Abstract
Despite constant threat of oxidative damage, sequence drift in mitochondrial and chloroplast DNA usually remains very low in plant species, indicating efficient defense and repair. Whereas the antioxidative defense in the different subcellular compartments is known, the information on DNA repair in plant organelles is still scarce. Focusing on the occurrence of uracil in the DNA, the present work demonstrates that plant mitochondria possess a base excision repair (BER) pathway. In vitro and in organello incision assays of double-stranded oligodeoxyribonucleotides showed that mitochondria isolated from plant cells contain DNA glycosylase activity specific for uracil cleavage. A major proportion of the uracil–DNA glycosylase (UDG) was associated with the membranes, in agreement with the current hypothesis that the DNA is replicated, proofread and repaired in inner membrane-bound nucleoids. Full repair, from uracil excision to thymidine insertion and religation, was obtained in organello following import of a uracil-containing DNA fragment into isolated plant mitochondria. Repair occurred through single nucleotide insertion, which points to short-patch BER. In vivo targeting and in vitro import of GFP fusions showed that the putative UDG encoded by the At3g18 630 locus might be the first enzyme of this mitochondrial pathway in Arabidopsis thaliana.
INTRODUCTION
Electrons escaping from the respiration and photosynthesis transport chains in mitochondria and chloroplasts, as well as exogenous sources like UV irradiation or environmental stresses, give rise to reactive oxygen species (ROS). These can damage the biomolecules in the cell, and especially the DNA (1). The mitochondrial and the chloroplast genomes are particularly threatened in this respect because they lie in close proximity to the ROS-generating electron transport chains. Cells have developed several lines of defense to counteract the deleterious effects of DNA oxidation. The first defense is to scavenge the ROS through enzymic pathways (e.g. peroxidases) or nonenzymic antioxidant compounds (e.g. ascorbate) (2). The second defense line is to fix the oxidative damage through one of the DNA repair pathways (e.g. base excision repair or BER, nucleotide excision repair or NER) (3). Genomic and functional investigations have characterized DNA repair in the plant nuclear compartment (1) but these pathways remain to be investigated in plant mitochondria and in chloroplasts.
Insufficient protection and/or absence of repair would be expected to lead to rapid sequence drift in plant mitochondrial DNA (mtDNA) and chloroplast DNA (cpDNA), especially following accumulation of oxidative damage. Although there are occasional exceptions (4), genomic data show that on the contrary plant mtDNA and cpDNA coding sequences evolve very slowly, a striking difference versus animal or yeast mtDNA (5). Thus, the data suggest that plant organelles possess very efficient antioxidant defenses and/or a strong ability to repair DNA lesions. Both plant mitochondria and chloroplasts have been shown to possess a number of enzymes involved in ROS scavenging, as well as nonenzymic antioxidant molecules (2). In contrast, there is very limited evidence documenting DNA repair pathways in plant organelles. Analysis of replication efficiencies in unirradiated and UV-irradiated Arabidopsis thaliana leaves suggested the existence of organelle DNA photorepair in plants (6). On the other hand, investigating nuclear control of mtDNA rearrangements in A. thaliana highlighted the Msh1 gene encoding a homolog of the Escherichia coli mismatch repair and DNA recombination component MutS fused with a homing endonuclease (7). The Msh1 protein was shown to target to both mitochondria and chloroplasts (7,8). Homologs of bacterial RecA also exist in plant organelles and appeared to be involved in mtDNA repair in the moss Physcomitrella patens (9) or in surveillance of mitochondrial genome stability in A. thaliana (10).
A major consequence of oxidative damage is the occurrence of uracil in the DNA, which can be due to two different processes (11). The first one is the misincorporation of dUTP during DNA replication, despite the presence of dUTPase activities which hydrolyze dUTP in the dNTP pool (12). The second possibility is the oxidative (13) or enzymatic (14) deamination of cytosine in situ, leading to a C : G to T : A transversion during replication. Uracil–DNA glycosylases (UDGs) are the initial enzymes in BER pathways repairing uracil in the DNA (15). Genomic analyses, combined with predictions of subcellular localization, identified a cluster of nuclear genes encoding putative mitochondrial DNA metabolizing enzymes in A. thaliana (16). These included the At3g18630 locus (http://www.arabidopsis.org/) encoding a potential homolog of the human cytosolic and mitochondrial UDG provided by the UNG gene (17). Starting from this indication, and in the absence of further data, we have investigated the potential of plant mitochondria to repair uracil in the DNA and thereby to prevent its often mutagenic consequences. Developing a combination of in vitro and in organello approaches, we show that plant mitochondria eliminate uracil in the mtDNA through a short-patch BER mechanism. Organelle targeting analyses suggest that the UDG candidate encoded by the At3g18630 locus mediates the first step of this process.
MATERIALS AND METHODS
Cell cultures
Arabidopsis thaliana cell suspensions, ecotype Columbia, line T87-C33 (18), were cultivated at 25°C in Gamborg B5 medium (Duchefa, Haarlem, The Netherlands) supplemented with 20 g/l sucrose and 1 mg/l 2,4 D, final pH 5.7. Cells were grown under complete darkness. Cell cultures were harvested 5 days post-inoculation in fresh medium.
Oligodeoxyribonucleotide repair substrates
Modified and unmodified oligodeoxyribonucleotides were purchased from Sigma-Aldrich. To prepare the repair substrates (Figures 1 and 2), single-stranded DNA oligonucleotides (50 pmol) were radioactively labeled with T4 polynucleotide kinase and [γ32P]ATP (3000 Ci/mmol). Nonincorporated ATP was eliminated on Sephadex G-50 spin columns and 500 pmol of the relevant complementary oligonucleotide were added. The mixture was incubated at 70°C for 10 min in 100 mM Tris–HCl pH 7.6, 1 mM NaCl, 10 mM EDTA, and slowly cooled down to room temperature. Annealed hybrids were separated from remaining single-stranded oligonucleotides on nondenaturing 15% (w/v) polyacrylamide gels in TBE (Tris–borate 90 mM, EDTA 2 mM, pH 8.0), eluted in 0.5 M ammonium acetate, 10 mM magnesium acetate, 0.1% (w/v) SDS, 0.1 mM EDTA (19), phenol-extracted and twice ethanol-precipitated.
Figure 1.
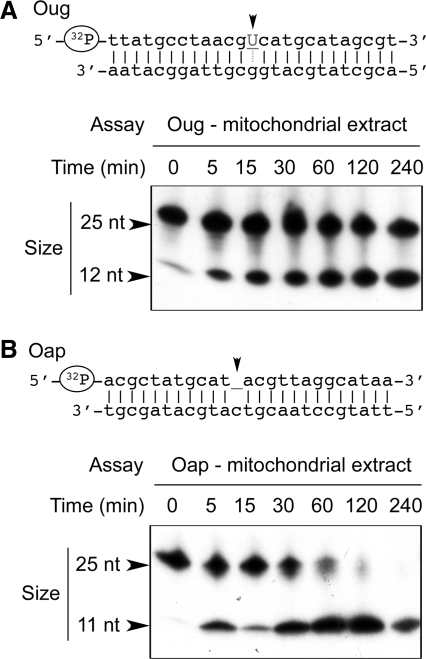
Plant mitochondria possess UDG and AP endonuclease activity. Double-stranded oligodeoxyribonucleotides Oug (A) and Oap (B), [32P]-labeled at the 5′-end of the strand carrying the lesion, were incubated in the presence of enriched A. thaliana mitochondrial enzymatic extract (25 µg of protein) for increasing times. To evaluate the extent of lesion-specific incision, the samples were subsequently denatured and analyzed by electrophoresis on polyacrylamide gels in the presence of 7 M urea. The migration of size markers is indicated by arrows.
Figure 2.
