/ebp Beta 3′ UTR (basepairs 1328–1837 accession no. NM_005194) were: forward 5′-aaaaaactcgagaaaactttggcactggggca-3′ (incl. a XhoI site), reverse 5′-aaaaaagcggccgcggctttgtaaccattctcaaa-3′ (incl. a NotI site). The miR-155 target site in the c/ebp Beta 3′ UTR was mutated by deleting the 8 nt miR-155 seed match sequence (AGCAUUAA at nucleotide positions 554–561 in the c/ebp Beta 3′ UTR) using the QuikChange Site-Directed Mutagenesis kit according to manufacturer's instructions (Stratagene). The pCDNA3.1 expression construct for the truncated rat c/ebp Beta isoform LIP (amino acids 153–297) was kindly provided by Dr. M.A. Chidgey and has been described elsewhere (19).
Western blot analysis
Raw264.7 and spleen proteins were extracted using RIPA lysis buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate) and electrophoresed on NuPAGE Bis Tris 4–12% gels (Invitrogen) using 100 µg protein per sample. The proteins were transferred to a nitrocellulose membrane using iBlot (Invitrogen) according to manufacturer's instructions. ECL advanced western kit (GE Healthcare Life Sciences) was used for blocking, antibody dilution and detection according to the manufacturer. A primary monoclonal mouse-anti-c/ebp Beta antibody (SC-7962, Santa Cruz Biotechnology), a primary rabbit-anti-PU.1 (#2258, Cell signaling Technology), a primary mouse-anti- Tubulin-alpha Ab-2 (MS-581-P, Neomarkers) and HRP-conjugated secondary rabbit anti-mouse (P0447, DAKO) or swine anti-rabbit antibodies (P0399, DAKO) were used according to the manufacturer's instructions.
Quantitative RT-PCR
The dissected mice spleens were immediately stored in RNA later (Ambion). Total RNA from spleens or Raw264.7 cells was extracted with Trizol reagent according to the manufacturer's instructions (Invitrogen), except that the precipitated RNA pellet was washed in 80% ethanol and not mixed. The miR-155 levels were quantified using the mirVana real-time RT-PCR detection kit (Ambion) following the manufacturer's instructions, except that 200 ng total RNA was used in the reverse transcription (RT) reaction. A 2-fold total RNA dilution series from LPS-treated mouse spleen RNA or LPS-treated Raw264.7 RNA served as standard to ensure a linear range (Ct versus relative copy number) of the amplification. The RT reaction was diluted ten times in water and 10 µl aliquots were subsequently used for RT-PCR amplification according to the manufacturer's instructions (Ambion). mRNA quantification of selected genes was done using standard TaqMan assays (Applied Biosystems). The reverse transcription reaction was carried out with random decamers, 0.5 μg total RNA and the M-MLV RT enzyme from Ambion according to protocol. First-strand cDNA was subsequently diluted 10 times in nuclease-free water before addition to the RT-PCR reaction mixture. The Applied Biosystems 7500 Real-Time PCR instrument was used for amplification, except that the Applied Biosystems 7900 Real-Time PCR instrument was used for amplification of 96 transcripts included on the low density mouse immune array from Applied Biosystems.
In vivo experiments
C57BL/6J female mice (Taconic Europe A/S) kept on a regular chow diet (Altromin no 1324, Brogaarden) and with an average body weight of ∼20 g (±2 g) at the start of the experiment were used in all studies. The experiments were carried out following the Danish Committee for Animal Experiments guidelines. The animal cages were illuminated to give a 12-h light/dark cycle. The temperature was 21°C ± 2°C and relative humidity 55 ± 10%. The LNA compounds were formulated in physiological saline (0.9% NaCl) to a final concentration allowing the mice to receive a tail vein injection volume of 10 ml/kg. The animals were dosed for three consecutive days with LNA-antimiR, LNA control or saline (vehicle control), receiving daily doses of 25 mg/kg and sacrificed 24 h after last dose. Saline-formulated bacterial LPS was administered by intraperitoneal injections at 0.5 mg/kg and the mice were sacrificed either 2 or 24 h post-LPS treatment. Immediately after sacrificing the animals, spleen samples were dissected.
