Abstract
VA RNAI is a non-coding adenoviral transcript that counteracts the host cell anti-viral defenses such as immune responses mediated via PKR. We investigated potential alternate secondary structure conformations within the PKR-binding domain of VA RNAI using site-directed mutagenesis, RNA UV-melting analysis and enzymatic RNA secondary structure probing. The latter data clearly indicated that the wild-type VA RNAI apical stem can adopt two different conformations and that it exists as a mixed population of these two structures. In contrast, in two sequence variants we designed to eliminate one of the possible structures, while leaving the other intact, each formed a unique secondary structure. This clarification of the apical stem pairing also suggests a small alteration to the apical stem–loop secondary structure. The relative ability of the two apical stem conformations to bind PKR and inhibit kinase activity was measured by isothermal titration calorimetry and PKR autophosphorylation inhibition assay. We found that the two sequence variants displayed markedly different activities, with one being a significantly poorer binder and inhibitor of PKR. Whether the presence of the VA RNAI conformation with reduced PKR inhibitory activity is directly beneficial to the virus in the cell for some other function requires further investigation.
INTRODUCTION
VA RNAI is a non-coding adenoviral RNA transcript that inhibits the anti-viral double-stranded RNA (dsRNA)-activated protein kinase (PKR). As a first line of defense against viral infection and under other conditions of cell stress (1,2), PKR exerts its negative regulatory effect on protein translation through phosphorylation of the eukaryotic translation initiation factor 2 (eIF2). This modification at serine 51 of the eIF2 α-subunit dramatically increases its affinity for the guanine nucleotide exchange factor (eIF2B) such that it is sequestered and the GTP-bound form of eIF2 sufficiently depleted to halt expression of most mRNAs (3–5). Inhibition of PKR by VA RNAI relieves this block and thus allows continued production of viral proteins on the host cell's translational machinery (6–8).
All adenoviruses produce VA RNAI but there is significant variation in sequence and length (149–174 nucleotides) between different viruses (9). However, all VA RNAI molecules are highly structured with three major domains for which specific functions have been determined: the terminal stem, central domain and apical stem. The largely double-stranded apical stem comprises the functional binding site for PKR (10–13), while the adjacent central domain contains the structural determinant(s) that make VA RNAI an inhibitor rather than activator of PKR (11,14–18). This latter domain is proposed to have a complex tertiary structure that is critical for inhibition (19) but so far few specific details are known about the mechanism of inhibition. Finally, the terminal stem contains essential transcription signals but is entirely dispensable for the PKR kinase inhibition activity of VA RNAI. Interestingly, however, deletion of the complete terminal stem produces a shortened VA RNAI molecule (termed TSΔ21 RNA) that is fully active in in vitro assays of PKR inhibition (20) and the same deletion is made in vivo by the RNase Dicer (21). Terminal stem fragments generated by Dicer are incorporated into RISC complexes (21), suggesting that each VA RNAI transcript may be able to block both innate immune response via PKR and saturate RNAi.
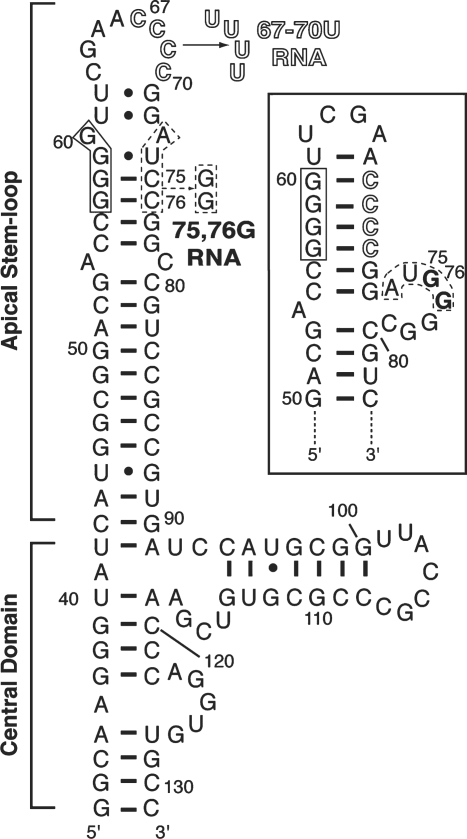
As the primary binding site for the two dsRNA-binding motifs that comprise the N-terminal domain of PKR, the structural requirements for a functional VA RNAI apical stem have been investigated in some detail (10–13). From these studies it can be broadly concluded that sequence is unimportant but a largely uninterrupted A-form helix of around eight base pairs or more is required for full activity. A secondary structure model for VA RNAI was proposed (14,15) and refined on the basis of comparative sequence analysis and further RNA structure probing (9,17,19,22). However, a potential mixture of two alternative structures was suggested (Figure 1) where both conformations are largely but not entirely consistent with the available structure probing data (19). Here we describe apical stem sequence variants that were designed to promote formation of one or the other of these two possible conformations to investigate this potential structural heterogeneity and its implications for VA RNAI action against PKR.
Figure 1.