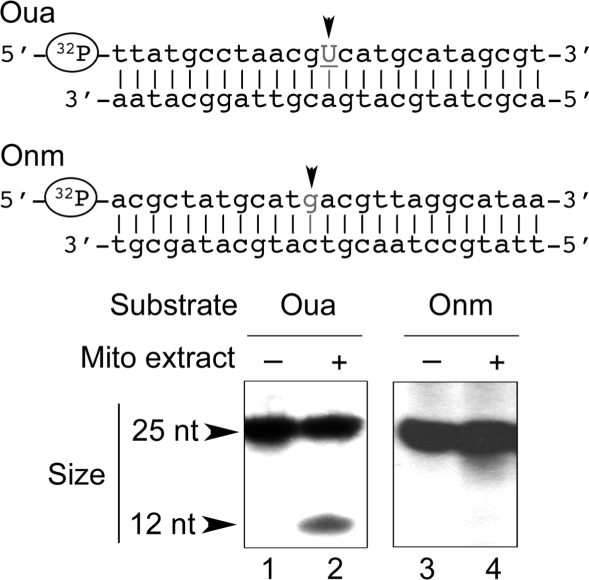
Plant mitochondrial UDG activity is independent from the sequence context. Double-stranded oligodeoxyribonucleotides Oua and Onm, [32P]-labeled at the 5′-end of one of the strands as indicated, were incubated in the absence of extract (lanes 1 and 3) and in the presence of enriched A. thaliana mitochondrial extract (Mito, lanes 2 and 4, 25 µg of protein) for 1 h. Analysis of lesion-specific incision was as in Figure 1.
Prior to repair tests with plant extracts or organelles, the presence of an uracil in the appropriate position in Oug and Oua was confirmed with a commercial enzymatic preparation (Uracil-DNA Excision Mix, Epicentre Biotechnologies, Madison, WI, USA) in the reaction conditions recommended by the manufacturer. To verify the presence of the abasic site in Oap, we used 6His-tagged E. coli endonuclease VI which we cloned, overexpressed and purified. Reactions were run in 50 mM Tris–HCl pH 7.5, 50 mM MgCl2, 20 mM ZnCl2, 50 mM KCl, 1 mM DTT, 10 µg/ml bovine serum albumin.
Large size DNA repair substrates
The 920 bp uracil-containing DNAs were PCR-amplified with no dUTP (U1) or in the presence of 1 µM (U2), 10 µM (U3), 100 µM (U4) or 1 mM (U5) dUTP. The template was a composite gene construct available in our laboratory from another project and composed of the sequences for the Tobacco etch virus (TEV) translation leader (accession number M15239), the Turnip yellow mosaic virus (TYMV) coat protein and 3′-terminal tRNA-like structure (accession number X16378) and the Hepatitis delta virus (HDV) antigenomic cis-ribozyme (accession number AM779578). This construct contained no sequence which would have been related to mtDNA or to DNA replication, transcription or maintenance in mitochondria and was under the control of the Cauliflower mosaic virus (CaMV) 35S promoter. Amplification was with the direct primer 5′-CATTTGGAGAGGACCTCGAG-3′ corresponding to the 3′-end of the CaMV 35S promoter and the reverse primer 5′-CTCCCTTAGCCATCCGAGTG-3′ annealing to the 3′-end of the HDV cis-ribozyme sequence. We observed that dUTP concentrations higher than 1 mM inhibited the PCR reaction. The 920 bp U1 to U5 PCR products were purified by electrophoresis on 1% (w/v) agarose gels in TAE (Tris–acetate 40 mM, EDTA 2 mM, pH 8.0) and elution. To test their uracil content, they were radioactively labeled with T4 polynucleotide kinase and [γ32P]ATP. Labeled products were treated with the Uracil–DNA Excision Mix (Epicentre) in the reaction conditions recommended by the manufacturer.
Isolation of mitochondria and mitoplasts
Mitochondria were isolated from Solanum tuberosum tubers according to previously described methods based on continuous Percoll gradients (20,21). Arabidopsis thaliana mitochondria were purified from cell suspensions using discontinuous Percoll gradients as described by Sakamoto et al. (22). Mitochondrial preparations were routinely checked for their integrity and respiratory control (21). Solanum tuberosum and A. thaliana mitoplasts were generated by hypotonic swelling of mitochondria and were purified on sucrose gradients according to Heins et al. (23) and Werhahn et al. (24), respectively.
Organelle extracts
When they were to be used to prepare suborganellar fractions, purified mitochondria and mitoplasts were treated with proteinase K (10 µg for an amount of purified organelles corresponding to 5 mg protein) for 30 min on ice. Proteinase K was inhibited with 2 mM phenylmethylsulfonyl fluoride and organelles were centrifuged through a 27% (w/v) sucrose cushion in 20 mM Tris–HCl pH 7.5, 1 mM EDTA, 100 mM K2HPO4, 1 mg/ml bovine serum albumin.
To prepare enriched enzymatic extracts deprived of endogenous nucleic acids and small size compounds, proteinase K-treated organelles were resuspended in chilled 1.5× enzyme buffer [1× enzyme buffer: 50 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 10% (v/v) glycerol, 1 mM EDTA, 5 mM 2-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride] completed with 15 µg/ml α2-macroglobulin and 15 µg/ml leupeptin. Organelles were lysed by 3 freeze/thaw cycles and sonication (3 times 10 s at 4°C). The lysate was centrifuged for 40 min at 40 000g. The supernatant was brought to 0.15 M NaCl and loaded onto a DEAE-cellulose column equilibrated with 1× enzyme buffer containing 0.15 M NaCl. The flow-through fraction was submitted to gel-filtration on Sephadex G-50 spin columns equilibrated with 1× enzyme buffer. The excluded fraction was completed with 10 µg/ml α2-macroglobulin, 10 µg/ml leupeptin, 10% (v/v) propanediol and stored at −80°C.
To analyze the submitochondrial distribution of DNA repair activities, the original lysate was centrifuged for 45 min at 105 000g. The resulting supernatant was taken as the soluble fraction, whereas the pellet was resuspended in a volume of 1× enzyme buffer equivalent to that of the supernatant, so as to constitute the membrane fraction.
The absence of significant nucleo-cytosolic or plastidial contamination in mitochondrial and mitoplast extracts, or of cross-contamination between mitochondrial soluble and membrane fractions, was shown by western blot analyses (see Supplementary Figures 1 and 2). Transcription factors E2F and E2F-like were used as nuclear markers, pyruvate dehydrogenase was used as a mitochondrial marker and the chlorophyll a/b-binding protein of the light harvesting complex of photosystem II (LHC2) served as a chloroplast marker. Subunit 9 of complex I of the respiratory chain (NAD9) was taken as a membrane marker for mitochondrial soluble fractions, whereas manganese superoxide dismutase (MnSOD) served as a soluble marker for membrane fractions. As it is considered to be exposed to the intermembrane space, NAD9 was also a marker for the efficiency of the proteinase K treatment on mitoplasts.