Isolation of B cells and monocytic/macrophage cell fractions from mice splenocytes
C57BL/6J female mice (Taconic Europe A/S) were injected intravenously with a FAM-labeled LNA control for three consecutive days, receiving daily doses of 25 mg/kg and the animals were sacrificed 24 h after last dose. Spleens were surgically removed and positive selection of monocytes/macrophages was carried out by MACS® Cell separation systems (Miltenyi Biotech) using magnetic beads conjugated with CD11b antibodies and the MACS® Cell separation columns according to the manufacturer's instructions (Miltenyi Biotec). B cells were isolated using magnetic beads conjugated with CD19 antibodies (Miltenyi Biotec). PE-conjugated CD11b and CD19 antibodies were added to isolated fractions to verify the identity of the isolated cells by FACS analysis. Fixed isolated cells were DAPI stained and transferred to microscope slides. Cellular uptake of the FAM-labeled LNA oligonucleotide was investigated by confocal microscopy.
RESULTS
LPS-mediated induction of miR-155 in cultured mouse Raw264.7 macrophages
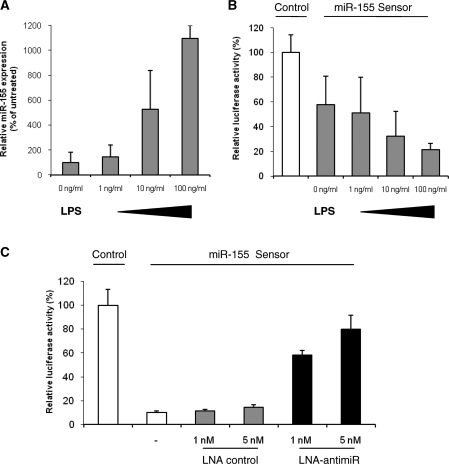
Since miR-155 has previously been shown to be upregulated during macrophage activation (8–10) we first investigated the expression of miR-155 in murine Raw264.7 macrophage cells upon LPS stimulation. Treatment of cultured mouse macrophages with LPS showed dose-dependent induction of miR-155 with more than 10-fold increase in miR-155 expression levels at a concentration of 100 ng/ml LPS after 18 h (Figure 1A). Consistent with these data, a luciferase reporter construct harbouring a perfect match miR-155 target site in the 3′ UTR of the Renilla luciferase gene showed a concentration-dependent repression of the luciferase reporter, which correlated with the increased expression of miR-155 in LPS-treated mouse Raw264.7 macrophages (Figure 1B). We have recently reported on effective miRNA silencing using complementary LNA-antimiR oligonucleotides in combination with transcriptome analysis as a useful approach to dissect the biological roles of individual miRNAs in vitro and in vivo (20,21). Hence, to enable further studies on miR-155 targets and miR-155 associated gene networks, we designed an LNA-antimiR complementary to mature miR-155 alongside an LNA control oligonucleotide having four mismatches in the miR-155 recognition sequence. The LNA-antimiR-155 showed concentration-dependent silencing of miR-155 in LPS-treated mouse Raw264.7 macrophages as shown by efficient derepression of the miR-155 sensor, whereas the LNA control oligonucleotide had no effect on the luciferase reporter activity at the same concentrations (Figure 1C). Since the LNA-antimiR resulted in potent and specific antagonism of miR-155 in cultured Raw264.7 cells at 5-nM concentration, we chose these experimental conditions for further studies in mouse macrophages.
Figure 1.
LPS-mediated induction of miR-155 in cultured mouse Raw264.7 macrophages. (A) Quantitative RT-PCR analysis of miR-155 expression in Raw264.7 cells stimulated with LPS for 18 h at the indicated concentrations. Values represent mean ± SD (n = 3). (B) Assessment of luciferase reporter activity in Raw264.7 cells transfected with either the Renilla/firefly luciferase psiCHECK2 control or psiCHECK2 harbouring the miR-155 perfect match target site in the 3′ UTR of the Renilla luciferase transcript (miR-155 sensor). After transfection, the Raw264.7 cells were stimulated with the indicated concentrations of LPS for 18 h. Values represent mean ± SD (n = 3). (C) Assessment of luciferase reporter activity in Raw264.7 cells cotransfected with LNA-antimiR or LNA control in combination with either the Renilla/firefly luciferase psiCHECK2 control (data not shown) or the miR-155 sensor. Raw264.7 cells were stimulated with 100 ng/ml LPS for 18 h. Values represent mean ± SD (n = 3).