Secondary structure and mutagenesis of VA RNAI. Sequence and secondary structure of the TSΔ21 variant of VA RNAI (20), shown in the extended conformation with the alternate asymmetric internal loop structure as boxed inset. The VA RNAI terminal stem is omitted but the nucleotide numbering shown corresponds to the full-length RNA (nts 1–155). Three tetranucleotide sequences involved in key base-pairing interactions for these alternate structures are highlighted along with mutations made to generate the 67–70U (outline font) and 75,76G RNAs (dashed line box).
MATERIALS AND METHODS
RNA in vitro transcription
Mutations 67–70U and 75,76G (Figure 1) in the VA RNAI apical stem were generated by QuikChange site-directed mutagenesis (Stratagene) in plasmids encoding either full-length wild-type VA RNAI (23,24) or TSΔ21 RNA (20). Plasmids encoding wild-type and mutant apical stem hairpin RNAs were created by direct ligation of chemically synthesized DNA oligonucleotides into the same plasmid (24). Run-off RNA in vitro transcription reactions were performed using T7 RNA polymerase under optimal conditions for VA RNAI (25). Denaturing polyacrylamide gel electrophoresis purification and recovery of RNA samples was accomplished as described previously (20,23).
RNA UV-melting analysis
Samples contained 20–25 μg RNA in a solution containing 10 mM MOPS buffer pH 7.0 and 50 mM KCl. UV-melting curves were collected on a Varian Cary 400 UV/Vis spectrophotometer with a six-cell multichanger. Sample temperatures were measured using an in-cell temperature probe. First derivatives of the melting curves, referred to as a ‘melting profiles’, were calculated using the program ‘OD Deriv’ (D.E. Draper, Johns Hopkins University).
RNA secondary structure probing
TSΔ21 RNA, and 67–70U and 75,76G mutants in the same terminal stem deletion context (TSΔ21) were dephosphorylated by treatment with calf intestinal alkaline phosphatase and 5′-end labeled using T4 polynucleotide kinase and γ-32P-ATP (MP Biomedicals). Excess γ-32P-ATP was removed using G-25 microspin columns (GE Healthcare) and the labeled RNAs gel purified, recovered by crush and soak of excised gel slices and quantitated by liquid scintillation counting.
Each 32P-labeled RNA was partially digested using RNases T1, V1 and A. The enzymes were purchased as components of structure probing kits (Ambion) and all reactions were performed according to the manufacturer's protocols. A single reaction mixture containing 100 000 cpm-labeled RNA was aliquoted and used to generate a serial dilution of each RNase to determine the optimal concentration for each enzyme. Experiments investigating the effect of PKR binding on apical stem structure were performed using methods adapted from a study of protein-induced RNA conformational switching (26). Three equal aliquots of 32P-labeled TSΔ21 RNA were incubated for 30 min in the presence of PKR (at 1:1 or 5:1 excess protein) or an equal volume of buffer as control. TSΔ21 RNA samples were recovered by phenol–chloroform extraction and ethanol precipitation and then probed with RNase A as before. To allow reading of the RNA sequences, alkaline hydrolysis (AH) and RNase T1 reactions in denaturing buffer were also performed. Three microliters of each reaction corresponding to ∼40 000 cpm was loaded per lane on a denaturing 10% polyacrylamide sequencing gel. The fixed and dried gel was imaged using both photographic film and a Typhoon 8600 phosphorimager.
PKR autophosphorylation inhibition assays
PKR was expressed from plasmid pET–PKR/PPase in E. coli Rosetta BL21(DE3) as described previously (20). Purified protein was dialyzed into 2× reaction buffer (100 mM Tris pH 7.8, 100 mM KCl, 10% glycerol, 5 mM dithiothreitol). RNA samples were exchanged into the same buffer at 5× stock concentrations. PKR (2 μl, ∼0.1 μg) was pre-incubated with VA RNAs (3 μl) at room temperature for 5 min. An equal volume of activating RNA solution (0.6 μg/ml poly(I)•poly(C), 40 μM ATP, 4 μM MgCl2 and 0.2 mCi/ml 32P ATP (6000 Ci/mmol; 10 mCi/ml in buffered solution; MP Biomedicals) was next added and the reaction incubated at room temperature for 10 min. The reaction was stopped by addition of 0.5 volumes of 3× SDS loading dye and heating at 90°C for 5 min. Control reactions were performed without poly(I)•poly(C) RNA to confirm the absence of any co-purifying activating RNAs in the VA RNAI preparations. Band intensities were quantitated using a Typhoon 8600 phosphorimager and Image J software following fractionation on 10% acrylamide SDS–PAGE gels.
Isothermal titration calorimetry
The thermodynamics of mutant VA RNAI–PKR interaction were measured using a VP-ITC microcalorimeter using procedures previously described for TSΔ21 RNA (20). RNA and protein samples were 3 μM (sample cell) and 30 or 60 μM (syringe), respectively in 10 or 50 mM Na2HPO4/NaH2PO4 pH 6.5, 100 mM NaCl and 5 mM β-mercaptoethanol. Titration curves were fit by a non-linear least-squares method in Microcal Origin software using a model for one or two binding sites. A model with one binding site was found to give the optimal fit in each case and was used to extract thermodynamic parameters Kd (binding constant), ΔH (enthalpy of binding) and N (stoichiometry). Similar fits were obtained from titration data sets at both PKR concentrations and the average value of each parameter is shown in Table 1.
Table 1.