In vitro incision and repair assays
Incision assays in 10 µl involved 5 pmol of double-stranded oligodeoxyribonucleotide repair substrate, [32P]-labeled at the 5′-end of one of the strands as indicated in Figures 1 and 2, and an amount of enriched organelle enzymatic extract corresponding to 25 µg protein. When analyzing submitochondrial distribution of DNA repair activities, we used a volume of soluble fraction containing 25 µg protein and the same volume of the corresponding resuspended membrane fraction (see above). UDG activity was tested in 50 mM Tris–HCl pH 7.5, 2 mM EDTA, 50 mM KCl. When looking specifically for AP endonuclease activity, we used 50 mM Tris–HCl pH 7.5, 50 mM MgCl2, 20 mM ZnCl2, 50 mM KCl, 1 mM DTT, 10 µg/ml bovine serum albumin. Reactions were incubated for 1 h (unless running kinetics) at 32°C. To test complete repair in vitro, reaction media involving enriched enzymatic organelle extracts were subsequently completed with 10 mM dNTPs (dATP, dCTP, dGTP, dTTP), 1 mM ATP, 8 mM MgCl2 and post-incubated for 30 min to 2 h. Following incision/repair assays, samples were completed with 1 vol. of 20 mM EDTA, 0.05% (w/v) bromphenol blue, 0.05% (w/v) xylene cyanol, 95% (v/v) formamide (stop buffer), denatured for 5 min at 95°C, chilled for 5 min on ice and analyzed by electrophoresis on 15% (w/v) polyacrylamide/7 M urea denaturing gels in TBE. Synthetic oligodeoxyribonucleotides corresponding to the expected cleavage products of the incision assays were also radioactively labeled with T4 polynucleotide kinase and [γ32P]ATP and were used as size markers on the gels, together with the single-stranded full-length oligodeoxyribonucleotides.
In organello DNA repair assays
Import of oligonucleotide repair substrates into isolated plant mitochondria was carried out in 40 mM potassium phosphate pH 7.0, 0.4 M sucrose (21). Samples of 200 µl containing 5 pmol of double-stranded oligodeoxyribonucleotide repair substrate (Onm, Oug or Oua), [32P]-labeled at the 5′-end of one of the strands as indicated in Figures 1 and 2, and an amount of purified mitochondria corresponding to 300 µg protein, were incubated at 25°C for various times up to 1 h under mild shaking. Following addition of 200 µg of DNaseI and 10 mM MgCl2, the incubation was continued for 20 min in the same conditions. Mitochondria were subsequently washed three times by resuspension in 1 ml of 10 mM potassium phosphate pH 7.5, 0.3 M sucrose, 10 mM EDTA, 10 mM EGTA, 0.1% (w/v) bovine serum albumin, 5 mM glycine and centrifugation for 5 min at 10 000g. The final pellets were extracted with one volume of 10 mM Tris–HCl pH 7.5, 1 mM EDTA, 1% (w/v) SDS, and one volume of phenol. The nucleic acids recovered in the aqueous phase were ethanol-precipitated, redissolved in 10 µl water, completed with 10 µl stop buffer, denatured for 5 min at 95°C, chilled for 5 min on ice and analyzed as previously by electrophoresis on 15% (w/v) polyacrylamide/7 M urea denaturing gels in TBE.
To further test in organello complete repair of oligonucleotide substrates, [32P]-labeled double-stranded oligodeoxyribonucleotides were imported for 30 min. Mitochondria were then submitted to DNaseI digestion and washing, as described above, before resuspension in 200 µl of 330 mM sucrose, 90 mM KCl, 10 mM MgCl2, 12 mM Tricine, 5 mM KH2PO4, 1.2 mM EGTA, 2 mM DTT, 2 mM ADP, 10 mM sodium succinate, 50 µM dNTPs (dATP, dCTP, dGTP, dTTP), pH 7.2 (DNA synthesis buffer) (21). Resuspended organelles were postincubated for 1–2 h at 25°C under mild shaking. Nucleic acids were subsequently extracted and analyzed as described above.
To investigate repair of large size DNA substrates, isolated plant mitochondria corresponding to 300 µg of proteins were preincubated for 20 min at 25°C in 200 µl of 40 mM potassium phosphate pH 7.0, 0.4 M sucrose completed with 2 µl (20 µCi) of either [α32P]dTTP or [α32P]dCTP (3000 Ci/mmol). The 920 bp uracil-containing unlabeled DNAs U1 to U5 (see above, 10 ng) were then added and the suspensions were further incubated for 45–60 min at 25°C to allow for import and possible repair. Mitochondria were subsequently submitted to DNaseI digestion and washing, as described above. Washed organelles were resuspended in 300 µl of DNA synthesis buffer (see above). At that stage, one half of each sample was analyzed directly as representative of the import step, whereas the other half was postincubated for 45–60 min at 25°C under mild shaking to allow for further repair. Nucleic acids were extracted from all samples following the above described procedures. The recovered nucleic acids were ethanol-precipitated, redissolved in 30 µl Tris–HCl 10 mM pH 7.5, 1 mM EDTA and gel-filtered through 1 ml Sephadex G-50 spin columns to eliminate the remaining unincorporated [α32P]dTTP or [α32P]dCTP. The flowthrough was completed with 30 µl stop buffer, denatured for 5 min at 95°C, chilled on ice for 5 min and analyzed by electrophoresis on 1% (w/v) agarose gels in TAE. Agarose gels were transferred in 0.4 M NaOH onto a nylon membrane (Hybond-XL, GE Healthcare Life Sciences) for autoradiography. For each experiment, DNA import efficiency was evaluated in parallel assays run in the absence of labeled dTTP or dCTP and taking as a substrate DNAs U1 to U5 labeled with T4 polynucleotide kinase and [γ32P]ATP.
In vivo targeting analysis of eGFP fusion proteins
Construction of the pGII35S-GFP and pGII35SNUDG-GFP plasmids designed to express the eGFP alone and the eGFP fused to the N-terminal part of the At3g18630-encoded protein, respectively, is described in the Supplementary Materials and methods section. pGII35S-GFP and pGII35SNUDG-GFP were used to transform the Agrobacterium tumefaciens LBA4404 strain (Life Technologies) together with the pSoup plasmid carrying the RepA gene required for the replication of pGreen-derived plasmids in agrobacteria (25). Transformation was performed by infiltration of Nicotiana benthamiana leaves (26) with a suspension of A. tumefaciens carrying pGII35S-GFP or pGII35SNUDG-GFP. After 24–48 h, protoplasts were prepared (27) from the infiltrated leaves, treated with the mitochondrion-specific dye MitoTracker® Orange CM-H2TMRos (Invitrogen) and analyzed using a Zeiss LSM510 laser scanning confocal microscope. For eGFP and chlorophyll imaging, excitation at 488 nm was obtained with an argon laser and for MitoTracker imaging, excitation at 543 nm was obtained with a helium/neon laser. Band pass 505–530 nm, long pass 585 nm and band pass 585–615 nm emission filters were used to simultaneously collect the signals for eGFP, chlorophyll and MitoTracker, respectively. Pictures were handled with the ImageJ (http://rsbweb.nih.gov/ij/) and Photoshop (Adobe) softwares. Complementary control experiments were run with plasmid COXIV-GFP (28) encoding the mitochondrial targeting sequence of yeast cytochrome oxidase subunit IV fused in-frame with the GFP.
RESULTS
Plant mitochondria contain uracil-DNA glycosylase and AP endonuclease
The first step in the repair of a point-modification in the DNA is the elimination of the damaged nucleotide. In the BER mechanism, the abnormal base is cleaved off by a glycosylase and only the remaining abasic nucleotide (AP site for apurinic and apyrimidinic site) is eliminated by lyase and/or AP endonuclease activities (15). During a NER process, a bulge forms at the level of the lesion in the DNA and a fragment carrying the damaged base is cut out by structure-specific endonucleases (29). We used double-stranded DNA oligonucleotides carrying a single uracil on one strand to characterize specific recognition and cleavage activities in plant mitochondrial enzymatic fractions. As mentioned above, uracil can form in the DNA upon oxidative deamination of cytosine or derive from misincorporation of dUTP during replication. We thus prepared two uracil-containing double-stranded DNA oligonucleotides. In the first one called Oug (Figure 1), uracil was paired with guanine, i.e. the situation which occurs upon cytosine deamination in the DNA. In the second substrate called Oua (Figure 2), uracil was paired with adenine, which was representative of dUTP incorporation into the DNA. These substrates were primarily aimed at characterizing putative mitochondrial UDG activity. However, UDGs are generally monofunctional enzymes that lack β-lyase activity (30). They can remove the base but not cleave the phosphodiester chain. The resulting AP site must thus be further processed by an AP endonuclease.