Translational repression of c/ebp Beta isoforms by miR-155
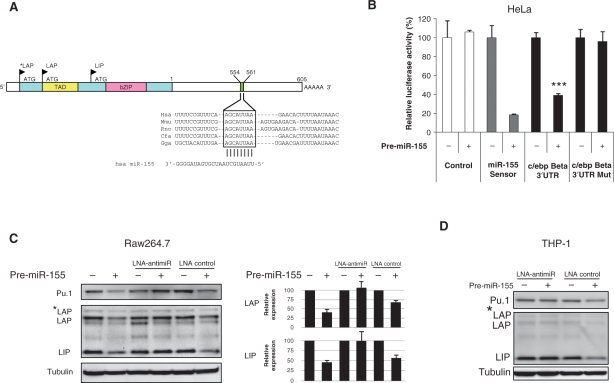
LPS stimulation of cultured mouse Raw264.7 cells leads to an inflammatory response in which miR-155 and more than 700 genes are up-regulated (22), including the transcription factor c/ebp Beta gene (14). Interestingly, the c/ebp Beta transcript is predicted by several computer algorithms (PicTar, miRanda, TargetScan) to be a miR-155 target due to the presence of a miR-155 target site in its 3′ UTR, which is highly conserved among five vertebrate species (Figure 2A). Indeed, we found that a luciferase reporter harbouring the c/ebp Beta 3′ UTR showed significant (P < 0.001, Student's t-test, two-sided) miR-155-dependent repression relative to a control reporter plasmid (Figure 2B), implying that c/ebp Beta is a direct target of miR-155, which is also consistent with two recent reports (23,24). Deletion of the highly conserved miR-155 seed match site (positions 554–561; AGCAUUAA) in the c/ebp Beta 3′ UTR abrogated the effect of miR-155. Combined these data support the notion that c/ebp Beta is post-transcriptionally regulated by miR-155 (Figure 2B and Supplementary Figure 1).
Figure 2.
Translational repression of c/ebp Beta isoforms by miR-155. (A) Schematic overview of the c/ebp Beta transcript. The miR-155 target site is indicated in the 3′ UTR with the 8-nt seed match boxed. TAD, transactivation domain; bZIP, basic region leucine zipper domain; ATG, translational start site; LAP, liver-enriched transcriptional activation protein; LIP, liver-enriched transcriptional inhibitory protein. (B) Assessment of luciferase reporter activity in HeLa cells cotransfected with pre-miR-155 in combination with either the Renilla/firefly luciferase psiCHECK2 control, the psiCHECK2 luciferase construct (miR-155 sensor) harbouring the miR-155 perfect match target site, the psiCHECK2 construct harbouring the c/ebp Beta 3′ UTR, or the psiCHECK2 c/ebp Beta 3′ UTR Mut construct harbouring a mutated miR-155 target site. Values represent mean ± SD (n = 3). The pre-miR-155 significantly represses the luciferase activity of the c/ebp Beta 3′ UTR sensor construct (*** indicates P < 0.001). (C) Western blot analysis of c/ebp Beta LAP*, LAP and LIP isoforms and Pu.1 in Raw264.7 cell lysates. Protein extracts from Raw264.7 cells cotransfected with 5 nM pre-miR-155 in combination with either LNA control (5 nM) or LNA-antimiR (5 nM) were subjected to western blot analysis. (Right) Quantification of the LIP and LAP protein bands shown on the western blot. Values represent mean ± SD (n = 3). (D) Western blot analysis of the c/ebp Beta LAP and LIP isoforms and Pu.1 protein in THP-1 cell lysates. Protein extracts from THP-1 cells cotransfected with pre-miR-155 (5 nM) in combination with either LNA control (5 nM) or LNA-antimiR (5 nM) were subjected to western blot analysis. Data are representative of three independent experiments.
The c/ebp Beta transcript encodes three isoforms, designated as LAP*, LAP and LIP, that are generated by differential translational initiation (Figure 2A). The short LIP form lacks the transactivation domain (TAD), but is still capable of binding to DNA and forms homo- or heterodimers through the basic region leucine zipper (bZIP) domain and therefore behaves as a dominant negative. The expression of the three isoforms is regulated in a complex manner, and even differential activation of the c/ebp Beta isoforms upon LPS stimulation has been reported (25).
