Abstract
Modified thrombin-binding aptamers carrying 2′-deoxyguanine (dG) residues with locked North- or South-bicyclo[3.1.0]hexane pseudosugars were synthesized. Individual 2′-deoxyguanosines at positions dG5, dG10, dG14 and dG15 of the aptamer were replaced by these analogues where the North/anti and South/syn conformational states were confined. It was found that the global structure of the DNA aptamer was, for the most part, very accommodating. The substitution at positions 5, 10 and 14 with a locked South/syn-dG nucleoside produced aptamers with the same stability and global structure as the innate, unmodified one. Replacing position 15 with the same South/syn-dG nucleoside induced a strong destabilization of the aptamer, while the antipodal North/anti-dG nucleoside was less destabilizing. Remarkably, the insertion of a North/anti-dG nucleoside at position 14, where both pseudosugar conformation and glycosyl torsion angle are opposite with respect to the native structure, led to the complete disruption of the G-tetraplex structure as detected by NMR and confirmed by extensive molecular dynamics simulations. We conclude that conformationally locked bicyclo[3.1.0]hexane nucleosides appear to be excellent tools for studying the role of key conformational parameters that are critical for the formation of a stable, antiparallel G-tetrad DNA structures.
INTRODUCTION
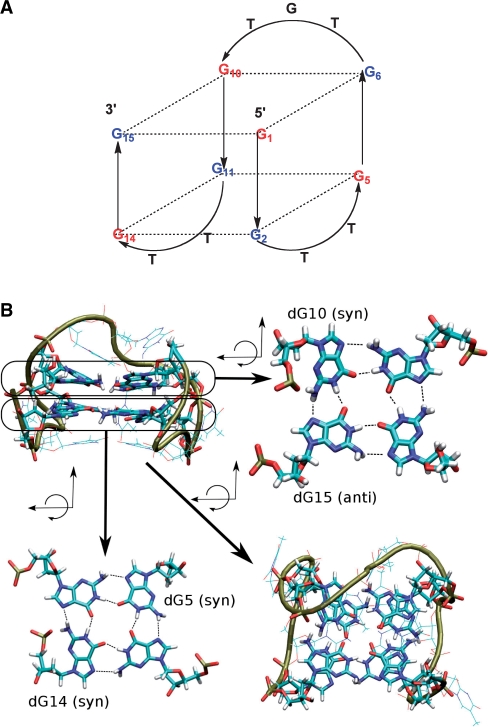
Aptamers are oligonucleotides that were originally derived from an in vitro evolution process known as SELEX (systematic evolution of ligands by exponential enrichment) (1,2), which selects them on the basis of their specific and tight binding affinity to a ligand of choice from a library of sequences. Through this approach, large number of aptamers with very high affinity have been developed for diagnostic, therapeutic and other technical applications (3), but there is still room for improvement in terms of increasing their binding properties and stability to nucleases (4). One of the most studied aptamers is the 15-base long thrombin-binding aptamer (TBA), 5′-G1G2T3T4G5G6T7G8T9G10G11T12T13G14G15-3′ (5). This oligonucleotide binds specifically to thrombin at nanomolar concentrations, and for this reason it has interesting anticoagulant properties (5,6). TBA is characterized (both in the thrombin bound and unbound forms) by a chair-like, antiparallel quadruplex structure consisting of two G-tetrads connected by two TT loops and a single TGT loop (7–11) (Figure 1). The antiparallel quadruplex structure of TBA has a distinctive denaturation–renaturation profile that is reversible and observable by different techniques, particularly by NMR experiments, which suggests that the denaturation of the quadruplex occurs by opening of the G–G base pairs that are not protected by a loop, followed by the opening the TGT loop (12). The conformational distribution of 2′-deoxyguanosines in the G-quartets of the TBA aptamer is well defined. All sugar puckers are South (S), while the guanines on the same G-quartet plane display alternating syn/anti conformations with respect to the glycosyl torsion angle (syn-dG at positions dG1, dG5, dG10 and dG14; anti-dG at positions dG2, dG6, dG11 and dG15, Figure 1). It is worth noting that despite the robust stability of the intramolecular quadruplex structure, alternative intermolecular tetraplexes are possible at high aptamer concentration, as detected by CD and electrophoresis migration experiments (13).
Figure 1.
(A) Schematic representation of the antiparallel folding of the thrombin-binding aptamer (TBA); blue represents guanines that are anti and red guanines that are syn. Positions of 5, 10, 14 and 15 were modified in this study. (B) Molecular representation: the upper left quadrant shows a lateral view of the TBA and the lower right quadrant a view from the top of the molecule [dG5(syn) and dG14(anti) in the lower quartet as well as dG10(syn) and dG15(syn) in the upper quartet are indicated].
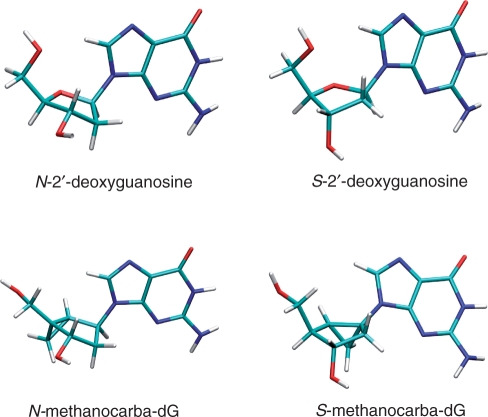
Historically, several modified TBAs have been synthesized containing different types of nucleobase modifications introduced by 2′-deoxyinosine (8), 2′-deoxy-6-thioguanosine (14) and 8-amino-2′-deoxyguanosine (15) residues. Other modifications included the insertion of 5′–5′ inversion sites (16,17), the incorporation of locked nucleic acids, LNA (18,19), 2′-deoxy-2′-fluoro-D-arabinonucleic acids, 2′-F-ANA (20) and ribonucleotides (21). Many of these modified nucleotides allowed a certain control of the sugar ring pucker, favouring either an RNA-like C3′-endo (North, N) conformation or a DNA-like C2′-endo (South, S) conformation. Use of modified LNAs (22–26) has helped to achieve complete conformational control due to the presence of a methylene bridge between the 2′-oxygen and the 4′-carbon (27). This bridge creates a rigid bicyclic structure where the embedded furanose ring is ‘locked’ in the N conformation. Prior to LNAs, the first synthesis of a conformationally locked nucleoside analogue was based on a carbocyclic bicyclo[3.1.0]hexane template (28). This template was later used in the construction of a single DNA strand carrying several N-locked methanocarba thymidine units which hybridized efficiently with its complementary RNA strand (29) and the synthesis of other oligonucleotides carrying N- and S-locked versions of 2′-deoxymethanocarba adenosine, methanocarba thymidine and abasic sites (30,31). An advantage of this methanocarba nucleoside system over LNAs is that both N- and S-locked platforms can be prepared by shifting the position of the fused cyclopropane ring (29) (Figure 2). Later, additional data suggested that the addition of N-locked methanocarba nucleosides to a standard B-DNA duplex could cause bending towards the minor groove (32,33), while incorporation of S-locked methanocarba nucleosides into a similar standard DNA produced a more complicated equilibrium between different duplex forms, where the major component was a duplex with a slightly higher thermodynamic stability than the unmodified duplex (34).
Figure 2.
Effect of the fused cyclopropane ring in locking the N- and S-conformations of the embedded cyclopentane ring of methanocarba-dG nucleosides closely mimicking the conformations of natural N- and S-2′-deoxyguanosines. For the sake of clarity only anti conformers are shown.