To have the possibility to characterize independently the AP endonuclease activity, we also used a distinct AP site-containing oligonucleotide called Oap (Figure 1). Another reason to test such a substrate was that AP sites should not be considered only as repair intermediates, as they can also form spontaneously or result directly from oxidation (31). Finally, an unmodified oligonucleotide called Onm was tested as a negative control (Figure 2). All oligonucleotides were double-stranded and 25-nt long. The strand carrying the modification was labeled at the 5′-end with T4 polynucleotide kinase. Uracil was standing at position 13 in Oug and Oua, whereas the AP site was at position 12 in Oap. Specific cleavage at the level of the modified nucleotide was thus expected to yield labeled single-stranded oligonucleotides of 12 and 11 nucleotides, respectively, upon denaturing of the samples.
The four DNA substrates were used for single strand incision assays in the presence of enriched A. thaliana mitochondrial enzymatic extracts deprived of endogenous nucleic acids and small size compounds (see ‘Materials and Methods’ section). Prior to extraction, mitochondria were protease-treated to eliminate contaminations from other subcellular compartments. The absence of significant cross-contaminations was assessed by western blot analyses (see ‘Materials and Methods’ section and Supplementary Figure 1). Arabidopsis thaliana mitochondrial enzymatic extracts efficiently promoted single-strand cleavage of Oug at the position of the uracil, as the corresponding shorter fragment appeared very rapidly (Figure 1A). The cut/uncut ratio increased with time to reach a plateau at about 50% after 1 h. Full cleavage was not obtained for these assays, which meant that either the UDG activity or the AP endonuclease activity was limiting. To distinguish between the two possibilities, the AP site-containing oligonucleotide Oap was tested in the same conditions (Figure 1B). As for Oug, the expected cleavage product appeared within minutes but in this case nearly full cleavage was reached after 2 h, implying that AP endonuclease activity was not limiting in our enriched enzymatic extracts.
To check the influence of the sequence context, we tested uracil removal from the DNA in the context of an U : A base pair, using the Oua substrate. The results were similar to those for Oug (Figure 2, lane 2). To ensure that all observed cuts were enzyme-related, Oug, Oap and Oua were incubated in the same conditions in the absence of protein extract. Cleavage was never detected in such control assays (e.g. Figure 2, lane 1). Conversely, the unmodified DNA substrate Onm was not cleaved by mitochondrial enzymatic extracts, confirming in turn that the observed cuts were specific for the modifications (Figure 2, lane 4). Similar observations were made with Onm, Oug, Oua, Oap and enriched mitochondrial enzymatic extracts from potato (S. tuberosum) tubers (see Supplementary Figure 3).
In further assays, A. thaliana and S. tuberosum mitochondria were submitted to hypotonic swelling, so as to break the outer membrane. The resulting mitoplasts were separated from the outer membrane fraction on sucrose gradients, protease-treated and in turn used to prepare enriched enzymatic extracts. The absence of significant membrane contamination in these extracts was checked by western blot analyses (see ‘Materials and Methods’ and Supplementary Figure 2). Incision assays with the Oug and Oua DNA substrates showed that, although the efficiency was somehow lower with Oua, UDG activity in enriched mitoplast enzymatic extracts was comparable to that in extracts from intact mitochondria (see Supplementary Figure 4), confirming the presence of the activity inside the organelles.
In all cases, the cleaved 5′ fragment was that expected from removal of the single modified nucleotide. This suggested that the observed activities were part of a BER pathway, as a NER mechanism would have led to the excision of a small fragment and thus to the deletion of further nucleotides upstream of the damaged position.
The UDG and AP endonuclease activities are distributed between soluble and membrane fractions in plant mitochondria
To characterize the submitochondrial distribution, we analyzed the uracil-cleaving activity associated with the mitochondrial membrane fraction recovered after lysis and high speed centrifugation of protease-treated, isolated A. thaliana and S. tuberosum mitochondria. The soluble fraction from the same samples was tested in identical conditions in the presence of the Oug and Oua DNA substrates. Cleavage of the AP site-containing substrate Oap was also analyzed. The absence of significant cross-contamination between membrane and soluble fractions was checked by western blot analyses (see ‘Materials and Methods’ section and Supplementary Figure 2).
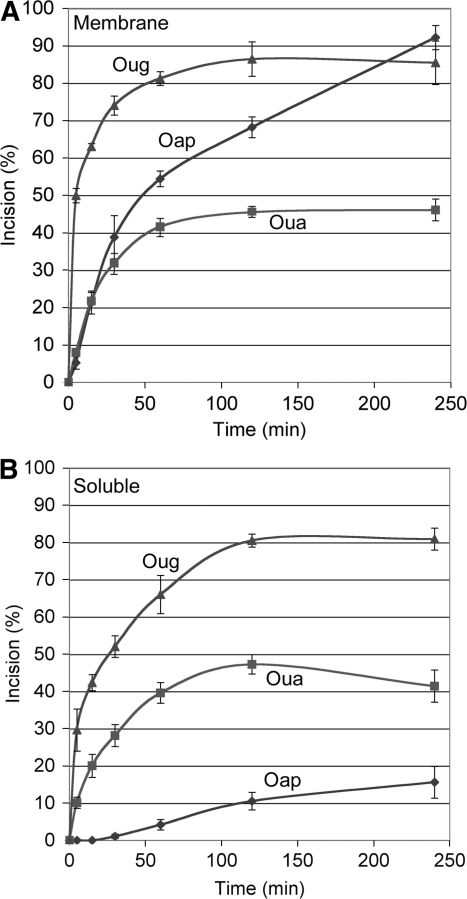
Incision assays with Oug and Oua established that the mitochondrial uracil cleavage activity was in part membrane-associated. Reaction kinetics spanning up to 4 h were run with the membrane and soluble fractions isolated from A. thaliana mitochondria and the results were quantified. The initial incision rate of Oug in the presence of the membrane fraction (Figure 3A) was actually about twice that measured with the soluble fraction (Figure 3B), although the reactions reached a similar final plateau between 80% and 90%. The incision kinetics of Oua obtained with the membrane and soluble fractions were very similar all along, ending with the same plateau at only about 45%. Strikingly, the initial incision rate of Oug was six times higher than that of Oua in the presence of the membrane fraction, whereas the ratio was about three with the soluble fraction. The results of these assays altogether imply that a uracil paired with a guanine, i.e. the product of DNA oxidation, is better cleaved by the putative mitochondrial UDG than a uracil paired with an adenine, which is normaly the consequence of uridine incorporation during DNA synthesis. It also seems that the membrane context further boosts the initial phase of the reaction. Similar conclusions could be drawn from parallel time course studies with submitochondrial fractions from S. tuberosum, although as a whole the activities detected were lower in that case. Previous studies identified a weak preference of the mammalian UDG encoded by the UNG gene for U : G mismatches over U : A base pairs, but the preference was sequence-dependent and not absolute (11,32,33).
Figure 3.
Plant mitochondrial UDG and AP endonuclease activities are partially membrane-bound. Double-stranded oligodeoxyribonucleotides Oug (triangles), Oua (squares) and Oap (lozenges), [32P]-labeled at the 5′-end of the strand carrying the lesion, were incubated for increasing times in the presence of a membrane fraction (A) or a soluble fraction (B) from A. thaliana whole mitochondria. The samples were subsequently denatured and fractionated by electrophoresis on polyacrylamide gels in the presence of 7 M urea. The corresponding autoradiograms were scanned and quantified with the ImageJ software (NIH), so as to estimate the percentage of lesion-specific incision at the different time points. Bars indicate standard deviation.