To assess miR-155-mediated regulation of the various c/ebp Beta isoforms, we investigated protein extracts from murine macrophages by Western blot analyses using a C-terminal specific c/ebp Beta antibody that recognizes all three isoforms. Transient transfection of a precursor miR-155 (pre-miR-155) at 5-nM concentration into mouse Raw264.7 macrophages resulted in significant repression of the c/ebp Beta isoforms LIP and LAP, whereas concomitant transfection with equimolar LNA-antimiR into the cells effectively antagonized the miR-155-mediated c/ebp Beta repression compared to the LNA control transfected cells (Figure 2C). We also observed repression of another direct miR-155 target, the Ets family transcription factor Pu.1 in Raw264.7 cells (Figure 2C) (11), implying that both c/ebp Beta and Pu.1 are targeted by miR-155 in the Raw264.7 macrophage model system. We obtained similar results with undifferentiated human monocytic THP-1 cells, in which transfection with pre-miR-155 resulted in repression of the c/ebp Beta isoforms (Figure 2D). Taken together, our data suggest that the levels of individual c/ebp Beta isoforms are regulated by ectopically expressed miR-155.
miR-155 regulates c/ebp Beta in Raw264.7 cells during LPS stimulation
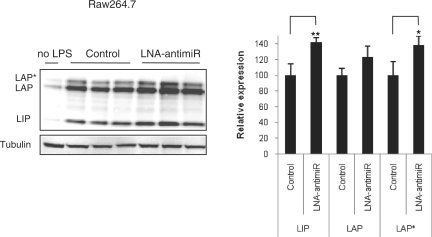
We next investigated the effect of endogenous miR-155 on c/ebp Beta expression during LPS stimulation by treating mouse macrophage Raw264.7 cells with LNA-antimiR followed by exposure of the cells to LPS. Western blot analysis using the C-terminal specific c/ebp Beta antibody showed that the levels of both the LAP and LIP isoforms of c/ebp Beta were increased after treatment with LPS, in accordance with a previous report (14) (Figure 3). Transfection of the LNA-antimiR (5 nM) into the Raw264.7 macrophages prior to LPS stimulation resulted in derepression of the c/ebp Beta isoforms (Figure 3), which thereby provides additional evidence that the levels of individual c/ebp Beta isoforms are regulated by endogenous miR-155 in mouse macrophages during LPS stimulation.
Figure 3.
Regulation of c/ebp Beta isoforms by endogenous miR-155 in Raw264.7 cells during LPS stimulation. Western blot analysis of c/ebp Beta LAP*, LAP and LIP isoforms in Raw264.7 cell lysates. Protein extracts from Raw264.7 cells either mock (control) or LNA-antimiR (5 nM) transfected before treatment with 100 ng/ml LPS for 18 h were subjected to western blot analysis. (Right) Quantification of the LIP, LAP and LAP* protein bands shown on the western blot. Values represent mean ± SD (n = 3) (** indicates P < 0.01; * indicates P < 0.05). Data represent three independent experiments.
miR-155 regulates c/ebp Beta in the splenocytes of LPS-treated mice
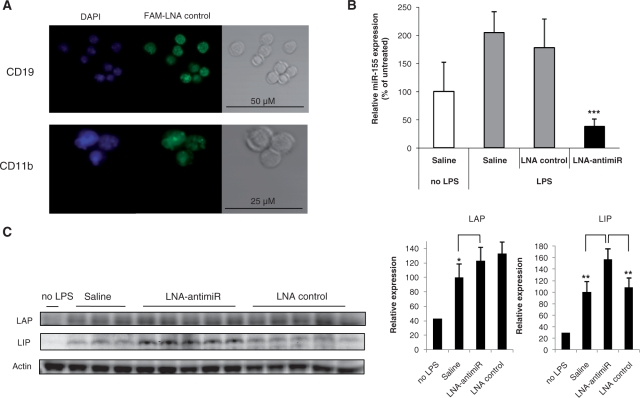
The expression of c/ebp Beta is induced during macrophage activation, whereas Pu.1 is expressed both in resting B cells and macrophages and in the spleen germinal center B cells (14,26). Since spleen contains populations of monocytes and macrophages together with B and T cells, we investigated miR-155-mediated regulation of c/ebp Beta in the splenocytes of LPS-treated mice in combination with LNA-antimiR based miR-155 silencing. We first asked whether these cells could be targeted by systemically administered LNA oligonucleotides in vivo by isolating B cells and monocytic cell populations from murine spleen after intravenous dosing with a 6-carboxyfluorescein (FAM)-labeled LNA oligonucleotide. Confocal microscopy of the murine B cells and monocytes/macrophages showed that the FAM-labeled LNA was readily taken up by these cells, indicating that miR-155 could be targeted in both cell types by an LNA-antimiR compound (Figure 4A). Intraperitoneal administration of LPS significantly induced the expression of miR-155 in splenocytes 2 h post-treatment, thereby corroborating our results obtained with LPS-stimulated murine macrophages (Figure 4B). The splenocytes of untreated mice showed low levels of c/ebp Beta proteins, whereas the levels of both the LAP and LIP isoform of c/ebp Beta were significantly increased 24 h after treatment with LPS (Figure 4C), in accordance with a previous report (14). Systemic administration of the LNA-antimiR in LPS-treated mice effectively antagonized miR-155 compared to the vehicle and LNA-mismatch-control-treated mice, respectively (Figure 4B). This resulted in significant derepression of both the LAP and LIP isoforms in comparison to the vehicle-treated control mice (Figure 4C), which is consistent with the notion that miR-155 negatively regulates c/ebp Beta proteins in vivo in mouse splenocytes during acute inflammatory response.