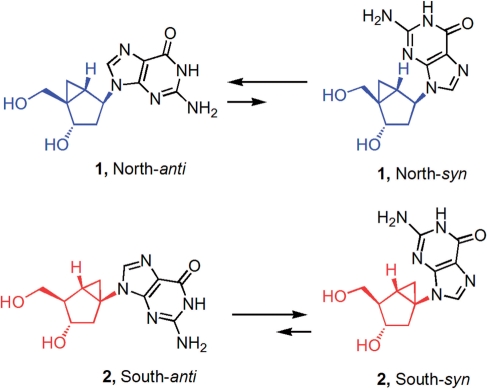
In this article, we describe the effects of replacing a single 2′-deoxyguanosine residue of the TBA (positions dG5, dG10, dG14 and dG15; see Figure 1) with methanocarba nucleosides locked in either the N- or S-conformation. These positions were selected to explore the combined effects of a constrained sugar pucker (N or S) and the corresponding biased glycosyl torsion angle (anti or syn) associated with a particular pseudosugar conformation. In conventional nucleosides, the glycosyl torsion angle is weakly associated with a specific sugar pucker: the syn conformation is associated with a S-sugar pucker, while for the anti conformation both N- and S-sugar puckers are equally probable (35). For methanocarba nucleosides, these associated preferences are enhanced due to the steric consequences introduced by the rigid pseudosugar ring and its effect on the rotation of the glycosyl bond. Indeed, both experimental and theoretical results [see (36) and results below] indicate that a N-pseudosugar conformation favours the anti glycosyl orientation, whereas the S-pseudosugar conformation favours the syn disposition of the base. Therefore, we anticipated that the introduction of methanocarba nucleosides with locked-N(anti) and locked-S(syn) conformations could fix the conformational state of these nucleosides and help us understand the impact of conformational restrictions on the antiparallel, G-quartet DNA structure of TBA (Figure 3).
Figure 3.
Biased anti-syn equilibrium of conformationally restricted North (blue) and South (red) methanocarba-dG nucleosides.
We thus prepared six modified TBA derivatives: (i) TBA-dG5S, (ii) TBA-dG10S, (iii) TBA-dG14S carrying the S-pseudosugar (same sugar puckering and syn glycosyl bond orientation as in unmodified TBA); (iv) TBA-dG15S carrying the same S-pseudosugar at position 15 (same sugar puckering as in native TBA, but opposite syn glycosyl conformation); (v) TBA-dG14N carrying the N-pseudosugar at position 14 (opposite sugar puckering and opposite glycosyl conformation relative to native TBA) and (vi) TBA-dG15N carrying the N-pseudosugar at position 15 (opposite sugar puckering but similar anti glycosyl conformation as in normal TBA) (5). The substitution at position 14 of the aptamer with a locked S-dG produced a modified aptamer with the same stability and global structure as the unmodified aptamer as confirmed by NMR and molecular dynamics simulations. Replacing position 15 for a similarly locked S-dG induced a strong destabilization of the aptamer, while the antipodal N-dG produced a somewhat lower destabilization; however, in both cases the global structure was reasonably preserved. In sharp contrast, the substitution at position 14 with N-dG produced a dramatic disruption in the aptamer structure, which was clearly observable by NMR and state-of-the-art molecular dynamics simulations. This last observation clearly shows that the combination of the wrong sugar pucker and the wrong anti conformation at this position is largely destabilizing to the entire tetraplex architecture.
The structural properties of these locked pseudosugars are therefore useful for exploring the role of the sugar pucker and the glycosyl bond orientation in the stabilization of G-quartet DNA structures.
MATERIALS AND METHODS
General methods of chemical synthesis
The general methods of synthesis and the specific preparation and characterization of all the compounds in Schemes 1 and 2 are included in the Supplementary Data.
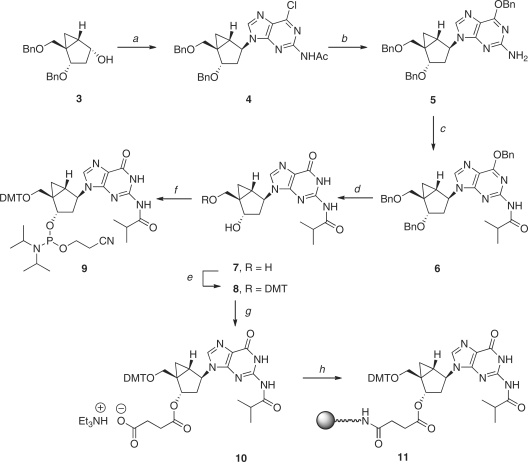
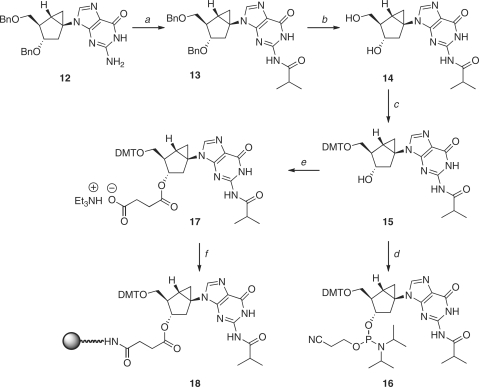
Scheme 1.
Reagents and conditions: (a) PPh3, DIAD, 2-acetamide-6-chrolopurine, THF, 0°C, 73%; (b) BnONa, BnOH, rt, 89%; (c) isobutyryl chloride, pyridine, rt, 84%; (d) BCl3, CH2Cl2, –78°C, 69%; (e) DMTr-Cl, pyridine, rt, 83%; (f) Diisopropylethylamine, 2-cyanoethyl diisopropylchlorophosphoramidite, CH2Cl2 (g) Succinic anhydride, Et3N, DMAP, CH2Cl2, rt, 81%; (h) i. LCAA-CPG, DCC, CH2Cl2; ii. pyridine:Ac2O (9:1, v/v), DMAP.
Scheme 2.
Reagents and conditions: (a) TMS-Cl, isobutyryl chloride, pyridine, rt, 84%; (b) H2, 10% Pd-C, MeOH, rt, 95%; (c) DMTr-Cl, Et3N, pyridine, rt, 83%; (d) Diisopropylethylamine, 2-cyanoethyl diisopropylchlorophosphoramidite, CH2Cl2; (e) Succinic anhydride, Et3N, DMAP, CH2Cl2, rt, 81%; (f) (1) LCAA-CPG, EDCl, Et3N, DMAP, pyridine (2) pyridine:Ac2O (9:1, V/V), DMAP.
Oligonucleotide synthesis and purification
The syntheses of the oligonucleotides TBA: d(5′-GGT TGG TGT GGT TGG-3′); TBA-dG15N: d(5′-GGT TGG TGT GGT TGG-3′), G: N-dG; TBA-dG15S: d(5′-GGT TGG TGT GGT TGG-3′), G: S-dG, TBA-dG14S: d(5′-GGT TGG TGT GGT TGG-3′), G: S-dG, TBA-dG14N: d(5′-GGT TGG TGT GGT TGG-3′), G: N-dG, dG5S: d(5′-GGT TGG TGT GGT TGG-3′), G: S-dG and dG10S: d(5′-GGT TGG TGT GGT TGG-3′), G: S-dG are described in the Supplementary Data.