Time course analyses of Oap cleavage in the presence of the submitochondrial fractions showed that AP endonuclease activity was also associated with the mitochondrial membranes but detected a somehow unexpected discrepancy. The initial incision rate of Oap was up to ten times lower than that of Oug in the presence of the A. thaliana membrane fraction. The reaction then proceeded efficiently, reaching over 90% cleavage after 4 h, but the rate remained lower than that of Oug cleavage all along the kinetics (Figure 3A). Moreover, incision of Oap with the soluble fraction started with a lag phase and remained low, ending at only about 15% after 4 h, i.e. way below the plateau levels of both Oug and Oua cleavage (Figure 3B). It thus appeared that the AP endonuclease activity did not match the UDG activity, although, as mentioned previously, UDGs usually lack a lyase activity and need the help of an AP endonuclease for phosphodiester chain incision. The Oap-cleaving activity was better balanced between the membrane fraction and the soluble fraction from S. tuberosum mitochondria, but again it did not match the UDG activity. This discrepancy was not detected in the above assays run with enriched enzymatic extracts because AP endonuclease was in large excess in such extracts, whereas UDG appeared to be limiting (Figure 1). Two hypotheses can be considered to account for these results. One can wonder whether the incision of Oug and Oua is more efficient because the UDG recruits the AP endonuclease, whereas the recognition of the naked AP site in Oap would be more difficult. On the other hand, it cannot be excluded that plant mitochondrial UDG uses an original cleaving mechanism, i.e. it possesses a lyase activity.
Plant mitochondria can carry out complete lesion repair in large uracil-containing DNA
The above experiments demonstrated that plant mitochondria possess enzymes able to excise uracil from the DNA and incise the phosphodiester chain. For full repair, these steps are expected to be followed by DNA polymerase-mediated reinsertion of the correct nucleotide and religation. In vitro assays with organelle protein fractions did not promote complete repair of uracil-containing oligodeoxyribonucleotides. To proceed further, we developed in organello incision and repair assays in intact plant mitochondria, based on their DNA import capacity (21).
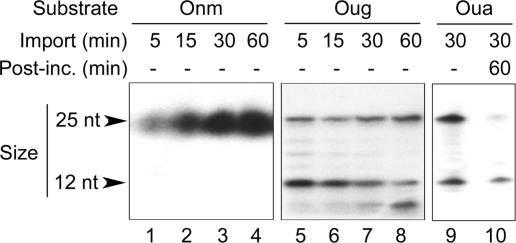
The ability of plant mitochondria to stably internalize short double-stranded oligodeoxyribonucleotides was confirmed using as an import substrate the unmodified DNA Onm. Figure 4 (lanes 1–4) shows that Onm was incorporated into A. thaliana isolated mitochondria as a function of time and remained intact in the organelles over the time range of the experiment. We then imported the Oug and Oua DNA substrates into A. thaliana or S. tuberosum mitochondria. Upon uptake, both of them were cleaved at the level of the uracil, as demonstrated by the appearence of the expected 12 nucleotide fragment (Figure 4, lanes 5–9). Hence, Oug and Oua were successively taken up by the mitochondrial DNA import pathway and specifically nicked in organello after recognition of the abnormal base, strengthening the observations made in vitro with enzyme extracts. However, even inside intact mitochondria placed in DNA synthesis conditions, such short modified substrates failed to be carried over to the repair synthesis enzymes (Figure 4, lane 10).
Figure 4.
A uracil-containing oligodeoxyribonucleotide is specifically incised upon import into isolated plant mitochondria. Lanes 1–8: double-stranded oligodeoxyribonucleotides Onm (1–4) and Oug (5–8), [32P]-labeled at the 5′-end of one of the strands as indicated in Figures 1 and 2, were imported into isolated A. thaliana mitochondria for increasing times. After DNase treatment, mitochondrial nucleic acids were extracted and analyzed as in Figure 1. Lanes 9 and 10: double-stranded oligodeoxyribonucleotide Oua, [32P]-labeled at the 5′-end of the strand carrying the lesion, was imported for 30 min into isolated S. tuberosum mitochondria. After DNase treatment, an aliquote of the mitochondria was kept for direct analysis (9), whereas the remaining organelles were post-incubated (Post-inc.) for 1 h in DNA synthesis conditions (10). Finally, mitochondrial nucleic acids were extracted and analyzed as in Figure 1.
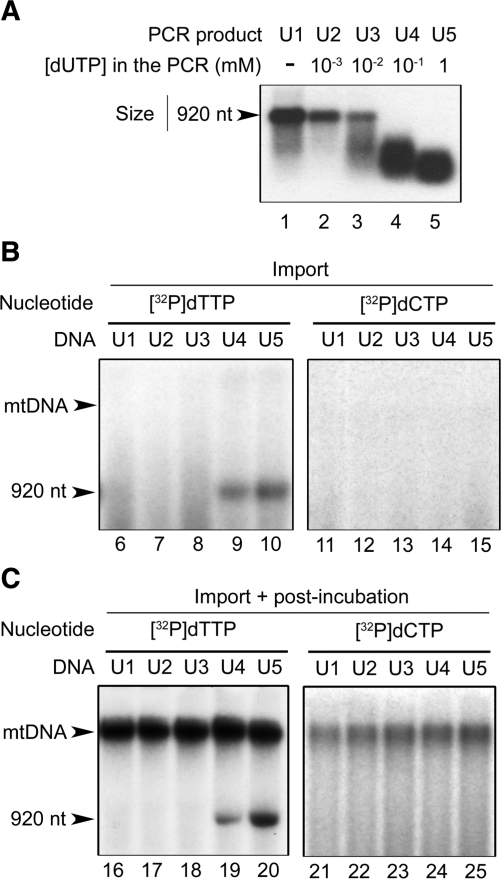
Following the observations made with short oligonucleotides, we developed further in organello assays with a long substrate. A 920 bp uracil-containing DNA fragment (see ‘Materials and Methods’ section for details) was prepared by PCR in the presence of different concentrations of dUTP, as it is known that Taq polymerase can insert an U in front of an A in the DNA (34). Prior to import assays, the presence of uracil in the amplified fragments was checked with a commercial uracil excision mix containing bacterial UDG and endonuclease IV (see ‘Materials and Methods’ section). Samples were denatured before analysis on agarose gels. No significant cleavage was observed with PCR products U1 and U2 amplified with no dUTP in the reaction medium or in the presence of only 1 µM dUTP (Figure 5A). Incorporation of uracil became detectable with DNA U3 amplified in the presence of 10 µM dUTP. Further increasing the dUTP concentration in the PCR reaction to 0.1 and 1 mM led to DNAs U4 and U5, respectively, which upon treatment with the uracil excision mix showed full cleavage and a decreasing size distribution of the cleavage products (Figure 5A). These patterns implied that in U4 and U5 all DNA molecules carried uracil, with a higher frequency in U5. In all cases, uracil was supposed to be incorporated opposite adenine, i.e. a situation representative of dUMP incorporation during DNA replication.
Figure 5.
Uridine can be exchanged for thymidine in large size DNA imported into isolated plant mitochondria. (A) A double-stranded DNA fragment of 920 bp was PCR-amplified in the presence of increasing concentrations of dUTP. To assess uracil incorporation, the resulting PCR products (U1–U5) were 5′-end-labeled, treated with a commercial uracil excision mix, denatured and analyzed by electrophoresis on agarose gels. The migration of the original full-length DNA fragment is indicated by an arrow. (B) The 920 bp PCR products U1–U5 containing increasing levels of uracil were imported for 45 min into isolated A. thaliana mitochondria preincubated for 20 min in the presence of [α32P]dTTP (lanes 6–10) or [α32P]dCTP (lanes 11–15). After import and DNase treatment, mitochondrial nucleic acids were extracted, denatured and analyzed by electrophoresis on agarose gels. (C) PCR products U1–U5 were imported for 1 h into isolated A. thaliana mitochondria preincubated for 20 min in the presence of [α32P]dTTP (lanes 16–20) or [α32P]dCTP (lanes 21–25). After import and DNase treatment, an aliquote of the mitochondria was kept for direct analysis (not shown, in this experiment label incorporation into the probes was not detectable yet at that stage), whereas the remaining organelles were post-incubated for 1 h in DNA synthesis conditions. Finally, mitochondrial nucleic acids were extracted from all samples, denatured and analyzed by electrophoresis on agarose gels. The migration of the original full-length DNA fragment and of the mtDNA are indicated by arrows.