Figure 4.
miR-155 regulates c/ebp Beta in the splenocytes of LPS-treated mice. (A) Fluorescence-microscopy analysis of LNA oligonucleotide uptake in mouse splenocytes sorted in either B-cell (CD19) or monocytic (CD11b) cell fractions. Mice were dosed intravenously with 25 mg/kg FAM-labeled LNA oligonucleotide for three consecutive days, and spleens were dissected the day after last dose. (B) Quantitative RT-PCR analysis of miR-155 expression in splenocytes of mice treated with LNA-antimiR or LNA control by three intravenous injections of 25 mg/kg over three consecutive days. Spleen samples were dissected 2 h after intraperitoneal injection of 0.5 mg/kg LPS. Values represent mean ± SD (n = 5). The LNA-antimiR compound effectively antagonizes miR-155 expression in LPS-treated mice compared to saline treated controls (*** indicates P < 0.001). (C) Western blot analysis of the c/ebp Beta LAP and LIP isoforms in splenocytes of mice treated with saline (n = 3), LNA-antimiR (n = 5) or LNA control (n = 5) by three intravenous injections of 25 mg/kg over three consecutive days. Spleen samples were dissected 24 h after intraperitoneal injection of 0.5 mg/kg LPS. (Right) Quantification of the LIP and LAP protein bands shown on the western blot. Systemically delivered LNA-antimiR compound derepresses the LIP isoform of c/ebp Beta compared to both saline- and LNA control-treated mice (**P < 0.01). Values represent mean ± SD.
miR-155 mediates regulation of G-CSF
Recent studies using knockout mice have implicated miR-155 in the regulation of cytokines and chemokines in T and B cells (6,7). Thus, to identify immune response genes whose expression could be modulated by miR-155 in activated macrophages, we carried out expression profiling of RNA samples extracted from LPS-stimulated mouse Raw264.7 cells using mouse immune real-time RT-PCR arrays (Supplementary table S1). To establish a link between miR-155 and the expression of immune response genes, the LPS-stimulated Raw264.7 cells were transfected with either LNA-antimiR-155 or LNA control. Interestingly, among the immune response genes induced by LPS treatment, expression of the Csf3 gene encoding G-CSF was significantly down-regulated in LNA-antimiR-treated cells compared to the untreated and LNA controls (P = 0.014 and P = 0.008, respectively, Student's t-test, two-sided), implying that the regulation of G-CSF expression is mediated by miR-155 (Figure 5A, Supplementary table S1).
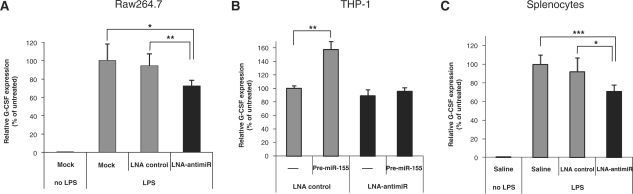
Figure 5.
