Abstract
The prototype foamy virus (PFV) is a nonpathogenic retrovirus that shows promise as a vector for gene transfer. The PFV (pre)genomic RNA starts with a long complex leader that can be folded into an elongated hairpin, suggesting an alternative strategy to cap-dependent linear scanning for translation initiation of the downstream GAG open reading frame (ORF). We found that the PFV leader carries several short ORFs (sORFs), with the three 5′-proximal sORFs located upstream of a structural element. Scanning-inhibitory hairpin insertion analysis suggested a ribosomal shunt mechanism, whereby ribosomes start scanning at the leader 5′-end and initiate at the downstream ORF via bypass of the central leader regions, which are inhibitory for scanning. We show that the efficiency of shunting depends strongly on the stability of the structural element located downstream of either sORFs A/A′ or sORF B, and on the translation event at the corresponding 5′-proximal sORF. The PFV shunting strategy mirrors that of Cauliflower mosaic virus in plants; however, in mammals shunting can operate in the presence of a less stable structural element, although it is greatly improved by increasing the number of base pairings. At least one shunt configuration was found in primate FV (pre)genomic RNAs.
INTRODUCTION
Translation initiation on most eukaryotic cellular mRNAs occurs via 5′-end-dependent ribosomal scanning, where 40S subunits equipped with the necessary translation initiation factors (eIFs) bind the capped 5′-end of the mRNA and scan linearly searching for the initiating AUG codon (1,2). In contrast, some viral and cellular mRNAs do not require cap recognition and use alternative mechanisms, such as internal initiation, for translation initiation. Internal initiation mechanisms, which are characterized by cap- and eIF4E-independent binding of 40S to an internal ribosome entry site (IRES) on mRNA, are used widely by viruses to capture host cell functions. Long structured IRESs and their protein requirements have been characterized in detail in picornaviruses, hepatitis C virus and in retroviruses (3,4). Another alternative mechanism of initiation—ribosomal shunt—usually depends on cap-dependent discontinuous scanning, where ribosomes are loaded onto mRNA at the 5′-cap structure, start scanning for a short distance before bypassing the large internal leader region and initiating at a downstream start site (5). Ribosomal shunt has been described in mammals for adenovirus late mRNAs (6,7), Sendai virus Y mRNAs (8), papillomavirus E1 mRNA (9), duck hepatitis B virus (DHBV; 10) and some cellular mRNAs (6,11). It seems to be used widely by plant pararetroviruses, e.g. Cauliflower mosaic virus (CaMV; 12) and Rice tungro bacilliform virus (RTBV; 13).
In CaMV, essential and sufficient shunting elements have been identified; a short upstream ORF (sORF) and a downstream stable hairpin structure were able to direct the ribosomal bypass through the central leader region, which is loaded with elements that would otherwise impede ribosome scanning (12,14–16). In adenovirus, cis-acting elements of the tripartite leader do not contain sORFs or significant structural elements, but contain elements of complementarity to the 3′-end of 18S rRNA that act to promote shunting in the host cell even under conditions when conventional cap-dependent translation initiation mechanisms are impaired (6).
Foamy viruses (FVs) are the only members of the retroviral subfamily Spumaretrovirinae, and their replication strategy differs in many respects from that of other retroviruses in the Orthoretrovirinae subfamily (17,18). A major difference is that FV particles contain an infectious DNA genome (19,20). FVs reverse transcribe their (pre)genomic RNA (pgRNA) mainly late in replication (21). FV pgRNA contains open reading frames (ORFs) for Gag, Pol and Env originating from the long terminal repeat (LTR) promoter, and ORFs for nonstructural proteins Bel1 and Bel2 that are derived from an internal promoter (Figure 1A; 22). The pgRNA is subject to intensive alternative splicing leading to multiple subgenomic RNAs, each of which seems to be dedicated to the translation of a single viral protein (23).
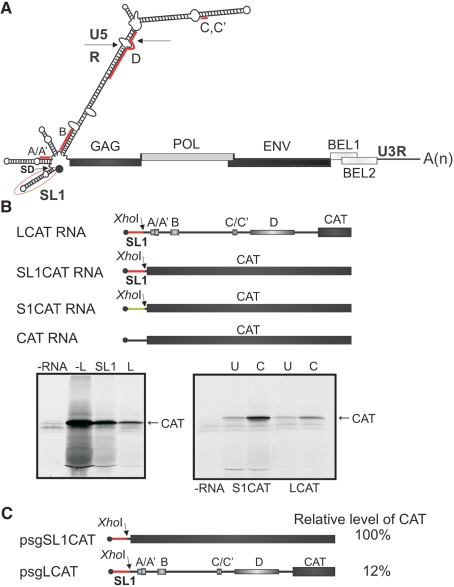
Figure 1.
The PFV RNA leader inhibits translation of a downstream ORF. (A) Schematic representation of PFV (pre)genomic RNA. Viral ORFs are shown as shaded boxes. The secondary structure of the PFV RNA leader as predicted by the ‘M fold’ program is presented. The small open reading frames (sORFs) in the leader are named and indicated by ‘thick red lines superimposed’ on the structure. A–D, sORFs; SD, splice donor; (A)n, poly(A) tail. The ‘circled in red section’ shows the 5′-proximal stem-loop (SL1). (B) Upper panel: mRNA constructs containing the chloramphenicol acetyl transferase (CAT) reporter ORF with three different leader sequences used for in vitro translation: LCAT RNA (the complete PFV leader); SL1CAT RNA; S1CAT (first 60 nt of the CaMV leader); and CAT RNA (50 vector nucleotides). Lower panels: (Left) in vitro translation of the constructs shown above in rabbit reticulocyte lysate (RRL); (right) translation efficiencies of capped (C) and uncapped (U) CAT mRNA containing either the S1 region (S1CAT RNA), or complete PFV leader (LCAT RNA) upstream of the CAT ORF. (C) 293T cells were transfected with a plasmid containing either the PFV 5′-proximal 50 nt (SL1 region; psgSL1CAT) or the PFV leader (psgLCAT) upstream of the CAT ORF. The psgGFP was used to monitor transfection efficiency. Levels of CAT expression relative to a GFP control after a 48-h incubation are indicated.
Prototypic FV (PFV) pgRNA contains an unusually long leader of 446 nt, which, according to computer predictions, folds into an extensive stem–loop structure that is potentially inhibitory for ribosomes scanning toward the first major ORF encoding Gag [see Figure 1A and ref. (24)].
This complex leader raises a question regarding the mechanism of translation initiation of the Gag ORF and essential leader elements required for initiation. The present study identifies cis-acting RNA elements in the PFV leader essential to achieve translation initiation via a ribosomal shunt mechanism.