PCR products U1 through U5 were imported into isolated A. thaliana mitochondria labeled beforehand with [α32P]dTTP. After the import step, mitochondria were DNase-treated, washed and postincubated in DNA synthesis medium. In these assays, incorporation of radioactive label into the otherwise unlabeled imported DNAs was expected to occur, reflecting the replacement of dUMP with [32P]dTMP in the course of a complete repair process. No incorporation of radioactivity was detected in U1, U2 and U3 which contained no uracil or only low uracil levels, but labeled U4 and U5 were indeed generated. Repair was obtained already during the uptake step in the most efficient experiments (Figure 5B, lanes 9 and 10) and in any case it occurred during postincubation in DNA synthesis conditions (Figure 5C, lanes 19 and 20). Label incorporation was always higher for U5 than for U4. These results perfectly matched the uracil content in the imported probes, as revealed earlier with the uracil excision mix (Figure 5A). That all five PCR products were able to be imported into mitochondria with the same efficiency was assessed in complementary uptake assays using as substrates DNAs U1 through U5 end-labeled with kinase and [γ32P]ATP.
During postincubation, label was incorporated into the high molecular weight mtDNA in all assays, confirming the DNA synthesis capacity of the mitochondria (Figure 5C, lanes 16–20). Therefore, to rule out the possibility that the observed [32P]dTTP incorporation be due to some random priming or nick-translation, the same experiments were carried out with [α32P]dCTP. No labeling of U4 and U5 was detected in this case (Figure 5B, lanes 14 and 15; Figure 5C, lanes 24 and 25). Formation of labeled U4 and U5 in the mitochondria was thus specific for [32P]dTTP and for the presence of uracil in the imported DNA. This implied that we had obtained in organello complete uracil repair, i.e. uracil excision, incision of the phosphodiester chain and incorporation of thymidine. Religation was evidenced by the homogeneous size of the labeled products at 920 nt despite denaturing of the samples.
Only the final product was labeled in these assays, so that the reaction intermediates were not visualized. However, that [α32P]dCTP could not substitute for [α32P]dTTP in turn meant that, in each modified position, only one nucleotide, i.e. the thymidine replacing the uridine, was incorporated into U4 and U5 during the in organello repair reaction. Like the other in vitro and in organello experiments presented above, this observation excluded a NER mechanism. On the other hand, the BER pathway involves DNA polymerase-mediated de novo insertion of either a single nucleotide (short-patch BER) or of several nucleotides (long-patch BER) (15). Our data also excluded the long-patch BER pathway because, like NER, such a mechanism would have directed the insertion of a short sequence extending from the incision site (29). It could thus be concluded that U4 and U5 were repaired in organello through a plant mitochondrial short-patch BER mechanism.
Genetic basis for a BER pathway in plant organelles: mitochondrial targeting of UDG
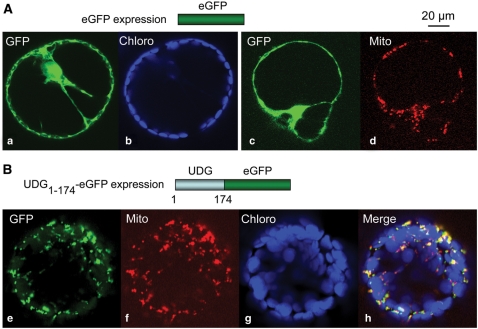
As mentioned, the At3g18630-encoded protein is a candidate for an organelle UDG (16). We thus transiently expressed in N. benthamiana leaves the first 174 amino acids of this polypeptide fused to the eGFP reporter and we analyzed the subcellular targeting of the fusion protein. eGFP alone was diffuse in the cytosol and in the nucleus (Figure 6A, panels a and c), matching neither the fluorescence of the chlorophyll (Figure 6A, panel b), nor that of the MitoTracker used to stain the mitochondria (Figure 6A, panel d). On the contrary, when the UDG1–174-eGFP fusion protein was expressed in N. benthamiana leaves, eGFP fluorescence colocalized with the mitochondria in protoplasts generated from the agro-infiltrated tissues (Figure 6B, panels e, f and h). These observations were further confirmed through in vitro import into isolated mitochondria (see the Supplementary Material). The in vivo and in vitro data altogether imply that the N-terminal region of the A. thaliana UDG encoded by the At3g18630 locus is a mitochondrial targeting and import sequence. This protein is thus likely to be the first enzyme in the plant mitochondrial BER pathway for uracil repair.
Figure 6.
The N-terminal region of the UDG encoded by the At3g18630 locus is a mitochondrial targeting sequence in vivo. Confocal microscopy analysis (lens 60×) of protoplasts derived from Agrobacterium-infiltrated N. benthamiana leaves expressing the eGFP alone (A) or the eGFP fused in-frame at its N-terminus to the first 174 amino acids of the At3g18630-encoded protein (B). eGFP fluorescence (green, GFP), chlorophyll autofluorescence (blue, Chloro) and MitoTracker fluorescence (red, Mito) were collected as detailed in ‘Materials and Methods’ section. (A) Images (a) and (b) correspond to one cell, images (c) and (d) to another cell. (B) Images (e), (f) and (g) correspond to the same cell and are merged in (h). The scale is indicated (top right).
DISCUSSION
Maintenance of the organellar genomes in a context of permanent oxidative pressure is a major challenge for eukaryotic cells. Despite that, it has long been considered that organelles in general are unable to repair their DNA, based on the original discovery that mammalian mitochondria do not eliminate pyrimidine dimers generated by UV irradiation (35). The alternative idea was that, due to their high polyploidy (some human oocytes can contain as much as 100 000 copies of the mtDNA), organelles would degrade and replace damaged molecules rather than repairing them. That mammalian and yeast mitochondria actually possess mechanisms to repair mtDNA damages other than pyrimidine dimers was mostly demonstrated from the late 1990s (36). Plant mitochondrial genetic systems present some fundamental differences to their animal counterparts. First, the pressure may seem less stringent on the very low-gene density of the 220–740 kb plant mitochondrial genomes (37) than on the compact and fully coding mammalian mtDNA of 16.5 kb (38). Second, homologous recombination seems to be a rare event in mammalian mitochondria (39), whereas it shapes the plant mtDNA (7). It was thus possible to speculate that dilution of the mutations in the vast non coding regions, recombination-based exchange between the multiple DNA copies and degradation of definitely damaged molecules would be sufficient to account for the integrity of the plant mitochondrial genome. The present work establishes that this is not the case. Our biochemical and functional approaches demonstrated the presence of a complete short-patch BER pathway in plant mitochondria for uracil repair. The observations we made can clearly not be explained by side effects of the organelle RNA editing mechanisms, as these involve no relevant activities. RNA editing in mitochondria of flowering plants is restricted to C to U changes likely to occur through deamination or transamination of the cytosine moiety. There is no removal of the base, no cleavage of the sugar-phosphate backbone, no nucleotide insertion [(40) and references therein].