Antagonism of miR-155 leads to down-regulation of the granulocyte colony-stimulating factor (G-CSF). (A) Quantitative RT-PCR analysis of G-CSF expression normalized to β2-microglobulin in Raw264.7 cells transfected with either LNA control (5 nM) or LNA-antimiR (5 nM) and stimulated with 100 ng/ml LPS for 6 h. Values represent mean ± SD (n = 5). The P-values (*P < 0.05; **P < 0.01) are shown for two-sided Student's t-test. (B) Quantitative RT-PCR analysis of G-CSF expression normalized to GAPDH after LPS-stimulation of THP-1 cells. THP-1 cells were cotransfected with 5 nM pre-miR-155 in combination with either LNA control (5 nM) or LNA-antimiR (5 nM) before LPS stimulation for 6 h. Values represent mean ± SD from one experiment performed in triplicate. The P-value (**P < 0.01) is shown for two-sided Student's t-test. (C) Quantitative RT-PCR analysis of G-CSF expression normalized to GAPDH in splenocytes of mice treated with LNA-antimiR or LNA control by three intravenous injections of 25 mg/kg over three consecutive days. Spleen samples were dissected 2 h after intraperitoneal injection of 0.5 mg/kg LPS. Values represent mean ± SD (n = 5). The P-values (*P < 0.05; ***P < 0.001) are shown for two-sided Student's t-test.
To validate this conclusion we assessed the effect of miR-155 on G-CSF expression in human monocytic THP-1 cells. Transient transfection of pre-miR-155 into LPS-stimulated THP-1 cells resulted in significant up-regulation of the G-CSF mRNA (P < 0.01, Student's t-test, two-sided), that reverted to control levels upon antagonism of miR-155 by LNA-antimiR (Figure 5B). Consistent with our data on mouse macrophages and human monocytes, we observed that the G-CSF mRNA was significantly down-regulated in the splenocytes of miR-155 antagonized LPS-treated mice (P = 0.0007 and P = 0.02, Student's t-test, two-sided) compared to saline and LNA-control-treated animals, respectively (Figure 5C). The miR-155-mediated effect on G-CSF mRNA in mouse splenocytes was robust as it was observed in three independent in vivo experiments (data not shown). Considered together, our data suggest that miR-155 mediates regulation of G-CSF expression during acute inflammatory response.
DISCUSSION
Inflammatory responses are initiated by monocytes that upon recognition of pathogens differentiate into macrophages, which then become activated to produce cytokines and chemokines. These inflammatory mediators help recruit effector cells to the site of infection and induce endothelial cell activation to increase vascular permeability (22). Several transcription factors are important for differentiation and activation of myeloid lineages, including the CCAAT/enhancer binding protein Beta (c/ebp Beta), which has been implicated in the regulation of proinflammatory cytokines during macrophage activation and the acute phase response (15,16). In this study, we show that c/ebp Beta is a bona fide target of miR-155 and that LPS treatment of murine macrophages strongly induces miR-155 expression, leading to translational repression of c/ebp Beta. Our observations are in good agreement with recent reports, showing that ectopically expressed miR-155 can target c/ebp Beta in vitro (9,24). Moreover we report here that functional inhibition of endogenous miR-155 in vitro leads to derepression of c/ebp Beta. Notably, we show for the first time that silencing of miR-155 by intravenously injected LNA-antimiR in LPS-treated mice results in marked derepression of the LAP and LIP isoforms of c/ebp Beta in mouse splenocytes, indicating that miR-155 acts as an important negative regulator of c/ebp Beta in vivo during acute inflammatory response.
A new discovery of this study is the finding that antagonism of miR-155 by LNA-antimiR leads to down-regulation of the G-CSF mRNA in vitro and in vivo. G-CSF is a master regulator of granulopoiesis produced by activated macrophages during acute inflammatory responses (27). Several studies have demonstrated that G-CSF is involved in inflammation and that it aggravates inflammatory diseases (28). G-CSF appears to play a central role in arthritis, where administration of G-CSF to mice has been shown to enhance collagen-induced arthritis (29), while blocking of G-CSF, on the other hand, markedly reduced disease manifestation in mice (30). Furthermore, G-CSF-deficient mice are protected from acute and chronic arthritis (30). In patients with active rheumatoid arthritis, G-CSF is elevated in the serum and synovial fluid and correlates with disease severity (31). Hence, G-CSF is considered as a potential therapeutic target for intervention of inflammatory joint diseases, such as rheumatoid arthritis (28). However, therapies that antagonize G-CSF may lead to neutropenia. Thus, partial antagonism of endogenous G-CSF, as described in this study for LNA-antimiR-mediated G-CSF down-regulation, might provide a positive outcome for arthritis patients without increased risk of severe neutropenia. Indeed, our findings warrant further investigations of the potential anti-inflammatory effect of miR-155 antagonism in relevant animal models of inflammatory disease, such as collagen-induced arthritis.