MATERIALS AND METHODS
Gene constructs for experiments in vitro
In pS1CAT most of the CaMV leader, between the XhoI–ClaI sites in the previously described as pLm-CAT (14), was replaced with double-stranded oligonucleotides covering the region including the first 60 leader nucleotides (S1 region). pLCAT, or pSL1CAT or pCAT were prepared from pS1CAT, where the S1 sequence downstream of the T7 promoter was replaced either by the complete PFV leader (nt 1–445) or its first 48 nt amplified from pHSRV13, respectively; the whole PFV-1 leader upstream of the T7 promoter was modified with new restriction sites (XhoI) at the position of the main splice donor site (49TTTGGT to CTCGAG, where 49—position of the wild-type leader) and NcoI between the leader and the start site of CAT ORF (A446 to C; Figure 1B) to facilitate the introduction of mutations.
pLD::CAT carries an insertion of A445 and sORF D stop codon mutation (T361/C) to create D::CAT fusion protein and was prepared by site-directed mutagenesis. In pD1::CAT most of the leader (sORF A to sORF C′) in the pLD::CAT, was replaced with double-stranded oligonucleotides covering the region including the start codon of sORF D to create D::CAT fusion ORF.
Constructions for experiments in vivo
psg5m was used for cloning of CAT ORF downstream of the PFV leader and its mutant derivatives; and the green fluorescent protein (GFP) ORF under the control of the SV40 promoter (25). psgLCAT was prepared from psg5m by subcloning of the corresponding PCR fragments: the PFV leader with a new restriction site (XhoI) between EcoRI and BamH1 restriction sites—from pLCAT and CAT sequence between BamH1 and BglII from pLm-CAT. Thus, psgLCAT containing the PFV leader sequence from the cap-site to position +446 inserted between the CMV promoter and the CAT reporter ORF was used as a reference construct for transient expression experiments in 293T cells.
An HindIII site was created at position 148 with one point mutation (A146 to g) or position 222 (222TATT to aAgcTT); or an Acc65I site—at position 186 by introducing two point mutations, 187AA to GT. Insertion of the stem (Kozak stem, KS) was done with the oligonucleotide (for HindIII site; 5′-AAGCTGGGGCGCGTGGTGGCGGCTGCAGCCGCCACCACGCGCCCCAGCTT-3′; the sequence in bold forms the stem–loop structure) in the context of the PFV leader construct (psgLCAT) by making use of XhoI, Acc65I and two HindIII restriction sites (Figure 2A).
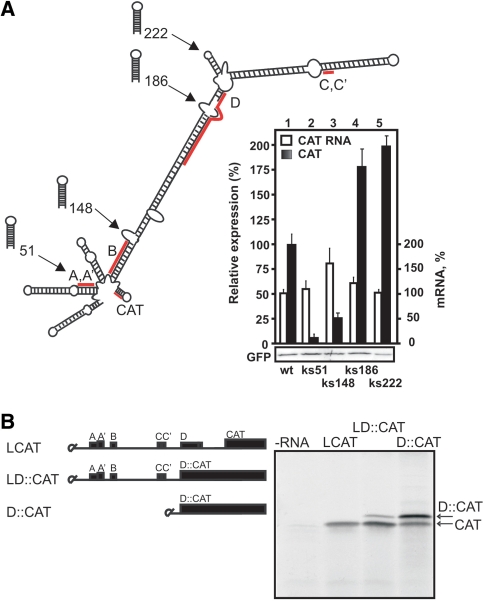
Figure 2.
The PFV RNA leader re-directs expression to a downstream AUG in vivo and in vitro via ribosomal shunt. (A) Effect of insertion of a stable stem–loop structure (ΔG < –43.9 kcal/mol) at the positions indicated along the leader on in vivo expression of the CAT ORF in 293T cells. 293T cells were co-transfected with either pLCAT or the Kozak stem (ks) containing construct and psgGFP (a transfection control). Expression levels of GFP analyzed by immunoblotting using anti-GFP antibodies are shown in the panel below. mRNA levels were measured using RT-PCR and are presented with the LCAT RNA level set as 100% (white bars). The results shown represent the means of three independent experiments. (B) Representative translation in RRL of capped RNA constructs containing a short (D::CAT) or full-sized leader (LD::CAT). In LCAT RNA, ORF D overlaps CAT ORF as in the wild-type situation with GAG ORF, whereas in the capped LD::CAT and D::CAT RNAs, ORF D is in-frame with the CAT ORF.
Mutations of the sORF A, or A′ or B (ATG to taG) around sORF A or A′ or B were introduced by PCR ligation between the XhoI and PstI sites in psgLCAT, yielding constructs (-A, -A′, -B). Mutations of the sORF A (ATG to ATGG), or A′ (ATG to ATGG) or to create fusion sORF A::B (ATG TTA CTA TtA TCC ATT AAC ACT CTG CTT AAG ATT GTA AGG GTG ATT GCA ATG CTT TCT GCA TAA; one point mutation is indicated) were introduced by PCR between XhoI and BamHI restriction sites, yielding constructs Astr, or A′str or A::B, respectively.
Substitutions of sORFs A and A′ with the original or mutated versions of the AdoMetDC uORF were achieved by PCR of TLMAGDIS, TLMAGRIS, TLMAGDI (16) with the 5′-primer of the desired sequence (sORFs A and A′ replaced by sORF MAGDIS or mutated versions) between the XhoI and PstI sites in psgLCAT resulting in clones, MAGDIS, MAGRIS and MAGDI (Figure 3A).
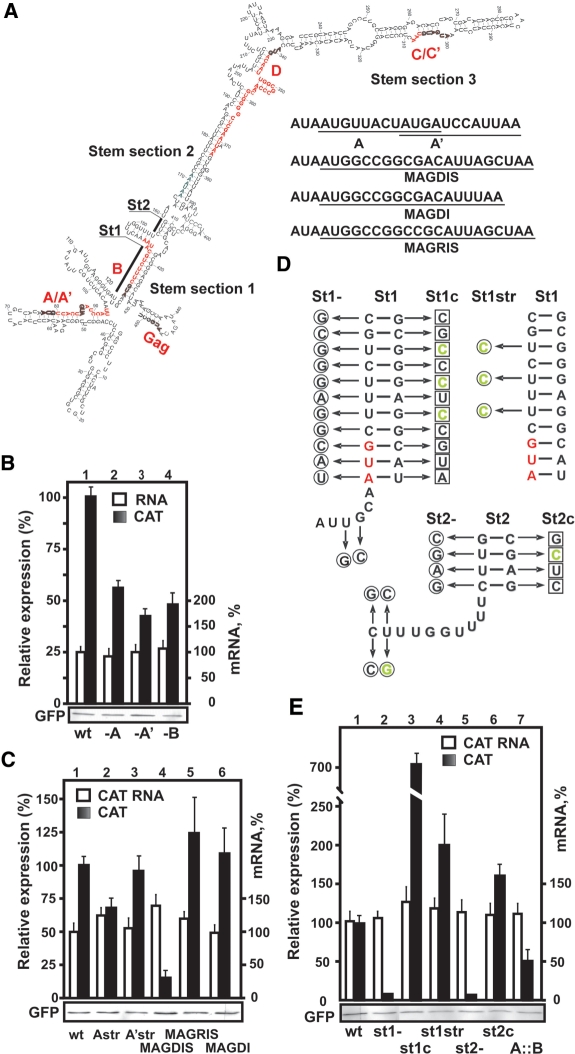
Figure 3.