ITC analysis of wild-type and mutant VA RNAI interaction with PKR
| RNA | Kd (nM) | N | ΔH (kcal/mol) | ΔS (cal/mol K) | ΔG (kcal/mol) | Ref. |
|---|---|---|---|---|---|---|
| Wild-type full-length VA RNAI | 79 | − | −11.3 | −4.8 | −9.8 | (28) |
| TSΔ21 | 83 | 1.16 | −6.7 | 10.4 | −9.8 | (20) |
| 67–70U | 47 ± 3.6 | 1.32 ± 0.08 | −23.4 ± 2.7 | −41.3 ± 10.7 | −10.9 ± 0.05 | − |
| 75,76G | 126 ± 1.5 | 1.45 ± 0.23 | −16.6 ± 2.8 | −25.2 ± 10.8 | −9.0 ± 0.06 | − |
| Averagea | 86 | −9.9 |
aAverage of values for 67–70U and 75,76G RNAs.
RESULTS
Design of apical stem sequence variants
The potential for alternate structures in the adenovirus VA RNAI apical stem arises because the tetranucleotide sequence 57GGGG60 can pair with two different 3′-sequences: 67CCCC70 or 73AUCC76 (Figure 1). Although pairing with the latter sequence might appear less optimal, the fully extended apical stem structure it creates is almost invariably used when drawing the VA RNAI secondary structure and is the closest to the lowest free energy structure predicted by M-fold (see the ‘Discussion’ section). We designed mutations at each 3′-tetranucleotide sequence to destabilize its base pairing with the 57GGGG60 sequence and thus promote formation of a unique conformation involving the other pairing. The mutation ‘67–70U' replaces the four loop cytidines with uridine (Figure 1), removing the potential for them to Watson–Crick G-C base pair with 57GGGG60 and thereby promoting the extended apical stem conformation with 67UUUU70 in the loop (Figure 1). Similarly, the mutation ‘75,76G' was designed to disrupt the two G–C base pairs formed by 73AUCC76 promoting the formation of an asymmetric internal loop containing these nucleotides and thus the shortened apical stem conformation (Figure 1, inset).
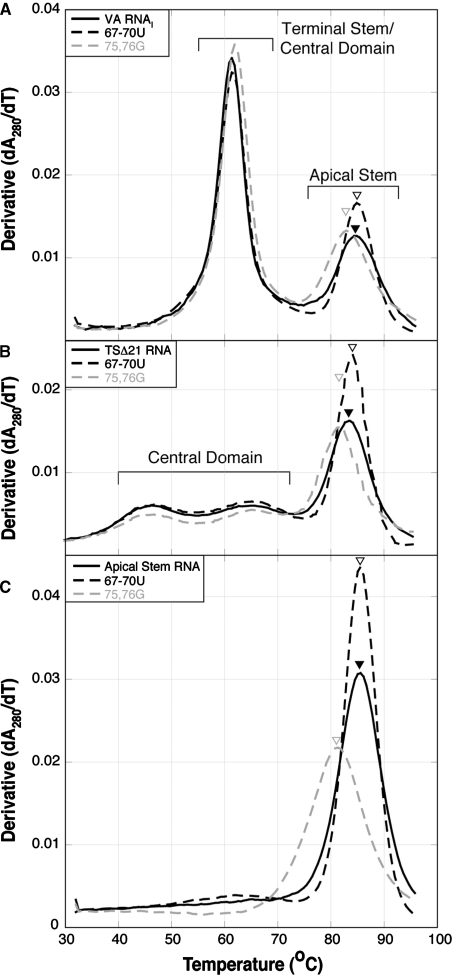
The 67–70U and 75,76G mutations were made in the context of full-length VA RNAI, the fully active terminal stem deleted variant TSΔ21 (20), and a 78 nt hairpin RNA corresponding to the isolated apical stem domain. We first used UV-melting analysis to assess the effect of these mutations on the secondary structure (folding and stability) of the VA RNAI apical stem mutants in each context (Figure 2). Each mutation alters the melting profile in only the apparent transition corresponding to apical stem unfolding (23), with little effect on the other region(s) of the profile. The 67–70U mutation does not significantly alter the apparent Tm for the apical stem unfolding (increased ∼1°C) but the transition is sharper suggesting a more cooperative unfolding event. In contrast, the 75,76G mutation decreased the apparent Tm by 2–5°C compared to the wild-type sequence for each RNA context. Strikingly, the effect of the 67–70U and 75,76G mutations is identical regardless of the RNA context in which it is made. Although the absolute values of hypochromicity for apical stem unfolding vary between RNA contexts, the relative differences between wild-type and mutant sequences and the changes in unfolding apparent Tm are entirely consistent. This result correlates well with our previous observation that the apical stem is an independent domain within the global structure of the RNA (23). The melting profiles of Figure 2C demonstrate that the apical stem adopts essentially the same structure in the full-length or truncated TSΔ21 RNA as it does in isolation. To further investigate the potential structural heterogeneity in this domain, we chose to use primarily the TSΔ21 context since the shorter sequence would simplify structure probing but still allow analysis of the functional properties of each mutant RNA.
Figure 2.
RNA UV-melting analysis of VA RNAI mutants. Melting profiles collected at 280 nm for (A) full-length VA RNAIs, (B) TSΔ21 VA RNAIs and (C) apical stem fragment RNAs, with wild-type (solid black line), 67–70U (dashed black line) and 75,76G (dashed gray line) sequences.