NMR spectroscopy
The samples of TBA derivatives for NMR measurements were dissolved in 500 μl H2O/D2O (9:1) or D2O containing 10 mM potassium phosphate buffer and 5 mM KCl (pH 6.9), corresponding to a G-quadruplex concentration of 0.8 mM. NMR spectra were performed at temperatures ranging between 5°C and 65°C on a Bruker AV-600 spectrometer, equipped with a z-gradient triple resonance cryoprobe and processed with TOPSPIN v. 1.3. Chemical shifts (δ) were measured in ppm. 1H NMR spectra were referenced to external DSS (2,2-dimethyl-2-silapentane-5-sulfonate sodium salt) set at 0.00 ppm. 1D and 2D spectra were recorded, in the case of D2O solutions, with water suppression obtained by presaturation pulse sequences, while, in the case of H2O/D2O (9:1) solutions, using a gradient-based solvent suppression. Two-dimensional homonuclear correlation spectra NOESY (37), and TOCSY (38,39) were acquired using standard pulse sequences in the phase-sensitive mode at 5°C (mixing times and spin lock values of 150 ms and 60 ms, respectively). The program Sparky (40) was used to assign the NOESY cross-peaks.
Thermal denaturation experiments of aptamers using UV spectroscopy
The thermal melting curves were performed following the absorption change at 295 nm from 20°C to 80°C with a linear temperature ramp of 0.5°C/min on a JASCO V-650 spectrophotometer equipped with a Peltier temperature control. All measurements were done in triplicate in 10 mM sodium cacodylate buffer and 100 mM KCl (pH 6.9). The concentration of the samples was 6 μM.
Thermal denaturation experiments of aptamers using circular dichroism (CD)
CD spectra were obtained following the change of ellipticity from 220 nm to 320 nm at different temperatures on a Jasco spectropolarimeter equipped with a Peltier temperature control used to set the temperature between 5°C and 75°C. The changes in ellipticity versus temperatures at λmax were plotted and used to obtain the melting temperature. All measurements were conducted in 10 mM potassium phosphate buffer and 5 mM KCl (pH 6.9). The concentration of the samples was 80 μM.
Computational methods
Conformational study of methanocarba 2′-deoxyguanosine derivatives
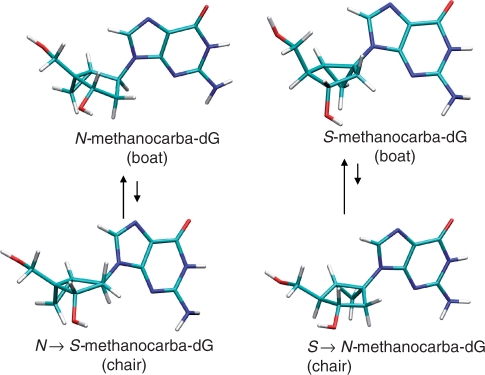
It is expected that the fusion of the cyclopropane ring through 4′ (using ribose numbering) leads to N-type conformers, while its fusion through 1′ leads to the antipodal S-type conformers. However, analysis of the structures show that such preferences could change due to the ‘boat’ ⇆ ‘chair’ equilibrium of the six member ring (Figure 4). Although electron diffraction microwave spectroscopy (41), X-ray crystallography (22,42,43), as well ab initio calculations (44–47) have demonstrated that the boat form is the preferred conformation in a free-standing bicyclo[3.1.0]hexane system, no definitive evidence on the thermodynamics of the ‘boat’ ⇆ ‘chair’ equilibrium of G-nucleosides in solution and in nucleic acid environment exists. To clarify this crucial point, we created models of 2′-deoxyguanosine, N-methanocarba-dG and S-methanocarba-dG in the syn and anti conformations and evaluated their relative energy with high-level quantum mechanical calculations. For 2′-deoxyguanosine, both N and S puckerings were considered, while for N- and S-methanocarba-dG both ‘boat’ and ‘chair’ conformers were studied. In the case of the ‘chair’ conformers, it is important to recognize that in shifting from ‘boat’ to ‘chair’, the puckering of the embedded cyclopentane ring inverts the pseudosugar pucker from N to S or vice versa (Figure 4). The rest of the internal parameters for the starting geometries (including the C4–C5′ rotation) were taken from those found in a typical G-quartet DNA structure and the starting models were optimized at the B3LYP/6-31G(d) level of theory (48). No restrictions were introduced in the calculations, except for N-methanocarba-dG, where the ‘chair’ conformation needed to be reinforced to avoid spontaneous conformational transitions. Solvent effects were introduced using B3LYP/MST calculations (49–54).
Figure 4.
Change in sugar puckering in the embedded cyclopentane ring when the bicyclo[3.1.0]hexane system is allowed to change from its stable ‘boat’ conformation to the unstable ‘chair’ conformation.
Parametrization of the methanocarba-2′-deoxyguanine derivatives for classical simulations
Classical calculations on the methanocarba-dG derivatives required suitable force-field parameters. Van der Waals, stretching and bending parameters were taken by homology with AMBER force field for nucleic acids (55). Atomic charges were derived from standard RESP/6-31G(d) methodology (56). Torsions not available in the latest version of the AMBER force field were derived by fitting to quantum chemical calculations at the B3LYP/6-31G(d) level. For this purpose, different ring conformations and glycosyl torsion angles were generated using appropriate dihedral restrictions. The corresponding energy differences were fitted by adding suitable torsional parameters to the standard AMBER force field (see below) using an ‘in house’ Monte Carlo fitting procedure (57). Parmbsc0-adapted (57) parameters for the methanocarba-guanine derivatives are available upon request.
Molecular dynamics (MD) simulations
MD simulations were used to evaluate the structural impact of the presence of methanocarba-dG (N and S) derivatives at the different positions in the aptamer, namely: TBA-dG14N, TBA-dG15N, TBA-dG14S and TBA-dG15S. We modelled five aptamers, one containing only the natural nucleotides and the remaining containing the different methanocarba-dG derivatives using as template the TBA structure derived by NMR (PDB code 148D (9). Starting geometries were neutralized by adding enough sodium ions (including those needed in the central G-channel) solvated with ∼3500 water molecules, optimized, thermalized and equilibrated using our standard multi-stage protocol (57) of doubling the length of individual steps to guarantee stability. Final structures were equilibrated for one additional nanosecond. A short 500 ps run was then used to derive 10 configurations (of the five structures) from which we generated (after velocity randomization and re-equilibration) 10 independent production trajectories that were extended for 10 ns. In cases where all trajectories sampled the same region of the conformational space, they were combined to generate a meta-trajectory. In cases where large conformational transitions happened in same cases, individual trajectories were treated independently.
Interactions between atoms were described by the parmbsc0 refinement of the parm99 force field (55) and TIP3P model (58). The new required parameters were derived as mentioned above. All simulations were carried out using the Gromacs-4 software (59), with periodic boundary conditions and the particle mesh Ewald method (60) for long-range electrostatic treatment and a cut-off radius of 14 Å for short-range repulsive and attractive dispersion interactions, modelled via a Lennard-Jones potential. The Settle algorithm (61) was used to constrain bond lengths and angles of water molecules, and P-LINCS (62) was used for all other bond lengths, allowing an integration time step of 2 fs. The temperature of the simulation was kept constant at 300 K using the uncoupled thermostat strategy of Bussi et al. (63). The pressure of the systems was kept constant by weak isotropic coupling to a pressure bath of 1 atm (64). All simulations were carried out at the BSC-MareNostrum supercomputer.