Identification of essential cis-acting shunting elements. (A) Schematic presentation of the leader indicating the main stem sections 1–3 and their mutated fragments (St1 and St2). Replacement of sORFs A and A′ by MAGDIS and variants is illustrated on the right. (B and C) Representative expression in 293T cells of DNA constructs containing alterations in the 5′-proximal sORFs: (B) [-A, -A′, -B], and (C) [wt, Astr, A′str, MAGDIS, MAGRIS and MAGDI). (D) Schematic representation of st1 and st2 of the PFV RNA leader. The sets of mutations are indicated by arrows; those introduced in st1- or in st2- are shown on the left panel in circles; the additional complementary mutations introduced in st1c or st2c to restore the formation of stem 1 are represented by squares. Mutations introduced to strengthen St1 (st1str) are shown on right panel in green. Mutations increasing base pairing in stem sections 1 and 2 are shown in green. (E) Effect of strength of base pairing in stem sections 1 and 2 on downstream CAT translation. Mutations destabilizing stem sections 1 (st1-) and 2 (st2-); restoring base pairing by compensatory mutations (st1c, st2c) or strong base pair insertion (st1str) and (st2str) were tested for their effect on CAT translation. Yields of CAT protein expressed as a percentage relative to the level of CAT detected upon transfection of LCAT mRNA are shown (black bars). mRNA levels are presented with the LCAT RNA level set as 100% (white bars). Expression levels of GFP analyzed by immunoblotting using anti-GFP antibodies are shown below (B), (C) and (E).
In st1- and st2- stem section 1 and stem section 2, respectively were either disrupted with several point mutations, or the bulges within stem section 1 were replaced by base-paired regions (st1str; Figure 3D). st1- and st2- were used to introduce compensatory mutations, yielding constructs st1c and st2c, respectively (Figure 3D). Error-free recombinant plasmids were identified by DNA sequencing.
Proviral expression clone mutant constructions
The infectious molecular clone pczHSRV2 containing PFV genome under the cytomegalovirus (CMV) immediate-early gene promoter (26) was kindly provided by Axel Rethwilm (Institut fûr Virologie and Immunobiologie, Universität Würzburg, Germany). Mutations within the PFV leader (A-, A′-, B- and st2-) were introduced into pczHSRV-2 leader using pGEM-HSRV-2. The PFV leader sequence from pczHSRV-2 was subcloned into pGEM vector (Promega) using MluI and SwaI restriction sites.
Point mutations in HSRV leader sequence were introduced in pGEM-HSRV-2 plasmid by site-directed mutagenesis (Stratagene, La Jolla, CA, USA). The recombinant plasmid was amplified by PCR with the pair of specific primers (see Supplementary Table 1). The mixture was then incubated with DpnI in order to eliminate the template. Further the HSRV-2 leader sequences with appropriate mutations were cloned back into pczHSRV-2 using MluI and SwaI restriction sites. Error-free recombinant plasmids were identified by DNA sequencing.
Transfection of 293T cells
The 293T cells (2.5 × 106) were co-transfected with 500 ng of psgSL1CAT or psgLCAT or leader mutant-containing derivatives and with 500 ng psgGFP and the Lipofectin reagent (Gibco-BRL) as specified by the manufacturer. Forty-eight hours post-transfection, cells were lysed in lysis buffer for protein analysis. CAT ELISA test was done as described in Roche manufacture protocol and GFP western blot was done with anti-GFP primary antibodies (kindly provided by David Gilmer, IBMP, Strasbourg, France).
Transfection of 293T cells with 500 ng of pczHSRV-2 or leader mutant pczHSRV-2 constructions was done together with 500 ng of pRL-TK (Promega) that encodes Renilla LUC as a transfection control. GAG accumulation was analyzed by western blot using polyclonal anti-GAG antibodies (kindly provided by Dirk Lindemann, Technische Unversität Dresden, Germany) and Renilla LUC activity tests (Promega) were performed 48 h after transfection. CAT or GAG ORF-containing mRNA accumulation was quantified after 48 h of cell incubation by Q-PCR.
In vitro transcription and translation
Capped and uncapped transcripts were generated by in vitro transcription with T7 polymerase and KpnI-linearized plasmids as described (16). Rabbit reticulocyte lysate (gel-filtered) was from Roche Boehringer Mannheim Biochemica; 0.05 pmol/μl of transcripts (Figure 1B) and 0.1 pmol/μl each of other transcripts (Figure 2B), as well as [35S]methionine [29.6 TBq/mmol (NEN, Boston, MA, USA)] were used for in vitro translation in RRL according to the manufacturer′s instructions. MgAc2 and KAc were added to a final concentration of 1.0 mM and 100 mM, respectively, and the reaction was incubated at 30°C for 40 min. Five microlitres of translation reaction mixtures were resolved on 15% SDS–polyacrylamide gels. Gels were dried, exposed to X-ray film (Fuji), and analyzed using a PhosphorImager PC (Molecular Dynamics, Sunnyvale, CA, USA). The capped and uncapped mRNAs used were relatively stable during 40 min of incubation in vitro.
Real-time PCR
The DNaseI-treated-RNA-extract pellet (4 μg) was reverse transcribed using Superscript III reverse transcriptase system (Invitrogen) and amplified by PCR with primers specific to the 3′-proximal sequence of CAT or GAG, respectively (Supplementary Table 1). Amounts of mRNA were calculated by subtraction of the value obtained from the reaction without reverse transcriptase from that with reverse transcriptase.
RNA secondary structure analysis was performed using the MFOLD program (Wisconsin Package, version 6.0, Genetics Group, Madison, WI, USA) at 37°C.