Enzymatic RNA secondary structure probing
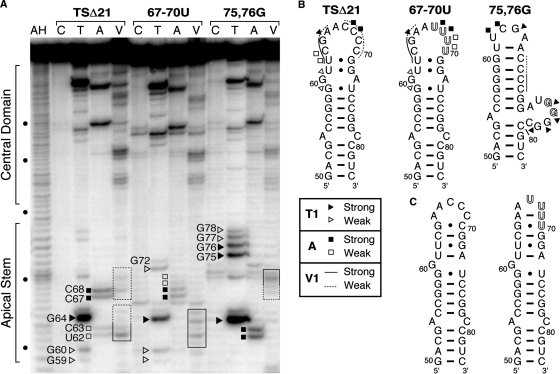
Wild-type, 67–70U and 75,76G VA RNAI transcripts (all in the TSΔ21 terminal stem deletion context) were 5′-end labeled with 32P and subject to partial digestion by RNAses T1, A and V1. These ribonucleases cleave 3′ of unpaired G nucleotides, 3′ of unpaired C or U nucleotides and in regions base paired or stacked nucleotides, respectively. An identical pattern of nuclease sensitivity was observed for the central domain of each RNA (Figure 3A). In contrast, several significant differences were observed in the region corresponding to the top of the apical stem that indicate each RNA contains distinct secondary structural features.
Figure 3.
Enzymatic RNA secondary structure probing. (A) Sequencing gel analysis of each RNA partially digested with RNases T1 (lane T; unpaired G), A (lane A; unpaired C or U) and V1 (lane V; stacked or base paired nucleotides) or untreated as control (lane C). Alkaline hydrolysis (AH) and denaturing RNase T1 (not shown) reactions were used to determine nucleotide positions (dots on the left side of the gel denote each 10th nucleotide). (B) Summary of structure probing data displayed on the proposed secondary structure of each apical stem. (C) Redrawing of the apical stem structure for the wild-type and 67–70U sequences (see ‘Discussion’ section).
Most strikingly, for TSΔ21 clear regions of apparent overlapping sensitivity to both single (ss) and double strand (ds)-specific nucleases are observed. RNase T1 produces weak cuts at G59/G60 and a very strong cut after G64 (Figure 3). These sensitivities correspond well to the locations of these guanines in the proposed secondary structure. However, two weak cleavages at G77/G78 correspond to a region that is predicted to be base paired. With RNase A, ss-specific cleavages are observed at U62/C63 (weak) and C67/C68 (strong). These observations are again reasonable, since these nucleotides are adjacent to or within the loop. However, both regions are also sensitive to the ds-specific RNase V1. Such mixed sensitivity might arise from unusual non-Watson–Crick base-paired structure or, alternatively, could indicate as proposed (19) that the wild-type apical stem exists as a mixed population of two conformations. The probing data for the two mutant RNA sequences provide clear demonstration of the latter.
For both mutant RNAs the RNase T1 cleavage at G64 is retained correlating with its position in the loop in both structures. The 67–70U RNA also retains sensitivity to RNase T1 at G60 but is no longer cut at G77/G78. For 75,76G RNA the reverse is observed: the G60 cleavage is completely absent, while those at G77/G78 are present and are stronger. Two additional strong cleavages are observed that correspond to the two residues altered from C to G (G75/G76). The G75–G78 cleavages clearly indicate the presence of the asymmetric loop containing these four nucleotides (Figure 3). A similar set of observations is made when comparing the RNase A ss-specific cleavage patterns at U62/C63 and C67/C68. Again, 67–70U RNA is cleaved at only C67/C68, while the bands corresponding to U62/C63 are absent. For 75,76G RNA the reverse is true with U62/C63 present and C67/C68 absent. Similarly, for the ds-specific RNase V1, only one set of cleavages is observed for each mutant where both were observed in TSΔ21. In 75,76G RNA the ds-cleavages overlapping with RNase A cleavage sites at C61–G63 are absent, correlating with the increase in intensity of the ss-specific bands, while the ds-specific cleavages at C67–G71 are retained. In contrast, as observed with both RNases A and T1, the opposite pattern of sensitivity changes is observed for 67–70U RNA: those at C67–G71 are now absent, while those at C61–G63 are retained. In fact, the ds-specific cleavage sites appear to extend further, with some sensitivity to RNase V1 observed further into the loop region. This indicates that base pairing may be more extensive than shown in the proposed secondary structure (Figure 3B,C). Collectively, the enzymatic probing data demonstrate that the wild-type apical stem does exist as a mixture of two distinct structural conformations. Furthermore, our designed sequence alterations have successfully altered their relative stabilities such that each mutant RNA adopts only one of these possible structures as a unique conformation.