RESULTS AND DISCUSSION
Conformational analysis
Conformational analysis of methanocarba-dG (N and S) derivatives at the ab initio level showed, as expected, that the structures displayed a clear ‘boat’ preference over the ‘chair’ conformation (Table 1) and, accordingly, the ‘puckering’ conformation appears strictly dictated by the placement of the cyclopropane ring in the bicyclo[3.1.0]hexane scaffold as experimentally suggested for related compounds (36,42). The only case where the boat ⇆chair equilibrium might have some impact is when the S-pseudosugar is forced to the anti conformation (Table 1), since then the boat/chair difference is not too large (3.1 versus 4.2 kcal/mol in aqueous solution). As experimentally suggested (36,42), in both the solid state and in solution the N-pseudosugar favours the anti conformation, while the S-pseudosugar is biased towards the syn conformation. In both cases, the difference found was quite significant (3–4 kcal/mol) indicating that the population of the minor conformer in solution would be negligible (Table 1). Thus, the S-pseudosugar template provides a conformation that is closer to that of 2′-deoxyguanosine at position 14 of the TBA. Since it is clear that S-pseudosugar is more rigid than the 2′-deoxyribose ring, S → N or syn → anti transitions are more difficult (Table 1), and hence, both pseudosugar and glycosyl conformations are considered virtually locked (Table 1).
Table 1.
Calculated relative energies of different conformers of N- and S-2′-deoxyguanosine and methanocarba-dG derivatives in the gas phase and in aqueous solution (see ‘Materials and Methods’ section)
| Sugar conformer | Glycosyl torsion | ΔE (vac) (kcal/mol) | ΔE (water) (kcal/mol) |
|---|---|---|---|
| methanocarba-dG-boat (N) | syn | 0.2 | 3.7 |
| methanocarba-dG-boat (N) | anti | 0.0 | 0.0 |
| *methanocarba-dG-chair (N → S) | syn | 5.3 | 7.1 |
| *methanocarba-dG-chair (N → S) | anti | 4.4 | 4.7 |
| methanocarba-dG-boat (S) | syn | 0.0 | 0.0 |
| methanocarba-dG-boat (S) | anti | 8.6 | 3.1 |
| *methanocarba-dG-chair (S → N) | syn | 6.6 | 4.8 |
| *methanocarba-dG-chair (S → N) | anti | 12.5 | 4.2 |
| 2′-deoxyguanosine N | syn | 2.4 | 2.7 |
| 2′-deoxyguanosine N | anti | 7.2 | 3.3 |
| 2′-deoxyguanosine S | syn | 0.0 | 0.0 |
| 2′-deoxyguanosine S | anti | 6.4 | 2.2 |
The conformation around the ‘C4′–C5′ bond’ found during optimization is shown (using by homology the ribose nomenclature). All values are referred to the most stable conformer found in our analysis, and they cannot be compared between different molecules. Star-labelled methanocarba-dG ‘chair’ derivatives were minimized under dihedral constraints to preserve the chair conformation. In these compounds the change from ‘boat’ to ‘chair’ inverts the pseudorotational position of the embedded cyclopentane from N to S and vice versa.
A word of caution is necessary when extrapolating the conformational preferences of isolated nucleosides to those inside the TBA. However, our theoretical results, as well as experimental data (36,42), strongly suggest that the preferred pseudosugar puckerings will be in the N region for N-methanocarba-2′-dG and in the S hemisphere for the S-methanocarba-2′-dG. The N compound is likely to be quite unstable in the syn region and will be found mostly in the anti conformation; the reverse is true for the S-compound. Incorporation of the modified methanocarba derivatives at positions of the G-DNA that do not match the conformation of the native structure are thus expected to have a negative impact on stability (and perhaps on the global structure) of the TBA (vide infra).
Synthesis of pseudonucleoside derivatives required for oligonucleotide synthesis
The syntheses of plain methanocarba nucleosides N-dG (1) and S-dG (2) have been reported (43,65). The required steps to prepare the corresponding N-dG phosphoramidite 9 and the CPG-loaded, N-dG solid support (11) are shown in Scheme 1. Under Mitsunobu conditions, the known precursor 3 (43) reacted with 2-acetamido-6-chloropurine to give the desired coupled product 4 in 73%. Displacement of the 6-chloro group with sodium benzylate provided compound 5; this reaction occurred with the simultaneous removal of the N-acetyl group. Protection of the free 2-amino group as the N-isobutyryl amide 6 was followed by the removal of all benzyl groups with BCl3 to give compound 7. From this point onward, conventional 5′-OH protection as the DMT ether 8 and phosphitylation with 2-cyanoethyl diisopropylchlorophosphoramidite provided the desired phosphoramidite building block 9. From compound 8, coupling with succinic anhydride gave the 3′-O-succinyl derivative 10 which was then loaded onto the CPG resin to give 11.
For the synthesis of the antipodal, S-dG phosphoramidite 16 and the CPG-loaded S-dG solid support (18), we started with the known 3′,5′-bis-O-benzyl protected compound 12 (Scheme 2) (65). As before, formation of the N-isobutyryl amide 13 and removal of all benzyl groups by catalytic hydrogenation provided the key intermediate 14, which was converted in a similar manner as before to the desired phosphoramidite building block (16) and the 3′-O-succinyl derivative 17, which was loaded onto the CPG resin to give 18.
In both cases, the phosphitylation reaction was slower than the comparable reaction with natural nucleosides and the yields were moderate. The resulting phosphoramidites 9 and 16 were stable and could be purified on silica gel and stored for a long time without the loss of reactivity.
Oligonucleotide synthesis
Oligonucleotide synthesis was performed using controlled pore glass (CPG) supports. The addition of the locked, methanocarba nucleoside phosphoramidites was performed manually using 10–15 mg of the appropriate monomers with an extended coupling time of 10 min. The rest of the sequences were assembled using an automated DNA synthesizer. After addition of the N-methanocarba-dG phosphoramidite, oxidation of phosphites was performed with a t-butylhydroperoxide (tBuOOOH) solution as described elsewhere (31) to avoid iodine degradation of the phosphodiester backbone at the pseudosugar position. Using these optimized synthesis conditions, all modified oligonucleotides were successfully synthesized and characterized by HPLC analysis of enzyme digestions and mass spectrometry. The isolated yields after HPLC purification and desalting were in the range of those obtained for unmodified oligonucleotides.
Thermal stability by CD and UV spectroscopy
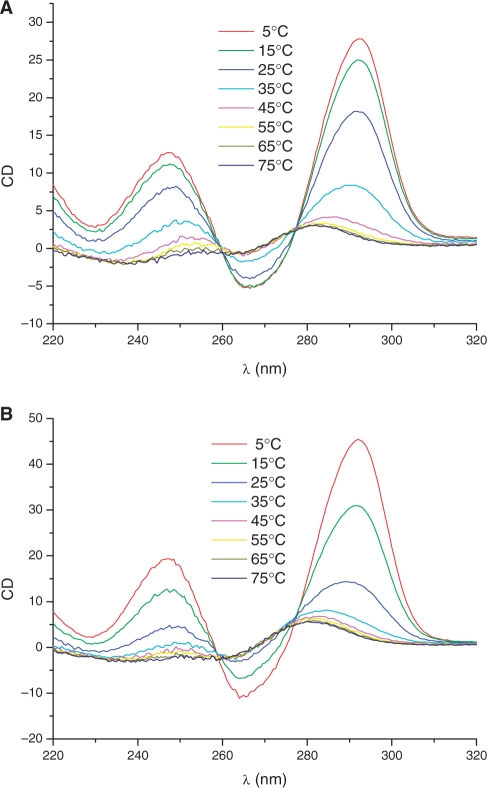
In order to define the effects of the modifications imposed by the rigid pseudosugar rings, melting curves of the modified samples were performed by CD and UV spectroscopy and compared with the unmodified sequence. The appearance of the CD spectra was consistent with an antiparallel G-quartet architecture characterized by a positive band at 248 nm, a positive maximum at 295 nm and a negative maximum at 265 nm (Figure 5). The CD spectrum of a parallel quadruplex in contrast presents a positive maximum near 265 nm. A reversal in topology from antiparallel to parallel folding can be induced by the selective substitution of 2′-deoxyguanosine for riboguanosine (20), which can be followed by changes in the CD spectra.