RESULTS
The PFV leader strongly inhibits downstream translation initiation, in a 5′-end dependent manner
The major splice donor site located at position +53 following the start of transcription and the first splice acceptor located in the Gag gene are used to generate the mRNA for Pol. Thus, the pgRNA itself is used as an mRNA for production of Gag. The PFV leader consists of the R-U5 region of 345 nt and an additional 101 nt preceding the Gag ORF. We note for the first time that the leader is loaded with multiple sORFs (A–D, Figure 1A). It potentially folds into an elongated hairpin stem–loop structure (ΔG < –116 kcal/mol; Mfold, GCG; Figure 1A). We predict that such a leader should exert a strong negative effect on translation initiation at the downstream Gag ORF if initiation of the latter occurs via a canonical cap-dependent scanning mechanism.
To investigate translation initiation at the Gag AUG, several different sequences were placed upstream of the chloramphenicol acetyl transferase (CAT) reporter gene (Figure 1B, upper panel): the full-length PFV leader (L); the first 48 nt of the leader up to the splice donor site (SL1); the first 60 nt of the CaMV leader (S1; 27); or 50 nt of the vector (–L). We used a rabbit reticulocyte lysate (RRL) cell-free translation system to compare translation efficiencies of capped mRNAs with these leaders produced in vitro (Figure 1B). The full-length PFV leader-containing mRNA was translated about 7-fold less efficiently compared to the mRNA with the SL1 leader, and 10-fold less efficiently when compared to the mRNA with the vector control leader (-L; Figure 1B, left panel). The SL1 region was 5-fold less efficient in CAT ORF translation than the -L region, probably due to the presence of a stem–loop-like secondary structure (Mfold, GCG) near the cap site. Analysis of capped versus uncapped PFV leader-containing mRNAs suggested that the cap structure facilitates CAT ORF translation in vitro, albeit less efficiently than the S1 region of the CaMV leader (Figure 1B, right panel; 28). The full-length leader or its truncated version (the SL1 region) were introduced into the psg5m vector (25) downstream of the cytomegalovirus (CMV) immediate-early gene promoter followed by the CAT ORF, and CAT mRNA levels and accumulation of CAT activity were compared after 48 h of transient expression in 293T cells. The results confirmed that the PFV leader down-regulates CAT ORF expression, diminishing it by about 88% as compared with the inhibitory SL1 (Figure 1C), indicating that alternative mechanisms are indeed involved in GAG ORF expression.
Ribosomes translating the ORF downstream of the leader bypass the central leader regions
To investigate the mechanism that allows translation initiation downstream of the PFV leader, and to discriminate between 5′-end and IRES-dependent initiation, we introduced a strong, scanning-inhibiting stem (‘Kozak stem’, ΔG < –43.9 kcal/mol; Mfold, GCG) at the beginning (51 nt, position of the cryptic splice site) or at several points within the central region (148 nt, 186 nt and 222 nt; Figure 2A) of the leader. Insertion of the Kozak stem (ks51) at the 5′-end of the leader caused strong repression of CAT reporter translation (Figure 2A, right panel), as expected for translation events depending on 5′-end entry of a scanning complex (29). Insertion of the Kozak stem at a position downstream of sORF B inhibited CAT ORF translation 2-fold, suggesting that the 5′-part of the leader, up to position 186, is used for scanning by ribosomes that will go on to translate CAT (Figure 2A). The same stem inserted at the top of the elongated hairpin (positions 186 or 222, Figure 2A) increased the level of CAT expression by about 2-fold, indicating that this region of the elongated hairpin was not scanned by ribosomes responsible for CAT ORF translation. Indeed, the latter can be loaded with insertions without inhibiting downstream translation. The presence of strong stems in the central part of the leader might further stabilize the strength of the elongated hairpin and thus increase the efficiency of ribosomal shunt (14). These results suggest that ribosomal shunt is used as a mechanism for CAT ORF translation initiation, whereby scanning proceeds from the 5′-end but nonlinearly bypasses the large central segment of the leader region.
We predicted that the AUG codon of sORF D within the central leader region and the AUG of the CAT reporter are located upstream and downstream of the shunt landing site, respectively. To analyze the relative efficiency of initiation events at the central sORF (sORF D) and the CAT ORF, we used a mutant version of the PFV leader, fusing sORF D lacking its stop codon (T367 to C) in frame with the CAT ORF (insertion A445; LD::CAT; Figure 2B, left panel). Ribosomes scanning via central regions and initiating at the start codon of sORF D (in a weak initiation context; UCCAUGA) should produce D::CAT fusion polypeptides, whereas shunting ribosomes will initiate downstream of sORF D at the AUG of the CAT ORF. To control initiation efficiency at sORF D, we prepared a truncated leader construct lacking sORFs A, A′, B, C and C′, and containing the AUG codon of sORF D fused in frame to the CAT ORF (Figure 2B, D::CAT). In this construct, the first AUG (AUG of sORF D) was recognized preferentially by ribosomes despite the weak initiation context, and a smaller fraction of ribosomes continued scanning to initiate at the AUG of the CAT ORF (Figure 2B, right panel). In contrast, the PFV leader redirects ribosomes to initiate preferentially at the AUG of the CAT ORF, further confirming shunting.
The 5′-proximal sORFs, A, A′ or B, and a corresponding downstream structural element activate shunting
The first two AUGs of the PFV leader open sORFs A and A′. These sORFs overlap by four nucleotides, both encode three-amino-acid (aa) polypeptides and terminate upstream of ‘stem section 1’. sORF B encodes a four-aa polypeptide and terminates upstream of ‘stem section 2′ (Figure 3A). To investigate the role of the 5′-proximal sORFs in translation of the CAT ORF, we introduced point mutations to knock out sORFs A, A′ or B (AUG to uaG; -A, -A′, -B; Figure 3B). Knockout of the start site of sORFs A, A′ or B reduced CAT expression by 40%, 60% and 50%, respectively, indicating their importance, and apparently redundant, role in the PFV shunting mechanism (Figure 3B). The decrease in CAT expression upon removal of sORF B was also suggestive of its role in PFV shunting. The moderate initiation contexts of sORFs A, A′ and B (AUAAUGU; ACUAUGA; GCAAUGC, respectively) could potentially support leaky scanning through the A, A′ and B AUG start sites.