PKR binding and inhibition properties of 67–70U and 75,76G RNAs
The ability of each mutant RNA to bind PKR was assessed using isothermal titration calorimetry (ITC). In contrast to wild-type VA RNAI (27) and TSΔ21 RNA (20), titration data for both mutant RNAs were best fit using a single binding site model to extract the thermodynamic parameters of binding (Table 1 and Supplementary Figure S1). The 67–70U RNA, with its extended apical stem structure, binds with moderately higher affinity, while 75,76G RNA binds more poorly compared to the wild-type apical stem structure in TSΔ21 RNA. The increase in Kd for 67–70U RNA is almost identical in magnitude to the decrease observed for 75,76G RNA resulting in an average value for the two RNAs comparable to the experimentally measured Kd for the full-length and TSΔ21 wild-type RNA sequences (Table 1). Similarly, despite significant variation in the values of ΔH and ΔS obtained from the fits, the average free energy change associated with binding (ΔG) of the two mutants is close to the ΔG measured for wild-type (28) and TSΔ21 RNA.
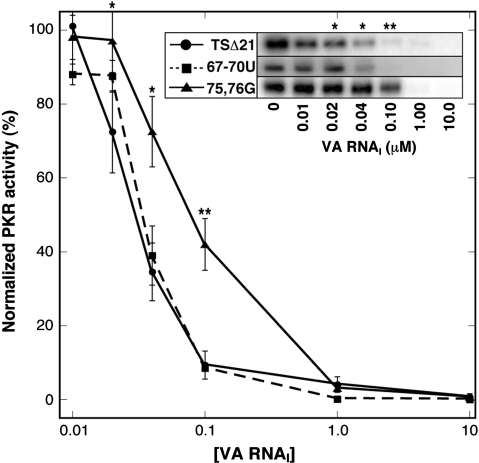
The level of PKR autophosphorylation correlates well with PKR activity against its primary cellular target eIF2 initiation factor (29) and can therefore be used as a simple assay of inhibitor RNA function. The ability of each mutant RNA to inhibit PKR autophosphorylation on addition of poly(I).poly(C) activator RNA was thus determined and compared to wild-type TSΔ21 RNA (Figure 4). The 67–70U RNA displayed activity comparable to that observed for wild-type TSΔ21 (Figure 4) and full-length wild-type VA RNAI (20) with complete inhibition above 0.04 μM inhibitor RNA. In contrast, 75,76G RNA is significantly less active requiring >0.1 μM for complete inhibition. An identical result was also obtained when comparing the ability of full-length wild-type VA RNAI and the two mutants made in the context of this larger RNA (data not shown). Thus the apical stem structures and PKR binding affinity of the mutant RNAs correlates with their activity against PKR.
Figure 4.
Inhibition of PKR kinase activity. PKR autophosphorylation following preincubation with wild-type, 67–70U and 75,76G VA RNAIs in the TSΔ21 context at the concentrations indicated, and subsequent kinase activation with poly(I).poly(C) dsRNA. The inset shows typical example of a gel for each RNA. At least three independent experiments were performed for each RNA and the normalized averaged PKR activity is plotted with standard errors of the mean at each concentration. Data for TSΔ21 and 75,76G were compared using Student's t-test (*P < 0.01; **P < 0.0025).
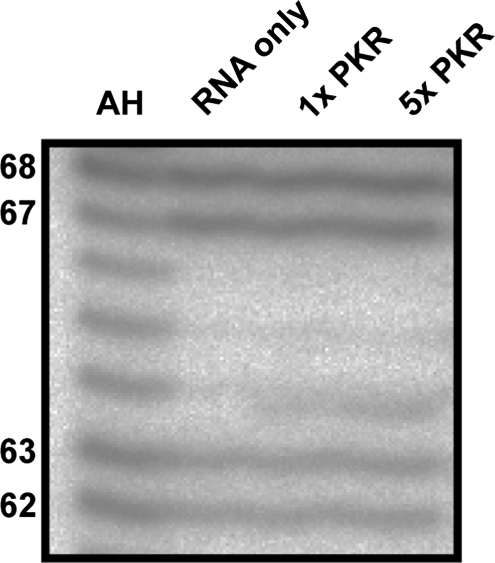
Finally, we asked whether the binding of PKR could influence the structure of the wild-type apical stem sequence, e.g. to increase the proportion of the extended conformation. Samples of RNA incubated without and with one and five molar ratios of PKR were probed with RNase A following extraction of the protein (Figure 5). No changes in the relative intensities of the characteristic ss nuclease sensitive sites at nucleotides 62/63 or 67/68 were observed. Promotion of the extended form of the apical stem by PKR would have been expected to increase the intensity of cleavage at 67/68 and decrease that at 62/63 to produce a pattern more similar to the 67–70U variant (Figure 3).
Figure 5.
PKR does not influence the relative amount of each conformation formed by the wild-type VA RNAI apical stem. RNase A probing of TSΔ21 RNA pre-incubated without (RNA only) and with PKR at one and five molar equivalents (1× PKR and 5× PKR, respectively). AH is alkaline hydrolysis reaction.
DISCUSSION
The propensity for RNA molecules to misfold into non-native structures of similar stability to the correct native fold is widely appreciated (30). This presumably arises through the relative lack of variation in the basic RNA nucleotide building blocks and the major energetic driving forces provided by base stacking and backbone phosphate group–ion interactions that are largely independent of sequence. Identifying conditions for obtaining correctly folded molecules can pose a significant hurdle for researchers interested in the unique secondary and tertiary structures of RNAs that may be required for their biological functions in vivo (30,31). However, there is also an increasing appreciation that RNA structural rearrangements may be exploited by nature, for example, regulate protein expression in organisms from bacteria to humans (26,32), act as switches in viral life cycles (33,34) and influence the initial steps of splicing (35,36).