Figure 5.
(A) CD spectra of TBA-d15GN at different temperatures. (B) CD spectra of TBA-d15GS at different temperatures.
In our experiments, CD spectra of native and modified TBA derivatives presented a maximum at 295 nm at all temperatures indicating an antiparallel quadruplex structure for all the TBA derivatives studied in this work. The melting temperatures for the TBA-dG5S, TBA-dG10S, TBA-dG14S, TBA-dG15N and TBA-dG15S were 44.7°C, 40.7°C, 45°C, 28.4°C and 17°C, respectively (Table 2), indicating a lower thermal stability of the two quadruplexes modified at position 15, but a similar stability as native TBA (Tm 46°C) for the quadruplexes carrying the same sugar pucker and syn glycoyl bond orientation present in the unmodified TBA (TBA-dG5S, TBA-dG10S, TBA-dG14S). TBA-dG14N instead gave a monotonous decrease of the CD signal at 295 nm without apparent Tm. A similar result was obtained when the melting curves were followed by UV (295 nm) at a concentration of 6 μM (UV experiment, Table 2) instead of 80 μM (CD experiment).
Table 2.
Melting temperatures of thrombin binding aptamers carrying methanocarba-dG nucleosides
| Sequence | Tm (°C), CD (80 μM)a | Tm (°C), UV (6 μM)b |
|---|---|---|
| TBA | 46.0 | 48.6 |
| TBA-dG15N | 28.4 | 35.0 |
| TBA-dG15S | 17.0 | 22.1 |
| TBA-dG14S | 45.0 | 47.3 |
| TBA-dG14N | n.d.c | n.d.c |
| TBA-dG5S | 44.7 | 47.3 |
| TBA-dG10S | 40.7 | 42.2 |
a5 mM KCl, 10 mM potassium phosphate buffer pH 6.9.
b100 mM KCl, 10 mM sodium cacodylate buffer pH 6.9.
cNot determined because curves were not cooperative and only a fraction of the transition (if any) were observed.
The combined results from the CD and UV studies (Table 2) indicate that relative to the Tm of the unmodified TBA (45–46°C), only TBA-dG5S, TBA-dG10S and TBA-dG14S, in which the modified methanocarba derivative mimics the native's S sugar pucker and the syn conformation of the guanine ring, generate G-quartets of similar stability to the unmodified TBA. The totally opposite arrangement represented by TBA-dG14N, where the TBA's conformational preferences at position 14 are completely reversed, was very destabilizing. The other combinations, representing a mismatch of either pseudosugar pucker or glycosyl torsion angle, relatively to the native TBA, induced different degrees of destabilization. The data support the claim that a reversal in the glycosyl torsion angle appears to be more critical for the stability of the G-quartet than a mismatch of the sugar pucker, since the drop in thermal stability was about 10°C more severe for TBA-dG15S than for TBA-dG15N. The structural impact that the introduction of both types of distortions has on the global architecture of the corresponding tetraplexes is discussed below in detail as ascertained by NMR experiments and MD simulations.
1H NMR studies
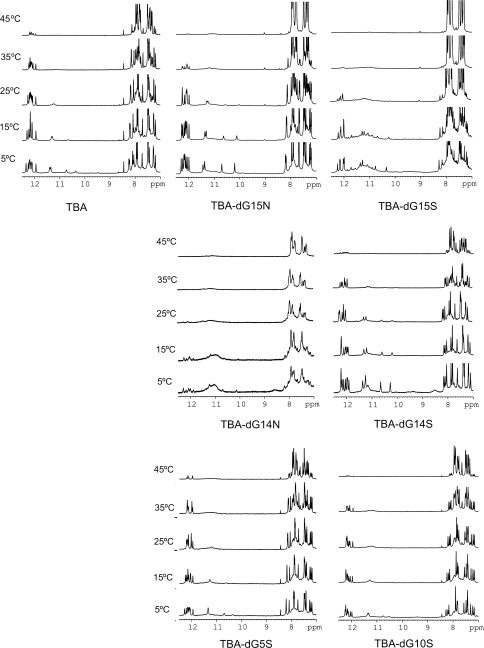
The imino proton region of the NMR spectra of the modified aptamers clearly indicates the formation of quadruplex structures. In the case of TBA-dG15N, TBA-dG14S, TBA-dG5S and TBA-dG10S, a single set of sharp and well-dispersed imino signals are observed between 10.0 and 12.5 ppm (Figure 6). These spectra are consistent with a single G-quadruplex structure similar to that of the unmodified aptamer (9). In contrast, the exceeding number of imino signals in the NMR spectra of TBA-dG15S at 5°C suggests the presence of several species in equilibrium. The broad band observed around 11.0 ppm may arise from a higher molecular weight species or from conformational heterogeneity due to multiple low populated conformers. Nevertheless, the set of sharp and more intense imino signals in the 1D spectra and the NOESY cross-peaks of this modified aptamer (data not shown) present features that closely resemble the spectra of the unmodified TBA, indicating that the major conformer of TBA-dG15S adopts a structure similar to the one of the unmodified TBA. The NMR spectra of TBA-dG14N at 5°C shown in Figure 6 also present a mixed of sharp signals and a large broad band around 11.0 ppm, but in this cases the sharp signals have a much lower intensity, indicating that this aptamer adopt multiple structures with no major prevailing conformer.
Figure 6.
Imino regions of the NMR spectra of the different TBA aptamers in H2O at different temperatures.
Unfolding transitions were followed by NMR. The melting behaviour of the four modified aptamers is consistent with the thermal stability data obtained by UV and CD spectroscopies. In the case of TBA-dG15N, TBA-dG14S, TBA-dG5S and TBA-dG10S, exchangeable protons are observed at temperatures above the denaturation midpoint, suggesting very slow dissociation kinetics (Figure 6). This effect is also observed in unmodified TBA, but it is more pronounced in the modified TBAs with locked nucleotides in stabilizing positions. This suggests that the presence of a locked nucleotide slows down the denaturation kinetics of the whole quadruplex. Interestingly, the broad signals around 11.0 ppm in TBA-dG15S can also be observed at temperatures quite above its Tm. A broad band is also observed in the case of TBA-dG15N at 45°C. It is tempting to assign these broad signals to the denaturation intermediate hypothesized by Mao and Gmeiner using hydrogen exchange experiments on unmodified TBA in the presence of Sr2+ cations (12). A deceleration of the denaturation kinetics might be a general phenomena of this kind of antiparallel quadruplex containing locked nucleotides, allowing in some cases the observation of NMR signals from denaturation intermediates. The fact that a single modification at the 3′ terminus of the TBA can affect its folding/unfolding process may have an influence in its binding to thrombin (18).