Previous studies have reported on impaired induction of G-CSF in LPS-stimulated macrophages derived from mice expressing only the dominant-negative LIP form of c/ebp Beta (33). Interestingly, we find that overexpression of the LIP isoform of c/ebp Beta in mouse macrophages results in down-regulation of the G-CSF transcript (Supplementary Figure 2). Thus, it is possible that c/ebp Beta, which is modulated by miR-155, acts in part by fine-tuning G-CSF expression levels, consistent with a c/ebp Beta binding motif in the promoter of G-CSF (15,32). However, since a single vertebrate miRNA may target up to ∼100–200 mRNAs, it is highly likely that functional antagonism of miR-155 in vivo results in more complex downstream effects that are associated with miR-155-mediated gene regulatory networks. Indeed, miR-155 was recently shown to regulate the PI3K–AKT signaling pathway during LPS stimulation of macrophages by targeting the inositol phosphatase SHIP1 (34). Nevertheless, given the importance of c/ebp Beta during macrophage activation and the role of G-CSF in stimulating granulopoiesis in the bone marrow, our data emphasize the potential of antagonizing miR-155 for modulating activation of macrophages and the number of circulating granulocytic cells with possible implications for treatment of inflammatory diseases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Danish National Advanced Technology Foundation, Danish Medical Research Council and the Lundbeck Foundation (to S.K.); Wilhelm Johannsen Centre for Functional Genome Research is established by the Danish National Research Foundation. Funding for open access charge: Santaris Pharma, Hørsholm, Denmark.
Conflict of interest statement. J.W., J.S., A.P., S.O., J.E., M.H., E.M.S., J.B.H. and S.K. are employed at Santaris Pharma. Santaris Pharma is a biopharmaceutical company engaged in the development of RNA based medicine.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Anja Konge, Charlotte Øverup, Diana Klüver, Jane Hinrichsen, Janni Juul Jørgensen, Lisbeth Bang, Otto Olsen and Rikke Sølberg for excellent technical assistance.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Kato M, Slack FJ. microRNAs: small molecules with big roles – C. elegans to human cancer. Biol. Cell. 2008;100:71–81. doi: 10.1042/BC20070078. [DOI] [PubMed] [Google Scholar]
- 2.Fabbri M, Garzon R, Andreeff M, Kantarjian HM, Garcia-Manero G, Calin GA. MicroRNAs and noncoding RNAs in hematological malignancies: molecular, clinical and therapeutic implications. Leukemia. 2008;22:1095–1105. doi: 10.1038/leu.2008.30. [DOI] [PubMed] [Google Scholar]
- 3.Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Curr. Opin. Hematol. 2008;15:352–358. doi: 10.1097/MOH.0b013e328303e15d. [DOI] [PubMed] [Google Scholar]
- 4.Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29:343–351. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Bi Y, Liu G, Yang R. MicroRNAs: novel regulators during the immune response. J. Cell Physiol. 2008;281:467–472. doi: 10.1002/jcp.21639. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van DS, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 8.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl Acad. Sci. USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 11.Vigorito E, Perks KL, breu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di VM, San-Martin BR, Heidkamp G, Schwickert TA, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorgoni B, Maritano D, Marthyn P, Righi M, Poli V. C/EBP beta gene inactivation causes both impaired and enhanced gene expression and inverse regulation of IL-12 p40 and p35 mRNAs in macrophages. J. Immunol. 2002;168:4055–4062. doi: 10.4049/jimmunol.168.8.4055. [DOI] [PubMed] [Google Scholar]
- 15.Natsuka S, Akira S, Nishio Y, Hashimoto S, Sugita T, Isshiki H, Kishimoto T. Macrophage differentiation-specific expression of NF-IL6, a transcription factor for interleukin-6. Blood. 1992;79:460–466. [PubMed] [Google Scholar]