We have recently investigated the domain structure and stability of the adenoviral non-coding RNA transcript VA RNAI and identified a minimal functional molecule lacking the entire terminal stem (20,23). These studies also indicated that the PKR-binding domain of VA RNAI (the apical stem–loop) exists as an essentially independent domain within the RNA structure, a finding confirmed by our current UV-melting analyses. Our nuclease sensitivity data with VA RNAI apical stem sequence variants have now experimentally demonstrated the presence of alternate conformations in the wild-type apical stem sequence. The pattern of RNase susceptibility for 67–70U RNA was in good agreement with the current secondary structure (Figure 1) though an extended sensitivity to ds-specific nuclease suggests that base pairing may be more extensive at the top of the apical stem (Figure 3C). For 75,76G RNA, the nuclease sensitivity pattern correlated precisely with the secondary structure model for the alternate apical stem conformation (Figure 3B). The structures predicted by M-fold (37) for nts 50–83 with wild-type and each mutant sequence also matched those we deduced from our nuclease sensitivity data and we therefore suggest a modification to the pairing of the apical stem loop as shown in Figure 3C.
VA RNAI mutants with the alternate apical stem structures displayed different activities in assays of PKR inhibition: 67–70U RNA exhibited wild-type inhibitory activity, whereas the 75,76G RNA, with a shorter helix interrupted by the internal loop, was significantly poorer. This decrease in activity correlated well with a decrease in PKR binding affinity measured by ITC. Thus the mixture of alternate conformations in the wild-type creates two functionally distinct populations of VA RNAI. Our ITC titration data for both mutant RNAs were optimally fit using a model for a single binding site (Table 1 and Supplementary Figure S1), in contrast to the wild-type sequence in both the full-length and TSΔ21 VA RNAIs, where the data are best fit using a two-site model, and the additional “low affinity” site attributed to non-specific binding (27). In our previous analysis of PKR–TSΔ21 interaction using ITC, we also observed two binding events and suggested these might have arisen through difficulty in analyzing data from RNA samples with a mixture of conformations. However, while the parameters corresponding to the functional interaction of RNA and protein were correctly reported (reproduced here in Table 1), they in fact clearly corresponded to the second observed binding event in the titration. The origin and significant of the “high affinity site” (Kd 0.5 nM) in the TSΔ21–PKR titration is currently not clear. Such issues were not observed in any of the replicate titrations for either variant RNA examined here. Therefore, while not entirely excluding the possibility that the mixture of structures in the wild-type apical stem might contribute to difficulty in the analysis of PKR–VA RNAI binding as we suggested previously, the present data suggest more obviously that these shortened VA RNAI molecules (i.e. in the TSΔ21 context) are bound by a single PKR molecule and lack significant secondary or non-specific binding site(s) which must presumably be present in the deleted terminal stem.
RNA molecules produced by in vitro transcription are often purified under denaturing conditions and the refolding process can produce samples with a mixture of conformations including misfolded structures. Where functional assays are possible such as aminoacylation of tRNA or ribozyme activity, such issues can be readily identified if not fully or easily resolved. Our data show conclusively that the apical stem of purified VA RNAI in vitro transcripts exists in two alternative conformations. While this might arise as a consequence of the denaturing purification procedure, several observations argue against this. VA RNAI is a remarkably stable molecule, particularly within the apical stem which has an apparent unfolding Tm of ∼85°C and does not unfold in denaturing gels (11,23). In agreement with these findings, M-fold analysis of the apical stem sequence predicted large folding free energies for both the extended (ΔG at 37°C, –28.0 kCal/mol) and shortened/bulge loop forms (ΔG –26.6 kCal/mol). The predicted free energies for the top of the apical stem containing the alternate structures (nts 50–83, see Figure 1A inset) are smaller, at –15.80 and –14.20 kCal/mol but still significant. Although there is only a relatively small difference in predicted folding free energy (ΔΔG 1.6 kCal/mol) between the alternate structures, these values indicate that interconversion between them, through unfolding of the apical stem, is unlikely. A common approach to resolving issues of RNA misfolding is to empirically identify an annealing regime that typically involve thermal unfolding and refolding, usually in the presence of Mg2+. VA RNAI, however, appears uncharacteristically ambivalent toward such procedures with no effect observed on UV-melting profiles or kinase inhibition assays performed without annealing or prior addition of Mg2+ (data not shown). Finally, co-transcriptional folding of VA RNAI in the cell would be expected to favor the less-active conformer with its shortened helix containing 73AUCC76 in the asymmetric internal loop. Thus it is likely that the competing effects of the kinetically favored shortened helix with bulge loop and thermodynamically favored extended conformation would result in the mixture of structures we observe both in vitro and in vivo.