sORFs A, A′ and B are too short for their translation products to be detected in vitro or in vivo. To prove that translation of the 5′-sORF preceded CAT ORF translation, we employed a sequence-specific sORF encoding the polypeptide ‘MAGDIS’ from the leader of the mammalian AdoMetDC mRNA, which conditionally causes stalling of translating ribosomes at its termination codon in a polyamine-responsive manner (16,30) leading to suppression of reinitiation at the associated downstream cistron. The inhibition is peptide sequence dependent; a D4R mutation (MAGRIS sORF) alleviates this suppression by 87%, and deletion of the Ser at position 6 (MAGDI sORF) removes suppression in HeLa cells (31). We replaced sORFs A and A′ by either the six-aa sORF MAGDIS, its mutated version MAGRIS, or the five-aa sORF MAGDI in the wild-type leader-containing construct (Figure 3A). In addition, to compare the effect of MAGDIS-derived sORFs in a strong initiation context with the wild-type sORFs A and A′, the start codons of sORF A or A′ were mutated to an optimal context (AUGU to AUGg and AUGA to AUGg for A and A′, respectively). The 293T cells were transfected with these constructs and levels of CAT expression after 48 h were analyzed (Figure 3C). Improving the initiation context of sORF A′ had no effect on CAT AUG initiation (Figure 3C, lane 3), suggesting that it was already well recognized in its authentic initiation context, perhaps due to its location near to the elongated hairpin. sORF A placed in a strong initiation context slightly reduced CAT ORF translation (Figure 3C, lane 2), which can be explained by the inefficient action of sORF A in ribosomal shunt due to the suboptimal location of its stop codon relative to the base of the stem (Figure 3A; 14,16).
In contrast, insertion of the MAGDIS sORF can efficiently suppress CAT ORF translation by more than 75% (Figure 3C, compare lanes 1 and 4), confirming that this sORF is translated, and translation of the 5′-proximal sORF is a prerequisite for reinitiation at the downstream CAT ORF. A single nucleotide mutation converted sORF MAGDIS into MAGRIS, which is translated and terminated, activating CAT ORF translation. sORF MAGDI also supported shunting (Figure 3C, lane 6). These results suggest strongly that translation and proper termination of an sORF precedes translation of the CAT ORF, and that CAT production occurs via a reinitiation mechanism.
The PFV leader can be folded into an elongated hairpin that can be divided into three stem sections separated by bifurcations (Figure 3A), with stem sections 1 and 2 located downstream of sORFs A/A′ and B, respectively. To determine whether these stem sections contribute actively to shunting, the base-paired regions within stem sections 1 and 2 were destroyed by the introduction of 13- and 6-point mutations, respectively, into the left arm of each stem (st1- and st2-, Figure 3D). These mutations diminished CAT ORF translation by 95%, while the level of mRNA accumulation in 293T cells was essentially unaffected (Figure 3E, compare lanes 1, 2 and 5). To discriminate between the effect of primary sequence and stability of the stem on shunting, the base-pairings within each stem were restored by introducing mutations into the right arm complementary to those introduced previously into the left arm (st1c and st2c; Figure 3D). These compensatory mutations not only restored the wild-type level of translation but considerably increased it, especially for stem 1 (Figure 3E, lanes 3 and 6). The stem configuration in st1c contains 8 GC pairs as compared to five GC pairs in the wild-type construct, thus it can better support shunting (13). To verify this hypothesis, three central U-G base pairs in the left arm of stem 1 were replaced by C-G pairs in stem section 1 (Figure 3D). The mutant construct (st1str) with 8 GC pairs in stem 1 was 2-fold more efficient in CAT ORF translation (Figure 3E). Again, mRNA levels were not significantly affected (Figure 3E; open bars). Thus, strengthening base pairings within stem section 1 or 2 is favorable for CAT ORF translation. Taking these results together, we conclude that translation of the CAT ORF via shunting relies on the stability of the leader hairpin rather than on its primary sequence. The CAT AUG is translated preferentially in mutants with stabilized stem sections as would be expected for initiation via shunting.
A shunt configuration is an important feature of PFV and is conserved among primate FVs
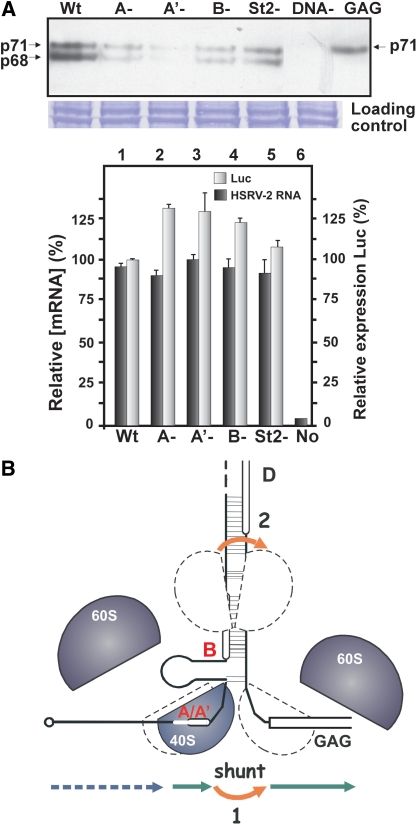
We used the proviral expression clone pczHSRV-2, which contains a complete viral genome but with the PFV main promoter replaced by the CMV promoter (26) to test the effect of specific point mutations within the leader region on GAG ORF translation. Mutations knocking out either sORF A (A-), A′ (A′-), or B (B-; AUG to uaG), and 4 nt substitutions within the right arm of st2 (GUUC to CGAG; st2-) were introduced separately into pczHSRV-2. Three independent experiments were performed for each mutant, analyzing cell extracts after 48 h of post-transfection incubation (Figure 4A). Two species of Gag with approximate sizes of 68 and 71 kDa (p68 was generated by a cleavage in the carboxyl terminus of PFV Gag) were detected in lysates of FV-infected cells (Figure 4A; the position of the full-length Gag is indicated on the right). Knockout of the start site of sORF A′ resulted in a significant reduction in GAG ORF expression, while knockout of either sORF A or B decreased GAG production by about 3-fold (Figure 4A). The effect of 5′-proximal sORFs was more pronounced in this context compared to that obtained in the experimental set up when the PFV leader was fused to the CAT ORF (Figure 3B). In contrast, partial disruption of stem section 2 was less inhibitory, decreasing GAG ORF expression by 2-fold (Figure 4A). None of these mutants affected RNA stability during the experiment. Although these results suggest that sORF A′ is a critical shunting element, sORF A and the second shunt configuration (sORF B and stem section 2) are also used by the virus to modulate shunting efficiency.
Figure 4.