Our study also brings the first hints on the genetic basis of uracil repair in plant mitochondria. The human UNG gene encodes both a mitochondrial form (UNG1) and a nuclear form (UNG2) of UDG. These have a common catalytic domain, but different N-terminal sequences generated by alternative splicing, which provides UNG1 with a mitochondrial targeting sequence (17). The results of the targeting experiments that we present here suggest that, as proposed earlier on the basis of sequence comparisons (16), the At3g18630 gene provides the mitochondrial UDG in A. thaliana. Further transcript pattern characterization and protein targeting experiments will determine whether this gene accounts for both the nuclear and the mitochondrial UDG, for instance through a mechanism similar to that described for the human UNG.
Oligonucleotide duplexes previously enabled to characterize complete BER reactions supported by protein extracts from mammalian mitochondria (41). In our in vitro and in organello studies, such substrates turned out to be too short to allow sequential recruitment of all the successive partners involved in the plant mitochondrial BER mechanisms. It seems likely in particular that they were unable to recruit the DNA polymerase. This may indicate significant differences between animal and plant mitochondrial BER proteins, especially in their interactions with each other and with the DNA substrate. Further fundamental differences seem to exist between the mammalian and plant mitochondrial mechanisms for uracil repair in the DNA. Whereas all our observations point to single nucleotide excision and reinsertion, i.e. short-patch BER, for the exchange of uridine for thymidine in plant mitochondria, repair of uracil can occur through both short- and long-patch BER in human mitochondria (42).
The mtDNA in yeast, mammalian or plant organelles is organized in discrete, membrane-associated protein–DNA complexes called nucleoids [(43) and references therein]. The data altogether suggest that mitochondrial nucleoids in eukaryotic cells are not only mtDNA compaction and segregation entities but also centers of mtDNA maintenance and expression. Stuart et al. (44) found that in mammalian mitochondria, the entire BER pathway, with the exception of AP endonuclease, is localized primarily in the membrane fraction. However, although they colocalize in the mitochondrial membrane, the different BER components do not seem to stably interact with each other and to build complete repair complexes in mitochondria, at least for the uracil repair pathway (45). Close colocalization would nevertheless enable efficient substrate handing over, avoiding exposure or release of sensitive and reactive repair intermediates (44–46). In agreement with these data, the submitochondrial localization studies that we present show that also in plant mitochondria the major part of the UDG activity is associated with the membranes. On the other hand, further differences appear between the mammalian and plant mitochondrial BER systems. Whereas AP endonuclease is soluble in mammalian organelles (44), the AP site-incising activity is essentially membrane-bound in plant mitochondria.
The in organello import-repair assays presented here confirm that DNA taken up by plant mitochondria can engage into complex genetic processes in the organelles. This opens the possibility to investigate complete repair of DNA modifications other than uracil or of mismatches. Furthermore, it was previously established that DNA incorporated into mitochondria can undergo transcription (21,47). It will be interesting to import a damaged DNA fragment carrying a sequence which can be transcribed in the organelles and to evaluate its repair efficiency [for instance with quantitative PCR (48)] versus that of an equivalent fragment which cannot be transcribed. Transcription-coupled repair (TCR) occurs in the nuclear compartment (49). Such analyses will give some hints as to whether DNA repair can be coupled to transcription in plant mitochondria, thus integrating DNA and RNA metabolisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French Centre National de la Recherche Scientifique (CNRS, UPR2357); Université Louis Pasteur (ULP), now part of the Université de Strasbourg. PhD fellowship from the French Ministry for Higher Education and Research (to P.B.); PhD fellowships provided by the Lebanese International University and the Lebanese Conseil National de la Recherche Scientifique (to N.I.); post-doctoral fellowship provided by the French Ministry for Higher Education and Research (to V.T.); the microscopy platform used in the present studies was co-financed by the CNRS, the ULP, the Ligue Nationale Contre le Cancer, the Association pour la Recherche contre le Cancer (ARC) and the Région Alsace. Funding for open access charge: IBMP, CNRS UPR2357.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank E. Meyer, C. Pujol, A.M. Duchêne, J. Gualberto, J. Canaday, P. Imbault, G. Houlne, D. Gagliardi and R. Lightowlers for help, discussions, suggestions or materials provided during the course of these studies.
Footnotes
Present addresses: Pierre Boesch, Mitochondrial Research Group, Institute for Ageing and Health, Newcastle University, Framlington Place, Newcastle upon Tyne, NE2 4HH, UK
Vladislav Tarasenko, Institute of Plant Physiology and Biochemistry, Russian Academy of Science, Ul. Lermontova 132, Irkutsk 664033, Russia
REFERENCES
- 1.Roldan-Arjona T, Ariza RR. Repair and tolerance of oxidative DNA damage in plants. Mutat. Res. 2009;681:169–179. doi: 10.1016/j.mrrev.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Møller IM, Jensen PE, Hansson A. Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 2007;58:459–481. doi: 10.1146/annurev.arplant.58.032806.103946. [DOI] [PubMed] [Google Scholar]
- 3.Kimura S, Sakaguchi K. DNA repair in plants. Chem. Rev. 2006;106:753–766. doi: 10.1021/cr040482n. [DOI] [PubMed] [Google Scholar]
- 4.Mower JP, Touzet P, Gummow JS, Delph LF, Palmer JD. Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol. Biol. 2007;7:135. doi: 10.1186/1471-2148-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muse SV. Examining rates and patterns of nucleotide substitution in plants. Plant Mol. Biol. 2000;42:25–43. [PubMed] [Google Scholar]
- 6.Draper CK, Hays JB. Replication of chloroplast, mitochondrial and nuclear DNA during growth of unirradiated and UVB-irradiated Arabidopsis leaves. Plant J. 2000;23:255–265. doi: 10.1046/j.1365-313x.2000.00776.x. [DOI] [PubMed] [Google Scholar]
- 7.Abdelnoor RV, Yule R, Elo A, Christensen AC, Meyer-Gauen G, Mackenzie SA. Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc. Natl Acad. Sci. USA. 2003;100:5968–5973. doi: 10.1073/pnas.1037651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen AC, Lyznik A, Mohammed S, Elowsky CG, Elo A, Yule R, Mackenzie SA. Dual-domain, dual-targeting organellar protein presequences in Arabidopsis can use non-AUG start codons. Plant Cell. 2005;17:2805–2816. doi: 10.1105/tpc.105.035287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Odahara M, Inouye T, Fujita T, Hasebe M, Sekine Y. Involvement of mitochondrial-targeted RecA in the repair of mitochondrial DNA in the moss, Physcomitrella patens. Genes Genet. Syst. 2007;82:43–51. doi: 10.1266/ggs.82.43. [DOI] [PubMed] [Google Scholar]
- 10.Shedge V, Arrieta-Montiel M, Christensen AC, Mackenzie SA. Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell. 2007;19:1251–1264. doi: 10.1105/tpc.106.048355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visnes T, Doseth B, Pettersen HS, Hagen L, Sousa MM, Akbari M, Otterlei M, Kavli B, Slupphaug G, Krokan HE. Uracil in DNA and its processing by different DNA glycosylases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009;364:563–568. doi: 10.1098/rstb.2008.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galperin MY, Moroz OV, Wilson KS, Murzin AG. House cleaning, a part of good housekeeping. Mol. Microbiol. 2006;59:5–19. doi: 10.1111/j.1365-2958.2005.04950.x. [DOI] [PubMed] [Google Scholar]