If cellular VA RNAI also exists in both conformations, what might be the implications for the virus? Our data clearly show that the shortened apical stem conformation is impaired in PKR-inhibitory function. Ultimately, at late stages of infection, where there are ∼108 copies of VA RNAI per cell, the pool of poorer inhibitors is unlikely to be detrimental. However, at earlier times when PKR is first activated in response to viral dsRNA and VA RNAI levels are considerably lower, this difference might be significant. It is possible that the propensity to form two conformations has arisen by chance and insufficient selective pressure exists to drive mutations that would direct the RNA toward a single high-activity conformation for PKR inhibition. More intriguing, however, is the possibility that the lower activity conformation of VA RNAI is directly beneficial to the virus for some other as yet undetermined function. For example, one potential benefit of sequence and structural plasticity in the apical stem might be to help maintain activity against a rapidly changing target molecule (39). Analysis of VA RNAs from other adenoviral serotypes by M-Fold (data not shown) and the gene structure of VA RNAII (38) suggest the potential for forming alternate conformations in this critical functional domain may be a common feature of VA RNAs. Just as recent data have uncovered a second function for VA RNAI in addition to PKR inhibition, where the terminal stem sequences contribute to viral suppression of cellular RNAi following cleavage by Dicer (20,21), it is thus conceivable that there exist further, as yet undiscovered, functions for VA RNAI for which the alternative structure is preferential and perhaps even promoted by interaction host cell or adenoviral proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Career Development Fellowship from The Wellcome Trust (ref. 061444) to G.L.C. and studentship support to A.M.W. and V.K.C. from the Egyptian Ministry for Higher Education and Medical Research Council (MRC, UK), respectively. Funding for open access charge: The Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
PKR expression plasmid pET-PKR/PPase was the generous gift of Dr. James Cole (University of Connecticut). The authors thank Dr. Christine Dunham for assistance with structure probing experiments and for comments on the manuscript.
REFERENCES
- 1.Clemens MJ, Elia A. The double-stranded RNA-dependent protein kinase PKR: Structure and function. J. Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 2.Meurs E, Chong K, Galabru J, Thomas NSB, Kerr IM, Williams BRG, Hovanessian AG. Molecular cloning and characterization of the human double stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 3.Krishnamoorthy T, Pavitt GD, Zhang F, Dever TE, Hinnebusch AG. Tight binding of the phosphorylated alpha subunit of initiation factor 2 (eIF2 alpha) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol. Cell Biol. 2001;21:5018–5030. doi: 10.1128/MCB.21.15.5018-5030.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pathak VK, Schindler D, Hershey JWB. Generation of a mutant form of protein synthesis initiation factor eIF2 lacking the site of phosphorylation by eIF2 kinases. Mol. Cell Biol. 1988;8:993–995. doi: 10.1128/mcb.8.2.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rowlands AG, Panniers R, Henshaw EC. The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor II. J. Biol. Chem. 1988;263:5526–5533. [PubMed] [Google Scholar]
- 6.Kitajewski J, Schneider RJ, Safer B, Munemitsu SM, Samuel CE, Thimmappaya B, Shenk T. Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2-alpha kinase. Cell. 1986;45:195–200. doi: 10.1016/0092-8674(86)90383-1. [DOI] [PubMed] [Google Scholar]
- 7.Soderlund H, Pettersson U, Vennstrom B, Philipson L, Mathews MB. New species of virus-coded low-molecular weight RNA from cells infected with Adenovirus type 2. Cell. 1976;7:585–593. doi: 10.1016/0092-8674(76)90209-9. [DOI] [PubMed] [Google Scholar]
- 8.O'Malley RP, Mariano TM, Siekierka J, Mathews MB. A mechanism for the control of protein synthesis by adenovirus VA RNAI. Cell. 1986;44:391–400. doi: 10.1016/0092-8674(86)90460-5. [DOI] [PubMed] [Google Scholar]
- 9.Ma YL, Mathews MB. Structure, function, and evolution of adenovirus-associated RNA: a phylogenetic approach. J. Virol. 1996;70:5083–5099. doi: 10.1128/jvi.70.8.5083-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghadge GD, Swaminathan S, Katze MG, Thimmapaya B. Binding of the Adenovirus VAI RNA to the interferon induced 68 kDa protein kinase correlates with function. Proc. Natl Acad. Sci. USA. 1991;88:7140–7144. doi: 10.1073/pnas.88.16.7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke PA, Pe'ery T, Ma YL, Mathews MB. Structural features of adenovirus 2 virus-associated RNA required for binding to the protein kinase DAI. Nucleic Acids Res. 1994;22:4364–4374. doi: 10.1093/nar/22.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellits KH, Pe'ery T, Mathews MB. Role of the apical stem in maintaining the structure and function of adenovirus virus-associated RNA. J. Virol. 1992;66:2369–2377. doi: 10.1128/jvi.66.4.2369-2377.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mellits KH, Kostura M, Mathews MB. Interaction of adenovirus VA RNAI with the protein kinase DAI – nonequivalence of binding and function. Cell. 1990;61:843–852. doi: 10.1016/0092-8674(90)90194-j. [DOI] [PubMed] [Google Scholar]