Expression of viral Gag depends on intact 5′-proximal sORFs and leader secondary structure. (A) A plasmid containing a proviral expression clone of PFV (czHSRV-2) under the control of the cytomegalovirus (CMV) immediate-early gene promoter was used to analyze the role of sORFs and secondary structure in translation initiation of the viral GAG ORF. (Left) Immunoblots of PFV GAG of lysates from 293T cells transfected with the following plasmids: either pczHSRV-2-wt or PFV leader mutants: A–, A′–, B–; st2–, or psgGAG (GAG) and non-transfected 293T cell extract (DNA-). pRL-TK containing Renilla LUC under the control of the thymidine kinase promoter was used as transfection control (LUC). Accumulation of GAG is shown after 48 h of incubation. The blots were incubated with primary antibodies against GAG and developed by staining with peroxidase-coupled goat anti-rabbit secondary antibodies. The Coomassie blue-stained gel is shown below, confirming equal loading of samples. (Right) LUC functional activity of the lysates (gray bars) described above was monitored and is shown together with mRNA levels analyzed by Q-PCR (black bars; LUC and HSRV-2 RNA values set at 100%). (B) Model showing redundancy in shunting strategy of PFV. Schematic presentation of the secondary structure stem of the PFV RNA leader with the GAG ORF. Broken outlines on the structure indicate the positions of 40S ribosomal subunits before and after shunting. Arrows show migration of scanning (dashed), translating (green) and shunting (red) ribosomes (40S and 60S subunits are shown as gray shapes, with outlines representing the subsequent path of the same 40S subunit). Scanning ribosomes enter the PFV RNA at the capped 5′-end and scan until they reach the 5′-proximal sORF start codon, where they initiate and translate this sORF. The translation event provides a specially modified shunt- and reinitiation-competent ribosome that bypasses the corresponding stem section and is ready to reinitiate close to the GAG AUG codon.
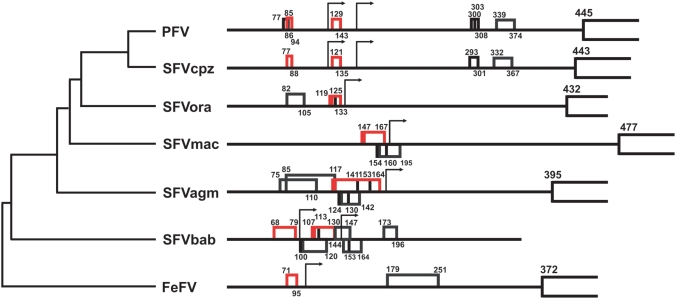
Computer-aided analysis of all sequenced primate foamy viruses helped reveal their structural organization and identified phylogenetically conserved features, i.e. cis-elements that might support ribosomal shunt. In most cases, the 5′-end of the leader can anneal to its 3′-sequence to form a stem–loop structure. In general, the pgRNA of primate foamy viruses begins with a long leader that carries several sORFs and is predicted to form a long elongated hairpin (5′-boundary of hairpin indicated by arrow, Figure 5). The (pg)RNA leader of SFVcpz (accession number NC_001364) is 443 nt in length and contains only four AUGs, with the most 5′-proximal sORF consisting of four codons as sORF A or A′ and terminating in front of the putative elongated hairpin. Two shunt configurations resembling those of the PFV (Y07725) leader can be seen in the leaders of SFVcpz and SFVbab (AF291395). In contrast, all AUGs are present upstream of the predicted elongated hairpin in SFVora (AJ544579), SFVmac (X54482) and SFVagm (NC_010820), suggesting only one single main shunt configuration. Remarkably, in the former case, at least one sORF has a strong initiation context to ensure its recognition. Surprisingly, even the distantly related feline foamy virus (NC001871) contains a putative shunt configuration within its leader.
Figure 5.
Comparison of primary structures of the pgRNAs of foamy viruses. The leader sequence preceding the GAG ORF is depicted by a thick line; sORFs are shown as boxes, with internal start codons indicated by vertical lines. Numbering within the leader is from the RNA 5′-end (except for SFVbab where the latter is unclear). The conserved sORFs preceding structured regions that can be used for shunting are in red. Arrows define the 5′- boundary of the elongated stem–loop structure. Leader sequences were derived from PFV proviral DNA; Simian foamy virus (SFVcpz); SFV-orangutan complete proviral genome, isolate bella (SFVora); SFV isolate OCOM1-5 long terminal repeat (SFVbab); SFV-3, (African green monkey complete genome; SFV-3agm); SFV-1 type 1 complete genome (SFV-1mac); Feline foamy virus complete genome (FeFV-1).
DISCUSSION
We have identified a mammalian retrovirus that exploits a ribosomal shunt mechanism activated by two leader cis-elements, i.e., an sORF and downstream elongated hairpin—a shunt configuration found earlier in Cauliflower mosac virus (CaMV). To our knowledge, this is the first example of a mammalian virus where the 5′-UTR regulates translation via sORF-stem-mediated shunting.
The peculiar feature of the PFV shunt is the redundancy of shunt configurations. The sORF-hairpin configuration is repeated twice in the PFV leader—1) sORFs A or A′ and stem section 1, and 2) sORF B and stem section 2—and both combinations can contribute to GAG synthesis (see model in Figure 4B). In CaMV we found two shunt configurations: configuration 1 represents the major shunting event, while configuration 2 represents a minor shunting event (32). In the case of PFV it seems that both configurations are used with roughly similar efficiencies, and both are essential for efficient GAG production (Figure 4A). It is interesting to note that some leaders of primate foamy virus isolates contain two shunt configurations (PFV, SFVcpz and SFVbab; see Figure 5), while others have only one (SFVora, SFVmac and SFVagm), with the loss of the B-like sORF being compensated for by the strong AUG context of the A-like sORF to ensure recognition of the latter (Figure 5). Overall, FV pgRNA leaders are structurally similar, each containing a structural configuration consisting of an sORF followed by a secondary structure element, and we therefore predict that sORF-dependent shunting is conserved, at least within primate FVs.
We used variations of the specialized sORF MAGDIS from the mammalian AdoMetDC mRNA (30), which can cause stalling of its translating ribosome at the stop codon preventing translation of the downstream ORF, to confirm that the 5′-proximal sORF is translated prior to shunting. Indeed, placing the MAGDIS sORF at the position of sORFs A/A′ almost blocks translation of the downstream CAT reporter, strongly indicating a translation event at this sORF, while a single aa change (MAGRIS sORF) that suppresses stalling activates the shunting event. Thus, translation of the GAG ORF is the second translation event and requires reinitiation. As previously suggested (16), the translation event at the sORF could render ribosomes less efficient in melting of stem sections, perhaps due to the removal of some helicase-like activities (eIF4A/eIF4B), which will favor bypass of the stem section. However, analysis of factor requirements for reinitiation after sORF translation in mammals suggested that eIF4F and eIF4A can remain associated with ribosomes terminating translation of the sORF (33). Thus, the role of the translation event in ribosomal bypass of the stem structure requires further analysis.