- 16.Scott LM, Civin CI, Rorth P, Friedman AD. A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood. 1992;80:1725–1735. [PubMed] [Google Scholar]
- 17.Hamilton JA. Rheumatoid arthritis: opposing actions of haemopoietic growth factors and slow-acting anti-rheumatic drugs. Lancet. 1993;342:536–539. doi: 10.1016/0140-6736(93)91653-4. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton JA. Colony-stimulating factors in inflammation and autoimmunity. Nat. Rev. Immunol. 2008;8:533–544. doi: 10.1038/nri2356. [DOI] [PubMed] [Google Scholar]
- 19.Smith C, Zhu K, Merritt A, Picton R, Youngs D, Garrod D, Chidgey M. Regulation of desmocollin gene expression in the epidermis: CCAAT/enhancer-binding proteins modulate early and late events in keratinocyte differentiation. Biochem. J. 2004;380:757–765. doi: 10.1042/BJ20040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 21.Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, Hansen JB, Hansen HF, Straarup EM, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H, Park CK, Ryu JY, Chang EJ, Lee Y, Kang SS, Kim HH. Expression profiling of lipopolysaccharide target genes in RAW264.7 cells by oligonucleotide microarray analyses. Arch. Pharm. Res. 2006;29:890–897. doi: 10.1007/BF02973911. [DOI] [PubMed] [Google Scholar]
- 23.O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin Q, McBride J, Fewell C, Lacey M, Wang X, Lin Z, Cameron J, Flemington EK. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J. Virol. 2008;82:5295–5306. doi: 10.1128/JVI.02380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su WC, Chou HY, Chang CJ, Lee YM, Chen WH, Huang KH, Lee MY, Lee SC. Differential activation of a C/EBP beta isoform by a novel redox switch may confer the lipopolysaccharide-inducible expression of interleukin-6 gene. J. Biol. Chem. 2003;278:51150–51158. doi: 10.1074/jbc.M305501200. [DOI] [PubMed] [Google Scholar]
- 26.Schebesta M, Heavey B, Busslinger M. Transcriptional control of B-cell development. Curr. Opin. Immunol. 2002;14:216–223. doi: 10.1016/s0952-7915(02)00324-2. [DOI] [PubMed] [Google Scholar]
- 27.Watari K, Asano S, Shirafuji N, Kodo H, Ozawa K, Takaku F, Kamachi S. Serum granulocyte colony-stimulating factor levels in healthy volunteers and patients with various disorders as estimated by enzyme immunoassay. Blood. 1989;73:117–122. [PubMed] [Google Scholar]
- 28.Eyles JL, Roberts AW, Metcalf D, Wicks IP. Granulocyte colony-stimulating factor and neutrophils–forgotten mediators of inflammatory disease. Nat. Clin. Pract. Rheumatol. 2006;2:500–510. doi: 10.1038/ncprheum0291. [DOI] [PubMed] [Google Scholar]
- 29.Campbell IK, Rich MJ, Bischof RJ, Hamilton JA. The colony-stimulating factors and collagen-induced arthritis: exacerbation of disease by M-CSF and G-CSF and requirement for endogenous M-CSF. J. Leukoc. Biol. 2000;68:144–150. [PubMed] [Google Scholar]
- 30.Lawlor KE, Campbell IK, Metcalf D, O'Donnell K, van NA, Roberts AW, Wicks IP. Critical role for granulocyte colony-stimulating factor in inflammatory arthritis. Proc. Natl Acad. Sci. USA. 2004;101:11398–11403. doi: 10.1073/pnas.0404328101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura H, Ueki Y, Sakito S, Matsumoto K, Yano M, Miyake S, Tominaga T, Tominaga M, Eguchi K. High serum and synovial fluid granulocyte colony stimulating factor (G-CSF) concentrations in patients with rheumatoid arthritis. Clin. Exp. Rheumatol. 2000;18:713–718. [PubMed] [Google Scholar]
- 32.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uematsu S, Kaisho T, Tanaka T, Matsumoto M, Yamakami M, Omori H, Yamamoto M, Yoshimori T, Akira S. The C/EBP beta isoform 34-kDa LAP is responsible for NF-IL-6-mediated gene induction in activated macrophages, but is not essential for intracellular bacteria killing. J. Immunol. 2007;179:5378–5386. doi: 10.4049/jimmunol.179.8.5378. [DOI] [PubMed] [Google Scholar]
- 34.O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl Acad. Sci. USA. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.