- 14.Furtado MR, Subramanian S, Bhat RA, Fowlkes DM, Safer B, Thimmappaya B. Functional dissection of Adenovirus VAI RNA. J. Virol. 1989;63:3423–3434. doi: 10.1128/jvi.63.8.3423-3434.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mellits KH, Mathews MB. Effects of mutations in stem and loop regions on the structure and function of Adenovirus VA RNA1. EMBO J. 1988;7:2849–2859. doi: 10.1002/j.1460-2075.1988.tb03141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghadge GD, Malhotra P, Furtado MR, Dhar R, Thimmapaya B. In vitro analysis of Virus Associated RNA-I (VAI RNA) – inhibition of the double stranded RNA activated protein kinase PKR by VAI RNA mutants correlates with the in vivo phenotype and the structural integrity of the central domain. J. Virol. 1994;68:4137–4151. doi: 10.1128/jvi.68.7.4137-4151.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pe'ery T, Mellits KH, Mathews MB. Mutational analysis of the central domain of Adenovirus Virus-Associated RNA mandates a revision of the proposed secondary structure. J. Virol. 1993;67:3534–3543. doi: 10.1128/jvi.67.6.3534-3543.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahman A, Malhotra P, Dhar R, Kewalramani T, Thimmapaya B. Effect of single base substitutions in the central domain of Virus Associated RNA-I on its function. J. Virol. 1995;69:4299–4307. doi: 10.1128/jvi.69.7.4299-4307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma YL, Mathews MB. Secondary and tertiary structure in the central domain of adenovirus type 2 VA RNA(I) RNA. 1996;2:937–951. [PMC free article] [PubMed] [Google Scholar]
- 20.Wahid AM, Coventry VK, Conn GL. Systematic deletion of the adenovirus-associated terminal stem reveals a surprisingly active RNA inhibitor of double-stranded RNA-activated protein kinase. J. Biol. Chem. 2008;283:17485–17493. doi: 10.1074/jbc.M802300200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersson MG, Haasnoot PCJ, Xu N, Berenjian S, Berkhout B, Akusjarvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J. Virol. 2005;79:9556–9565. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma YL, Mathews MB. Comparative analysis of the structure and function of Adenovirus virus associated RNAs. J. Virol. 1993;67:6605–6617. doi: 10.1128/jvi.67.11.6605-6617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coventry VK, Conn GL. Analysis of adenovirus VA RNA(I) structure and stability using compensatory base pair modifications. Nucleic Acids Res. 2008;36:1645–1653. doi: 10.1093/nar/gkn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker SC, Avis JM, Conn GL. General plasmids for producing RNA in vitro transcripts with homogeneous ends. NAR Methods. 2003;31:e82. doi: 10.1093/nar/gng082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pe'ery T, Mathews MB. Synthesis and purification of single-stranded RNA for use in experiments with PKR and in cell-free translation systems. Methods. 1997;11:371–381. doi: 10.1006/meth.1996.0435. [DOI] [PubMed] [Google Scholar]
- 26.Ray PS, Jia J, Yao P, Majumder M, Hatzoglou M, Fox PL. A stress-responsive RNA switch regulates VEGFA expression. Nature. 2009;457:915–919. doi: 10.1038/nature07598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenna SA, Lindhout DA, Shimoike T, Puglisi JD. Biophysical and biochemical investigations of dsRNA-activated kinase PKR. Meth. Enzymol. 2007;430:373–396. doi: 10.1016/S0076-6879(07)30014-1. [DOI] [PubMed] [Google Scholar]
- 28.McKenna SA, Kim I, Liu CW, Puglisi JD. Uncoupling of RNA binding and PKR kinase activation by viral inhibitor RNAs. J. Mol. Biol. 2006;358:1270–1285. doi: 10.1016/j.jmb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Manche L, Green SR, Schmedt C, Mathews MB. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat. Rev. Cell Mol. Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 31.Conn GL, Draper DE. RNA structure. Curr. Opin. Struct. Biol. 1998;8:278–285. doi: 10.1016/s0959-440x(98)80059-6. [DOI] [PubMed] [Google Scholar]
- 32.Marzi S, Fechter P, Chevalier C, Romby P, Geissmann T. RNA switches regulate initiation of translation in bacteria. Biol. Chem. 2008;389:585–598. doi: 10.1515/bc.2008.055. [DOI] [PubMed] [Google Scholar]
- 33.Huthoff H, Berkhout B. Two alternating structures of the HIV-1 leader RNA. RNA. 2001;7:143–157. doi: 10.1017/s1355838201001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ooms M, Verhoef K, Southern E, Huthoff H, Berkhout B. Probing alternative foldings of the HIV-1 leader RNA by antisense oligonucleotide scanning arrays. Nucleic Acids Res. 2004;32:819–827. doi: 10.1093/nar/gkh206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolff T, Bindereif A. Conformational changes of U6 RNA during the spliceosome cycle: an intramolecular helix is essential both for initiating the U4-U6 interaction and for the first step of splicing. Gen. Dev. 1993;7:1377–1389. doi: 10.1101/gad.7.7b.1377. [DOI] [PubMed] [Google Scholar]
- 36.McManus CJ, Schwartz ML, Butcher SE, Brow DA. A dynamic bulge in the U6 RNA internal stem-loop functions in spliceosome assembly and activation. RNA. 2007;13:2252–2265. doi: 10.1261/rna.699907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akusjarvi G, Mathews MB, Andersson P, Vennstrom B, Pettersson U. Structure of genes for virus-associated RNAI and RNAII of adenovirus type-2. Proc. Natl Acad. Sci. USA. 1980;77:2424–2428. doi: 10.1073/pnas.77.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elde NC, Child SJ, Geballe AP, Malik HS. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature. 2009;457:485–488. doi: 10.1038/nature07529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.