The PFV leader is shorter than the leader of CaMV pgRNA and folds into a less stable elongated hairpin (ΔG < -116 kcal/mol versus ΔG < -162 kcal/mol in CaMV). Our previous analysis of shunting efficiency in vitro suggests that CaMV stem section 1 is more stable in RRL compared to wheat germ extract (WGE; 16) and note that other secondary structure elements, such as the iron response element, inhibit scanning more in WGE than in RRL (34). In contrast, the sORF-stem configuration mediating shunting that was discovered recently on the mRNA of the cellular inhibitor of apoptosis 2 (cIAP2; 11) represents the other extreme, with a very stable secondary structure (ΔG < -620 kcal/mol). This mRNA contains a long (2.78-kb) 5′-UTR with a predicted elongated hairpin that, together with an upstream sORF, directs scanning ribosomes to initiate at the downstream main ORF exclusively via shunting (11). Thus, at least in mammals, ribosomes can bypass stems of different annealing capacities via a sORF translation event, although stabilization of the stem by viral or cellular proteins is not excluded. In PFV, introduction of additional base pairings into the PFV hairpin leads to a dramatic increase in translation of the downstream CAT reporter ORF.
Our results suggest that initiation-competent ribosomes arrive at the 5′-end of the PFV (pg)RNA and start scanning downstream towards sORF A/A′. We assume that some ribosomes can bypass these sORFs by leaky scanning and continue to migrate along the leader via an initiation–reinitiation mechanism. In CaMV, those ribosomes are intercepted by sORFs within the central leader regions, followed by their dissociation upon the termination event and subsequent inability to melt the stem–loop structure (32). In the PFV leader, sORFs C/C′ or sORF D as well as stem sections 1 and 2 may play similar roles. Interestingly, the first 48 leader nucleotides (SL1) upstream of the main splice donor site form a stem–loop structure, which could affect loading of ribosomes onto the 5′-end of the pgRNA, and indeed SL1 alone inhibits downstream ORF translation in vitro (Figure 1B).
The PFV (pre)genomic RNA leader contains multiple regulatory signals required for viral replication (18). The U5 region ends with the binding site for the tRNA primer (PBS) for reverse transcription, and is followed by several dimerization elements (DLSs, I-III) up to the Gag start codon (35). The R region positioned up to +51 in the leader region has been implicated in imprinting events in the nucleus involved in modifying the cytoplasmic fate of PFV RNA (36). Here, we have identified signals within the PFV 5′-UTR controlling translation of the downstream GAG ORF, thus revealing an additional layer of control. A region in U5 appears to be involved in either Pol packaging or viral protease activation (37). Binding of Gag or Pol at later stages of infection could then sequester the RNA from polyribosomes into previrions for reverse transcription. Thus, the central structural region, including at least some of these cis-signals, would be bypassed by shunting ribosomes and thus not melted, leaving then available for the replication process and for possible interactions with viral proteins. In CaMV, a purine-rich sequence at the top of the leader structure that is conserved in some other plant pararetroviruses is thought to constitute part of an encapsidation signal representing a stable stem–loop structure that can interact physically with CaMV Gag (38). Whatever the mechanism, we predict that sorting of genomic RNA between translation and replication processes plays an important role in the PFV life cycle.
FVs cause persistent but benign infections in their natural hosts, and have an extremely broad tropism, making them of great interest in the field of development of FV-derived vectors for vaccines and gene therapy (18). Knowledge of leader cis-elements required for translation versus replication and their interplay will help to dissect those signals, which will be essential to the design of effective gene transfer vehicles.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Fondation Recherche médicale (FRM, France) (INE20031114123 to L.R.); and Association française contre les myopathies (AFM, France) (N°11199 to L.R.). Funding for open access charge: IBMP, CNRS UPR 2357.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully thank A. Rethwilm for PFV proviral expression clone, D. Lindemann for anti-GAG antibodies. Thanks to H. Rothnie and M. Pooggin for critical reading of the manuscript.
Footnotes
Present address: Konstantina Katsarou, Laboratory of Molecular Virology, Hellenic Pasteur Institute, Vas.Sofias 127, Ave; 11521 Athens, Greece
REFERENCES
- 1.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 2.Pestova T, Lorsch JR, Hellen CUT. The mechanism of translation initiation in eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. NY: Cold Spring Harbor; 2007. pp. 87–128. [Google Scholar]
- 3.Jackson R. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 2005;33:1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- 4.Balvay L, Lopez LM, Sargueil B, Darlix JL, Ohlmann T. Translational control of retroviruses. Nat. Rev. Microbiol. 2007;5:128–140. doi: 10.1038/nrmicro1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryabova LA, Pooggin MM, Hohn T. Translation reinitiation and leaky scanning in plant viruses. Virus Res. 2006;119:52–62. doi: 10.1016/j.virusres.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 6.Yueh A, Schneider RJ. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev. 2000;14:414–421. [PMC free article] [PubMed] [Google Scholar]
- 7.Xi Q, Cuesta R, Schneider RJ. Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting. Genes Dev. 2004;18:1997–2009. doi: 10.1101/gad.1212504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Breyne S, Simonet V, Pelet T, Curran J. Identification of a cis-acting element required for shunt-mediated translational initiation of the Sendai virus Y proteins. Nucleic Acids Res. 2003;31:608–618. doi: 10.1093/nar/gkg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Remm M, Remm A, Ustav M. Human papillomavirus type 18 E1 protein is translated from polycistronic mRNA by a discontinuous scanning mechanism. J. Virol. 1999;73:3062–3070. doi: 10.1128/jvi.73.4.3062-3070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sen N, Cao F, Tavis JE. Translation of duck hepatitis B virus reverse transcriptase by ribosomal shunting. J. Virol. 2004;78:11751–11757. doi: 10.1128/JVI.78.21.11751-11757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sherrill KW, Lloyd RE. Translation of cIAP2 mRNA is mediated exclusively by a stress-modulated ribosome shunt. Mol. Cell Biol. 2008;28:2011–2022. doi: 10.1128/MCB.01446-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fütterer J, Kiss-László Z, Hohn T. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell. 1993;73:789–802. doi: 10.1016/0092-8674(93)90257-q. [DOI] [PubMed] [Google Scholar]