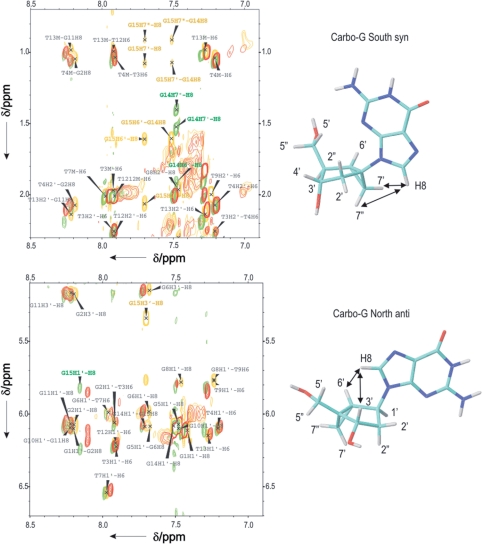
The structures of TBA-dG15N and TBA-dG14S were studied in more detail by bidimensional NMR spectroscopy. The close similarity in their structures and the unmodified TBA is apparent in the NOESY spectra (Figure 7). Assignment of most of the exchangeable and non-exchangeable protons in these two aptamers was carried out on the basis of previously reported assignments of TBA (9) (see Supplementary Data for assignment tables). The pattern of NOEs observed indicated that the global backbone conformation is very similar. The strong intensities of intraresidue NOE cross peaks between the H8 protons of G and H1′ protons confirmed the syn conformation for the glycosyl angle of dG1, dG5, dG10 and dG14. The NOESY spectra of TBA and TBA-dG14S are nearly identical with minimal changes in the chemical shifts of residues around the modification. However, more pronounced changes are observed in TBA-dG15N, like some different NOEs between dG15 and T9 relative to the unmodified aptamer. This could be explained by the rigid conformation of the pseudo sugar that produces a slight local deformation in the G-quartet structure with a different orientation of the T9.
Figure 7.
Selected regions of NOESY spectra of unmodified TBA (red), TBA-dG15N (yellow) and TBA-dG15S (green). Locked-nucleotide resonances are labelled in their respective colour, according to the numbering scheme shown. The conformation of the glycosyl angle can be determined from some key strong NOEs, indicated with arrows.
Assignment of the bicyclo[3.1.0]hexane pseudosugars could be also performed for TBA-dG15N and TBA-dG14S. Overlaid NOESY spectra of these two samples and the unmodified TBA are shown in Figure 7. Bicyclo-hexane spin systems were identified in the TOCSY spectra in D2O. The two additional resonances (H7′/H7′′ according to the naming scheme shown in Figure 7) and the high field shift of these protons (∼1.0–1.5 ppm) allowed for distinguishing the bicyclo-hexane from the deoxyriboses spins systems. Starting from H7′/H7′′ resonances, the remaining bicyclo-hexane protons were assigned by combining through-bond TOCSY connections with through-distances cross-peaks derived from NOESY experiments recorded at a short mixing time. The glycosyl angle of the locked nucleotides can be determined from the intraresidual H8-sugar NOE intensities. Thus, the very strong NOEs between H7′/7′′ and H8 of G14S in TBA-dG14, as well as the medium or weak intensity of the H6′–H8 NOE (see green spectra in Figure 7), clearly indicates the glycosidic conformation is syn. On the other hand, the strong H6′–H8 and H3′–H8 NOEs and the medium or weak H1′–H8 cross-peak (yellow spectra in Figure 7) show that the glycosidic conformation of G15N in TBA-dG15N is anti. This analysis could not be carried out in the case of TBA-dG15S and TBA-dG14N, since the presence of multiple species impedes the complete assignment of the NMR spectra.
Molecular dynamics analysis of natural and modified aptamers
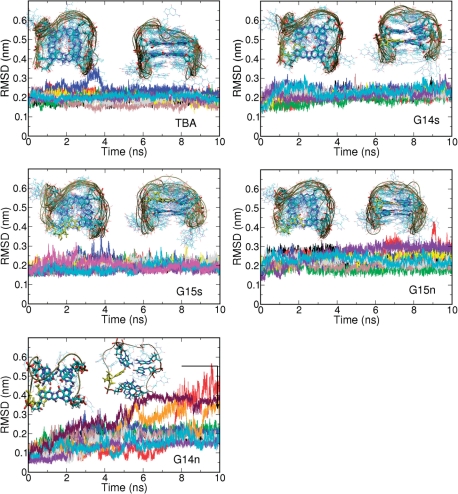
As found in previous MD simulations, which extended to the microsecond range (57), the unmodified TBA behaved like a very stable structure during the 10 × 10 ns simulations (0.1 μs effective simulation time), sampling conformational regions very close to those expected from the NMR structures (Figure 8). One or two ions remained inside the central channel during all the trajectories and the G–G pattern of hydrogen-bond interactions was maintained without distortions for the entire set of simulations. The TBA-dG14S trajectories are in practice identical to those of the unmodified TBA, confirming that the introduction of the S-methanocarba-2′-dG at a position where 2′-dG is found in the S-syn conformation does not have any negative structural impact. The 10 trajectories of TBA-dG15N and the 10 trajectories of TBA-dG15S also sampled conformations not far from those of the unmodified aptamer, in good agreement with NMR measures. Small distortions were observed (data not shown) in the χ angle of the bonding partners dG1 and dG10 and the stacking partner dG14, and in the puckering of dG15. In the case of TBA-dG15N, there were small changes in the χ angle around the neighbours of dG15 (dG1, dG10 and dG14) and the puckering of dG14. It is possible that longer simulations (micro to millisecond long) could lead to the disruption of some of these aptamers (for example TBA-dG15S which shows a Tm below our working temperature), but in general all structures appeared to be reasonably stable, confirming the unmistakable plasticity of the G-DNA.
Figure 8.
Evolution of the RMSD (from NMR structure) in MD simulations performed for different variants of the thrombin-binding aptamer (TBA) containing only 2′-deoxyguanosine or methanocarba-dG nucleotides at selected positions. In cases where the structure is not corrupted, the frontal and top view of the system as observed in the 10 ns simulations is displayed. For the TBA-dG14N system an example of the structure obtained after unfolding is displayed.
Trajectories for the TBA-dG14N aptamer showed a quite different scenario, since ultra-fast unfolding (<10 ns) was observed in 3 of the 10 trajectories, illustrating the large instability of the structure (Figure 8) already predicted from experimental measurements. The unfolded trajectories led to the corruption of the G-quadruplex structure with separation of the terminal guanines, which originally form half of the guanine quartet, in agreement with the mechanism already suggested by NMR experiments (vide supra). During our MD simulations, the unfolded conformation retained the antiparallel hairpin-like structure due to the still existing hydrogen bond interaction between non-terminal guanines and the stabilizing stacking interactions, but the similarity to the original tetrad is completely lost and the structure sampled a much wider configurational space, thus behaving similar to a random coil. Clearly, the MD simulations strongly support the NMR, CD and UV derived models, providing an atomistic view of the structural impact of biasing conformational preferences on the scaffold of the TBA aptamer.
CONCLUSIONS
Conformationally locked N and S 2′-deoxyguanosine nucleosides built on a bicyclo[3.1.0]hexane template (methanocarba nucleosides) were incorporated into TBA aptamers at four specific positions where the 2′-deoxyguanosines display different conformational preferences. Contrary to the situation in B-DNA, where the methanocarba substitutions had a small effect on the global stability of the duplex (32–34), the stability of the TBA aptamer was very dependent on the type and location of the methanocarba-dG nucleotides. The substitution of the native S(syn)dG in TBA with an equivalent, locked S(syn)dG at positions 5, 10 and 14 maintained stability except for a slight decrease on Tm at position 10. Positions 5 and 10 are located in the same bottom G-quartet, but position 10 is located in the upper G-quartet. This result indicates that both quartets are not equivalent and our S(syn)dG analogue accommodates better on the lower quartet.
The destabilization observed when the native S(anti)dG at position 15 was replaced with a S(syn)dG was greater than when it was replaced by a N(anti)dG, supporting the concept that the glycosyl conformation is more restrictive for TBA stability than the sugar puckering. Finally, the substitution of S(syn)dG at position 14 by a locked N(anti)dG led to a dramatic destabilization of the TBA, which according to NMR and MD simulations begins with the structural disruption of the terminal G–G pairs, followed quite closely by the formation of other global conformations that were previously assigned to denaturation intermediates but never observed in the unmodified TBA.