- 13.Duncan BK, Miller JH. Mutagenic deamination of cytosine residues in DNA. Nature. 1980;287:560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- 14.Shen JC, Rideout WM, III, Jones PA. High frequency mutagenesis by a DNA methyltransferase. Cell. 1992;71:1073–1080. doi: 10.1016/s0092-8674(05)80057-1. [DOI] [PubMed] [Google Scholar]
- 15.Sung JS, Demple B. Roles of base excision repair subpathways in correcting oxidized abasic sites in DNA. FEBS J. 2006;273:1620–1629. doi: 10.1111/j.1742-4658.2006.05192.x. [DOI] [PubMed] [Google Scholar]
- 16.Elo A, Lyznik A, Gonzalez DO, Kachman SD, Mackenzie SA. Nuclear genes that encode mitochondrial proteins for DNA and RNA metabolism are clustered in the Arabidopsis genome. Plant Cell. 2003;15:1619–1631. doi: 10.1105/tpc.010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nilsen H, Otterlei M, Haug T, Solum K, Nagelhus TA, Skorpen F, Krokan HE. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 1997;25:750–755. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B. A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiol. Biochem. 1992;30:123–128. [Google Scholar]
- 19.Maxam AM, Gilbert W. A new method for sequencing DNA. Proc. Natl Acad. Sci. USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neuburger M, Journet EP, Bligny R, Carde JP, Douce R. Purification of plant mitochondria by isopycnic centrifugation in density gradients of Percoll. Arch. Biochem. Biophys. 1982;217:312–323. doi: 10.1016/0003-9861(82)90507-0. [DOI] [PubMed] [Google Scholar]
- 21.Koulintchenko M, Konstantinov Y, Dietrich A. Plant mitochondria actively import DNA via the permeability transition pore complex. EMBO J. 2003;22:1245–1254. doi: 10.1093/emboj/cdg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakamoto W, Spielewoy N, Bonnard G, Murata M, Wintz H. Mitochondrial localization of AtOXA1, an Arabidopsis homologue of yeast Oxa1p involved in the insertion and assembly of protein complexes in mitochondrial inner membrane. Plant Cell Physiol. 2000;41:1157–1163. doi: 10.1093/pcp/pcd045. [DOI] [PubMed] [Google Scholar]
- 23.Heins L, Mentzel H, Schmid A, Benz R, Schmitz UK. Biochemical, molecular, and functional characterization of porin isoforms from potato mitochondria. J. Biol. Chem. 1994;269:26402–26410. [PubMed] [Google Scholar]
- 24.Werhahn W, Niemeyer A, Jansch L, Kruft V, Schmitz UK, Braun H. Purification and characterization of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis. Identification of multiple forms of TOM20. Plant Physiol. 2001;125:943–954. doi: 10.1104/pp.125.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- 26.Rubino L, Weber-Lotfi F, Dietrich A, Stussi-Garaud C, Russo M. The open reading frame 1-encoded (‘36K’) protein of Carnation Italian ringspot virus localizes to mitochondria. J. Gen. Virol. 2001;82:29–34. doi: 10.1099/0022-1317-82-1-29. [DOI] [PubMed] [Google Scholar]
- 27.Nagy JI, Maliga P. Callus induction and plant regeneration from mesophyll protoplasts of Nicotiana sylvestris. Z. Pflanzenphysiol. 1976;78:453–455. [Google Scholar]
- 28.Menand B, Maréchal-Drouard L, Sakamoto W, Dietrich A, Wintz H. A single gene of chloroplast origin codes for mitochondrial and chloroplastic methionyl-tRNA synthetase in A. thaliana. Proc. Natl Acad. Sci. USA. 1998;95:11014–11019. doi: 10.1073/pnas.95.18.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dip R, Camenisch U, Naegeli H. Mechanisms of DNA damage recognition and strand discrimination in human nucleotide excision repair. DNA Repair. 2004;3:1409–1423. doi: 10.1016/j.dnarep.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Krokan HE, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem. J. 1997;325(Pt 1):1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 32.Eftedal I, Guddal PH, Slupphaug G, Volden G, Krokan HE. Consensus sequences for good and poor removal of uracil from double stranded DNA by uracil-DNA glycosylase. Nucleic Acids Res. 1993;21:2095–2101. doi: 10.1093/nar/21.9.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slupphaug G, Eftedal I, Kavli B, Bharati S, Helle NM, Haug T, Levine DW, Krokan HE. Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry. 1995;34:128–138. doi: 10.1021/bi00001a016. [DOI] [PubMed] [Google Scholar]
- 34.Slupphaug G, Alseth I, Eftedal I, Volden G, Krokan HE. Low incorporation of dUMP by some thermostable DNA polymerases may limit their use in PCR amplifications. Anal. Biochem. 1993;211:164–169. doi: 10.1006/abio.1993.1248. [DOI] [PubMed] [Google Scholar]
- 35.Clayton DA, Doda JN, Friedberg EC. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc. Natl Acad. Sci. USA. 1974;71:2777–2781. doi: 10.1073/pnas.71.7.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stuart JA, Brown MF. Mitochondrial DNA maintenance and bioenergetics. Biochim. Biophys. Acta. 2006;1757:79–89. doi: 10.1016/j.bbabio.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 37.Kubo T, Newton KJ. Angiosperm mitochondrial genomes and mutations. Mitochondrion. 2008;8:5–14. doi: 10.1016/j.mito.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 38.Saccone C, Gissi C, Reyes A, Larizza A, Sbisa E, Pesole G. Mitochondrial DNA in metazoa: degree of freedom in a frozen event. Gene. 2002;286:3–12. doi: 10.1016/s0378-1119(01)00807-1. [DOI] [PubMed] [Google Scholar]
- 39.Elson JL, Lightowlers RN. Mitochondrial DNA clonality in the dock: can surveillance swing the case? Trends Genet. 2006;22:603–607. doi: 10.1016/j.tig.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 40.Takenaka M, Verbitskiy D, van der Merwe JA, Zehrmann A, Brennicke A. The process of RNA editing in plant mitochondria. Mitochondrion. 2008;8:35–46. doi: 10.1016/j.mito.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 41.Stierum RH, Dianov GL, Bohr VA. Single-nucleotide patch base excision repair of uracil in DNA by mitochondrial protein extracts. Nucleic Acids Res. 1999;27:3712–3719. doi: 10.1093/nar/27.18.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akbari M, Visnes T, Krokan HE, Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair. 2008;7:605–616. doi: 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 43.Dai H, Lo YS, Litvinchuk A, Wang YT, Jane WN, Hsiao LJ, Chiang KS. Structural and functional characterizations of mung bean mitochondrial nucleoids. Nucleic Acids Res. 2005;33:4725–4739. doi: 10.1093/nar/gki783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stuart JA, Mayard S, Hashiguchi K, Souza-Pinto NC, Bohr VA. Localization of mitochondrial DNA base excision repair to an inner membrane-associated particulate fraction. Nucleic Acids Res. 2005;33:3722–3732. doi: 10.1093/nar/gki683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akbari M, Otterlei M, Pena-Diaz J, Krokan HE. Different organization of base excision repair of uracil in DNA in nuclei and mitochondria and selective upregulation of mitochondrial uracil-DNA glycosylase after oxidative stress. Neuroscience. 2007;145:1201–1212. doi: 10.1016/j.neuroscience.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 46.Nakabeppu Y. Regulation of intracellular localization of human MTH1, OGG1, and MYH proteins for repair of oxidative DNA damage. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:75–94. doi: 10.1016/s0079-6603(01)68091-7. [DOI] [PubMed] [Google Scholar]
- 47.Koulintchenko M, Temperley RJ, Mason PA, Dietrich A, Lightowlers RN. Natural competence of mammalian mitochondria allows the molecular investigation of mitochondrial gene expression. Hum. Mol. Genet. 2006;15:143–154. doi: 10.1093/hmg/ddi435. [DOI] [PubMed] [Google Scholar]
- 48.Santos JH, Mandavilli BS, Van Houten B. Measuring oxidative mtDNA damage and repair using quantitative PCR. Methods Mol. Biol. 2002;197:159–176. doi: 10.1385/1-59259-284-8:159. [DOI] [PubMed] [Google Scholar]
- 49.Laine JP, Egly JM. When transcription and repair meet: a complex system. Trends Genet. 2006;22:430–436. doi: 10.1016/j.tig.2006.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.