- 13.Fütterer J, Potrykus I, Bao Y, Li L, Burns TM, Hull R, Hohn T. Position-dependent ATT initiation during plant pararetrovirus rice tungro bacilliform virus translation. J. Virol. 1996;70:2999–3010. doi: 10.1128/jvi.70.5.2999-3010.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dominguez DI, Ryabova LA, Pooggin MM, Schmidt-Puchta W, Fütterer J, Hohn T. Ribosome shunting in cauliflower mosaic virus. Identification of an essential and sufficient structural element. J. Biol. Chem. 1998;273:3669–3678. doi: 10.1074/jbc.273.6.3669. [DOI] [PubMed] [Google Scholar]
- 15.Pooggin MM, Hohn T, Fütterer J. Role of a short open reading frame in ribosome shunt on the cauliflower mosaic virus RNA leader. J. Biol. Chem. 2000;275:17288–17296. doi: 10.1074/jbc.M001143200. [DOI] [PubMed] [Google Scholar]
- 16.Ryabova LA, Hohn T. Ribosome shunting in the cauliflower mosaic virus 35S RNA leader is a special case of reinitiation of translation functioning in plant and animal systems. Genes Dev. 2000;14:817–829. [PMC free article] [PubMed] [Google Scholar]
- 17.Murray SM, Linial ML. Foamy virus infection in primates. J. Med. Primatol. 2006;35:225–235. doi: 10.1111/j.1600-0684.2006.00171.x. [DOI] [PubMed] [Google Scholar]
- 18.Rethwilm A. Foamy virus vectors: an awaited alternative to gammaretro- and lentiviral vectors. Curr. Gene Ther. 2007;7:261–271. doi: 10.2174/156652307781369092. [DOI] [PubMed] [Google Scholar]
- 19.Yu SF, Sullivan MD, Linial ML. Evidence that the human foamy virus genome is DNA. J. Virol. 1999;73:1565–1572. doi: 10.1128/jvi.73.2.1565-1572.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy J, Rudolph W, Juretzek T, Gärtner K, Bock M, Herchenröder O, Lindemann D, Heinkelein M, Rethwilm A. Feline foamy virus genome and replication strategy. J. Virol. 2003;77:11324–11331. doi: 10.1128/JVI.77.21.11324-11331.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moebes A, Enssle J, Bieniasz PD, Heinkelein M, Lindemann D, Bock M, McClure MO, Rethwilm A. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J. Virol. 1997;71:7305–7311. doi: 10.1128/jvi.71.10.7305-7311.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Löchelt M, Flügel RM, Aboud M. The human foamy virus internal promoter directs the expression of the functional Bel 1 transactivator and Bet protein early after infection. J. Virol. 1994;68:638–645. doi: 10.1128/jvi.68.2.638-645.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muranyi W, Flügel RM. Analysis of splicing patterns of human spumavirus by polymerase chain reaction reveals complex RNA structures. J. Virol. 1991;65:727–735. doi: 10.1128/jvi.65.2.727-735.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J, Mergia A. Mutational analysis of the 5′ leader region of simian foamy virus type 1. Virology. 2000;274:203–212. doi: 10.1006/viro.2000.0423. [DOI] [PubMed] [Google Scholar]
- 25.Green S, Issemann I, Sheer E. A versatile in vivo and in vitro eukaryotic expression vector for protein engineering. Nucleic Acids Res. 1988;16:369. doi: 10.1093/nar/16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt M, Herchenröder O, Heeney J, Rethwilm A. Long terminal repeat U3 length polymorphism of human foamy virus. Virology. 1997;230:167–178. doi: 10.1006/viro.1997.8463. [DOI] [PubMed] [Google Scholar]
- 27.Fütterer J, Gordon K, Sanfaçon H, Bonneville JM, Hohn T. Positive and negative control of translation by the leader sequence of cauliflower mosaic virus pregenomic 35S RNA. EMBO J. 1990;9:1697–1707. doi: 10.1002/j.1460-2075.1990.tb08293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt-Puchta W, Dominguez D, Lewetag D, Hohn T. Plant ribosome shunting in vitro. Nucleic Acids Res. 1997;25:2854–2860. doi: 10.1093/nar/25.14.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozak M. Bifunctional messenger RNAs in eukaryotes. Cell. 1986;47:481–483. doi: 10.1016/0092-8674(86)90609-4. [DOI] [PubMed] [Google Scholar]
- 30.Raney A, Law GL, Mize GJ, Morris DR. Regulated translation termination at the upstream open reading frame in s-adenosylmethionine decarboxylase mRNA. J. Biol. Chem. 2002;277:5988–5994. doi: 10.1074/jbc.M108375200. [DOI] [PubMed] [Google Scholar]
- 31.Mize GJ, Ruan H, Low JJ, Morris DR. The inhibitory upstream open reading frame from mammalian S-adenosylmethionine decarboxylase mRNA has a strict sequence specificity in critical positions. J. Biol. Chem. 1998;273:32500–32505. doi: 10.1074/jbc.273.49.32500. [DOI] [PubMed] [Google Scholar]
- 32.Ryabova LA, Pooggin MM, Dominguez DI, Hohn T. Continuous and discontinuous ribosome scanning on the cauliflower mosaic virus 35 S RNA leader is controlled by short open reading frames. J. Biol. Chem. 2000;275:37278–37284. doi: 10.1074/jbc.M004909200. [DOI] [PubMed] [Google Scholar]
- 33.Pöyry TA, Kaminski A, Jackson RJ. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev. 2004;18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paraskeva E, Gray NK, Schlager B, Wehr K, Hentze MW. Ribosomal pausing and scanning arrest as mechanisms of translation regulation from cap-distal iron-responsive elements. Mol. Cell Biol. 1999;19:807–816. doi: 10.1128/mcb.19.1.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linial ML. Foamy viruses are unconventional retroviruses. J. Virol. 1999;73:1747–1755. doi: 10.1128/jvi.73.3.1747-1755.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell RA, Zeng Y, Erlwein O, Cullen BR, McClure MO. The R region found in the human foamy virus long terminal repeat is critical for both Gag and Pol protein expression. J. Virol. 2001;75:6817–6824. doi: 10.1128/JVI.75.15.6817-6824.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiktorowicz T, Peters K, Armbruster N, Steinert AF, Rethwilm A. Generation of an improved foamy virus vector by dissection of cis-acting sequences. J. Gen. Virol. 2009;90:481–487. doi: 10.1099/vir.0.006312-0. [DOI] [PubMed] [Google Scholar]
- 38.Guerra-Peraza O, de Tapia M, Hohn T, Hemmings-Mieszczak M. Interaction of the cauliflower mosaic virus coat protein with the pregenomic RNA leader. J. Virol. 2000;74:2067–2072. doi: 10.1128/jvi.74.5.2067-2072.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.