Taken together our theoretical and experimental results establish that both N- or S-methanocarba nucleosides represent uniquely discriminating probes to study the role of North/South puckering and syn/anti conformations on the stability of the antiparallel TBA quadruplex and possibly other aptamers with a similar architecture. These locked analogues also have an impact in the quadruplex formation/denaturation kinetics and helped us visualize a denaturation intermediate whose existence had only been hypothesized.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Education [BFU2007-63287, NAN2004-09415-C05-5, BIO2006-01602, CTQ2007-68014-C02-02, Consolider Escience Project]; the Generalitat de Catalunya [2005/SGR/00693, 2006PIV00028]; the Instituto de Salud Carlos III [CIBER-BBN, CB06/01/0019]; the National Institute of Bioinformatics, the Marcelino Botin foundation, the COST project (G4-net, MP0802), and the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. Funding for open access charge: FUNMOL, FP7-NMP-213382-2.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Brody EN, Gold L. Aptamers as therapeutic and diagnostic agents. Rev. Mol. Biotech. 2000;74:5–13. doi: 10.1016/s1389-0352(99)00004-5. [DOI] [PubMed] [Google Scholar]
- 4.Kusser W. Chemically modified nucleic acid for in vitro selections: evolving evolution. Rev. Mol. Biotech. 2000;74:27–38. doi: 10.1016/s1389-0352(99)00002-1. [DOI] [PubMed] [Google Scholar]
- 5.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 6.Griffin LC, Tidmarsh GF, Bock LC, Toole JJ, Leung LL. In vivo anticoagulant properties of a novel nucleoside-based thrombin inhibitor and demonstration of regional anticoagulation in extracorporeal circuits. Blood. 1993;81:3271–3276. [PubMed] [Google Scholar]
- 7.Wang KY, McCurdy, Shea RG, Swaminathan S, Bolton PH. A DNA aptamer which binds to and inhibits thrombin exhibits a new structural motif for DNA. Biochemistry. 1993;32:1899–1904. doi: 10.1021/bi00059a003. [DOI] [PubMed] [Google Scholar]
- 8.Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl Acad. Sci. USA. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultze P, Macaya RF, Feigon J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 10.Padmanabhan K, Padmanabhan KP, Ferrara JD, Sadler JE, Tulinsky A. The structure of alfa-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J. Biol. Chem. 1993;268:17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- 11.Kelly JA, Feigon J, Yeates TO. Reconciliation of the X-ray and NMR structures of the thrombin-binding aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1996;256:417–422. doi: 10.1006/jmbi.1996.0097. [DOI] [PubMed] [Google Scholar]
- 12.Mao X, Gmeiner WH. NMR study of the folding-unfolding mechanism for the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) Biophys. J. 2005;113:155–160. doi: 10.1016/j.bpc.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Fialova M, Kypr J, Vorlickova M. The thrombin binding aptamer GGTTGGTGTGGTTGG forms a bimolecular guanine tetraplex. Biochem. Biophys. Res. Comm. 2006;344:50–54. doi: 10.1016/j.bbrc.2006.03.144. [DOI] [PubMed] [Google Scholar]
- 14.Marathias VM, Sawicki MJ, Bolton PH. 6-Thioguanine alters the structure and stability of duplex DNA and inhibits quadruplex DNA formation. Nucleic Acids Res. 1999;27:2860–2867. doi: 10.1093/nar/27.14.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez de la Osa J, Gonzalez C, Gargallo R, Rueda M, Cubero E, Orozco M, Aviñó A, Eritja R. Destabilization of quaduplex DNA by 8-aminoguanine. Chem. BioChem. 2006;7:46–48. doi: 10.1002/cbic.200500281. [DOI] [PubMed] [Google Scholar]
- 16.Martino L, Virno A, Randazzo A, Virgilio A, Esposito V, Giancola C, Bucci M, Cirino G, Mayol L. A new modified thrombin binding aptamer containing a 5’-5’ inversion of polarity site. Nucleic Acids Res. 2006;34:6653–6662. doi: 10.1093/nar/gkl915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pagano B, Martino L, Randazzo A, Giancola C. Stability and binding properties of a modified thrombin binding aptamer. Biophys. J. 2007;94:562–569. doi: 10.1529/biophysj.107.117382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Virno A, Randazzo A, Giancola C, Bucci M, Cirino G, Mayol L. A novel thrombin aptamer containing a G-LNA residue. Biorg. Med. Chem. 2007;15:5710–5718. doi: 10.1016/j.bmc.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 19.Bonifacio L, Church FC, Jarstfer MB. Effect of locked-nucleic acid on a biologically active G-quadruplex. A structure-activity relationship of the thrombin aptamer. Int. J. Mol. Sci. 2008;9:422–433. doi: 10.3390/ijms9030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng CG, Damha MJ. G-quadruplex induced stabilization by 2′-deoxy-2′-fluoro-D-arabinonucleic acids (2′F-ANA) Nucleic Acids Res. 2007;35:4977–4988. doi: 10.1093/nar/gkm520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang CF, Shafer RH. Engineering the quadruplex fold: nucleoside conformation determines both folding topology and molecularity in guanine quadruplexes. J. Am. Chem. Soc. 2006;128:5966–5973. doi: 10.1021/ja0603958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez JB, Marquez VE, Nicklaus MC, Mitsuya H, Barchi JJ. Conformationally locked nucleosides analogues. Synthesis of dideoxycarbocyclic nucleosides analogues structurally related to neplanocin C. J. Med. Chem. 1994;37:3389–3399. doi: 10.1021/jm00046a024. [DOI] [PubMed] [Google Scholar]
- 23.Obika S. Development of bridged nucleic acids analogues for antigene technology. Chem. Pharm. Bull. 2004;52:1399–1404. doi: 10.1248/cpb.52.1399. [DOI] [PubMed] [Google Scholar]
- 24.Koshkin AA, Rajwanski VK, Wengel J. Novel convenient syntheses of LNA[2.2.1]bicyclo nucleosides. Tetrahedron Lett. 1998;39:4381–4384. [Google Scholar]
- 25.Renneberg D, Boiliong E, Reber U, Schumperli D, Leumann CJ. Antisense properties of tricycle-DNA. Nucleic Acids Res. 2002;30:2751–2757. doi: 10.1093/nar/gkf412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Opalinska JB, Kalota A, Gifford LK, Lu PZ, Jen KY, Pradeepkumar PI, Barman J, Kim TK, Swider CR, Chatopadhyaya J, et al. Oxitane modified conformationally constrained, antisense oligodeoxyribonucleotides function efficiently as gene silencing molecules. Nucleic Acids Res. 2004;32:5791–5799. doi: 10.1093/nar/gkh893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jepsen JS, Sorensen MD, Wengel J. Locked nucleic acids: A potent nucleic acid analog in therapeutics and biotechnology. Oligonucleotides. 2004;14:130–146. doi: 10.1089/1545457041526317. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez JB, Marquez VE, Nicklaus MC, Barchi JJ. Synthesis of cyclopropane-fused dideoxycarbocyclic nucleosides structurally related to neoplanocin C. Tetrahedron Lett. 1993;34:6233–6236. [Google Scholar]
- 29.Marquez VE, Siddiqui MA, Ezzitouni A, Puss P, Wang JY, Wagner RW, Matteucci MD. Nucleosides with a twist. Can fixed forms of sugar ring pucker influence biological activity in nucleosides and oligonucleotides. J. Med. Chem. 1996;39:3739–3747. doi: 10.1021/jm960306+. [DOI] [PubMed] [Google Scholar]
- 30.Wang P, Nicklaus MC, Marquez VE, Brank AS, Christman JK, Banavali NK, MacKerrell AD., Jr Use of oligodeoxyribonucleotides with conformationally constrained abasic sugar targets to probe the mechanism of base flipping by HhaI DNA (Cytosine C5)-mehtyltransferase. J. Am. Chem. Soc. 2000;122:12422–12434. [Google Scholar]
- 31.Maier MA, Choi Y, Gaus H, Barchi JJ, Marquez VE, Manoharan M. Synthesis and characterization of oligonucleotides containing conformationally constrained bicyclo[3.1.0]hexane pseudosugar analogs. Nucleic Acids Res. 2004;32:3642–3650. doi: 10.1093/nar/gkh667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Z, Madeira M, Barchi JJ, Marquez VE, Bax A. Changes in DNA bending induced by restricting nucleotide ring pucker studied by weak alignment NMR spectroscopy. Proc. Natl Acad. Sci. USA. 2005;102:24–28. doi: 10.1073/pnas.0408498102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madeira M, Wu J, Bax A, Shenoy S, O’Keefe B, Marquez VE, Barchi JJ. Engineering DNA topology with locked nucleosides: A structural study. Nucleosides Nucleotides, Nucleic Acids. 2005;24:687–690. doi: 10.1081/ncn-200060256. [DOI] [PubMed] [Google Scholar]
- 34.Madeira M, Shenoy S, Van QN, Marquez VE, Barchi JJ. Biophysical studies of DNA modified with conformationally constrained nucleotides: comparison of 2′-exo (north) and 3′-exo (south) ‘locked’ templates. Nucleic Acids Res. 2007;35:1978–1991. doi: 10.1093/nar/gkm025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saenger W. Principles of Nucleic Acid Structure. New York, NY: Springer; 1984. [Google Scholar]
- 36.Marquez VE, Ben-Kasus T, Barchi JJ, Jr, Green KM, Nicklaus MC, Agbaria R. Experimental and structural evidence that herpes 1 kinase and cellular DNA polymerase(s) discriminate on the basis of sugar pucker. J. Am. Chem. Soc. 2004;126:543–549. doi: 10.1021/ja037929e. [DOI] [PubMed] [Google Scholar]
- 37.Kumar A, Ernst RR, Wutrich K. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Comm. 1980;95:1–6. doi: 10.1016/0006-291x(80)90695-6. [DOI] [PubMed] [Google Scholar]
- 38.Braunschweiler L, Ernst RR. Coherence transfer by isotropic mixing: application to proton correlation spectroscopy. J. Magn. Reson. 1983;53:521–528. [Google Scholar]
- 39.Redfield AG, Kunz SD. Quadrature Fourier NMR detection-simple multiplex for dual detection and discussion. J. Magn. Reson. 1975;19:250–254. [Google Scholar]
- 40.Goddard TD, Kneller DG. SPARKY 3. San Francisco, USA: University of California; 2004. [Google Scholar]
- 41.Mastryukov VS, Osina EL, Vilkov LV, Hildebrandt RL. The zero-point-average structure of bicyclo[3.1.0]hexane as determined by electron diffraction and microwave spectroscopy. J. Am. Chem. Soc. 1977;99:6855–6861. [Google Scholar]
- 42.Marquez VE, Ezzitouni A, Russ P, Siddiqui MA, Ford H, Feldman RJ, Mitsuya H, George C, Barchi JJ., Jr HIV-1 reverse transcriptase can discriminate between two conformationally locked carbocyclic AZT triphosphate analogues. J. Am. Chem. Soc. 1998;120:2780–2789. [Google Scholar]
- 43.Marquez VE, Russ P, Alonso R, Siddiqui MA, Hernández S, George C, Nicklaus M, Dai F, Ford H. Synthesis of conformationally restricted carbocyclic nucleosides. The role of the O4′-oxigen in the key hydration step of adenosine deaminase. Helv. Chim. Acta. 1999;82:2119–2129. [Google Scholar]
- 44.Mjöberg PJ, Almlöf JA. Conformational study of bicyclo[3.l.0]hexane and the 3-bicyclo[3.l.0]hexane carbonium ion: an ab initio SCF investigation. Chem. Phys. 1978;29:201–208. [Google Scholar]
- 45.Skancke PN. The molecular and electronic structure of bicyclo[l.l.0]butane, bicyclo[2.1.0]pentane, and bicyclo[3.1.0]hexane as obtained by ab initio SCF Calculations. J. Mol. Struct. 1982;86:255–266. [Google Scholar]
- 46.Siam K, Ewbank JD, Schafer L, Van Alsenoy C. A critical comparison of the ab initio geometry and zero-point-average structure of bicyclo[3.l.0lhexane. J. Mol. Struct. 1987;150:121–128. [Google Scholar]
- 47.Okazaki R, Niwa J, Kato S. An ab initio molecular orbital study of the conformations of bicyclo[3.l.0]hexane derivatives. Bull. Chem. Soc. Jpn. 1988;61:1619–1624. [Google Scholar]
- 48.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Vreven T, Kudin KN, Burant JC, et al. Gaussian 03, Revision D.02. Wallingford, CT: Gaussian, Inc.; 2004. [Google Scholar]
- 49.Orozco M, Bachs M, Luque FJ. Development of optimized MST/SCRF methods for semiempirical calculations: the MNDO and PM3 Hamiltonians. J. Comput. Chem. 1995;16:563–575. [Google Scholar]
- 50.Luque FJ, Zhang Y, Aleman C, Bachs M, Gao J, Orozco M. Solvent effects in chloroform solution: parametrization of the MST/SCRF continuum model. J. Phys. Chem. 1996;100:4269–4276. [Google Scholar]
- 51.Luque FJ, Aleman C, Bachs M, Orozco M. Extension of the MST/SCRF method to organic solvents: ab initio and semiempirical parametrization for neutral solutes in CC14. J. Comput. Chem. 1996;17:806–820. [Google Scholar]
- 52.Curutchet C, Orozco M, Luque FJ. Solvation in octanol: parametrization of the continuum MST model. J. Comput. Chem. 2001;22:1180–1193. [Google Scholar]
- 53.Miertus S, Tomasi J. Approximate evaluations of the electrostatic free energy and internal energy changes in solution processes. Chem. Phys. 1982;65:239–245. [Google Scholar]
- 54.Miertus S, Scrocco E, Tomasi J. Electrostatic interactions of a solute with a continuum. A direct utilization of ab initio molecular potentials for the prevision of solvent effects. Chem. Phys. 1981;55:117–129. [Google Scholar]
- 55.Cheatman TE, III, Cieplack P, Kollman PA. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 56.Bayly CI, Cieplack P, Cornell W, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for determining atom-centered charges: The RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- 57.Perez A, Marchan I, Svozil D, Sponer J, Cheatham TE, III, Laugthon CA, Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 59.Hess B, Kutzer C, van der Spoel D, Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 60.Dardem T, York D, Pedersen L. Particle mesh Ewald: an N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 1983;98:10089–10092. [Google Scholar]
- 61.Miyamoto S, Kollman PA. SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 62.Hess B. P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 63.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126 doi: 10.1063/1.2408420. 014101/1–014101/7. [DOI] [PubMed] [Google Scholar]
- 64.Berendsen HJC, Postma JPM, van Gunsteren WF, Dinola A, Hook JR. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 65.Ezzitouni A, Barchi JJ, Jr, Marquez VE. Conformationally locked carbocyclic nucleosides built on a bicyclo[3.1.0]hexane template with a fixed southern conformation. Synthesis and antiviral activity. J. Chem. Soc. Perkin Trans. 1997;I:1073–1078. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.