Abstract
Acetobacter species have been used for brewing traditional vinegar and are known to have genetic instability. To clarify the mutability, Acetobacter pasteurianus NBRC 3283, which forms a multi-phenotype cell complex, was subjected to genome DNA sequencing. The genome analysis revealed that there are more than 280 transposons and five genes with hyper-mutable tandem repeats as common features in the genome consisting of a 2.9-Mb chromosome and six plasmids. There were three single nucleotide mutations and five transposon insertions in 32 isolates from the cell complex. The A. pasteurianus hyper-mutability was applied for breeding a temperature-resistant strain grown at an unviable high-temperature (42°C). The genomic DNA sequence of a heritable mutant showing temperature resistance was analyzed by mutation mapping, illustrating that a 92-kb deletion and three single nucleotide mutations occurred in the genome during the adaptation. Alpha-proteobacteria including A. pasteurianus consists of many intracellular symbionts and parasites, and their genomes show increased evolution rates and intensive genome reduction. However, A. pasteurianus is assumed to be a free-living bacterium, it may have the potentiality to evolve to fit in natural niches of seasonal fruits and flowers with other organisms, such as yeasts and lactic acid bacteria.
INTRODUCTION
Acetic acid bacteria (AAB) is a widely divergent group within the alpha-proteobacteria and are isolated from a variety of natural fields such as fruits, flowers and fermented foods, and are rarely found in soils and insect guts (1–3). Recently, one species was identified as a human pathogen (4). Its metabolic uniqueness was also traditionally utilized to generate fermented food, especially vinegar, and also for industrial production of sorbose and dihydroxyacetone (5). AAB plays a crucial role in the quantity, taste, nutrition and hygienic quality of the productions, thus the reliability of strains in terms of functional properties and growth robustness is indispensable. AAB has a noticeable physiological instability and it is reported in at least two different terms of temporal acclimation and heritable adaptation (6–9).
Acetobacter strains, which are among the most popular AAB for the production of vinegar in many countries, may gradually acquire resistance against higher concentrations of acetic acid when properly adapted to the conditions. However, the rapid loss of the acquired phenotype is observed when the cells are maintained in environments without acetate as a selection pressure (7,8). This acquisition of acetic acid resistance must be an example of the temporal acclimation or physiologic adaptation, and one reason why new tanks for fermentation are continuously inoculated with AAB from old tanks, but not from a preserved seed AAB in traditional vinegar production.
Heritable deficiencies in various physiological properties of Acetobacter strains, such as ethanol oxidation, acetic acid resistance and bacterial cellulose synthesis, are observed at high frequencies. Very little is known about the genetic background for the instability but phenotypic modifications by transposon insertion were reported in ethanol oxidation and acetic acid resistance (6,9), and cellulose formation (10). Relatively copious transposons were identified in the genome of AAB strains, such as Gluconobacter oxydans 621H (11) and Granulibacter bethesdensis CGDNIH1 (12).
Genomic mutations are fundamental phenomena in the evolution of any organism. Bacteria may be more directly influenced by any mutations because of its minimized genomes or the density of information in its genomes. New information about the genomic structures and mutational events of bacteria have clarified various systems of the genomic mutations, such as a horizontal gene transfer of mobile gene units (e.g. transposon, plasmid and phage), hyper-mutable tandem repeat (HTR), genome-wide rearrangement and genome reduction (13), adding to the well-known mutation systems, such as nucleotide substitution, insertion and deletion and gene duplication (14).
In a harmful environment, mutations could provide beneficial alleles, but most mutations are likely to be neutral or deleterious, at least slightly, to organisms. Bacterial genomes accumulate mutations at certain rates even in well-fitted environments, and a natural environment causing stresses and stimuli may elevate the mutation rates (15,16). Therefore, DNA repair and proofreading systems must have evolved to minimize the rates of mutations and natural selection acts on the reliable maintenance and transmission of genome information (17).
Genome modulation for survival under certain stressful environments may cause other types of mutation, i.e. genome-wide rearrangement and genome reduction. Under nutrition depleted conditions, genome-wide rearrangements were experimentally observed with a certain frequency in Salmonella enterica (18) and Lactococcus lactis (19), suggesting the existence of a biological system acquired in evolution due to repeatedly facing harmful environments. Extreme genome reduction is mostly observed not only in endosymbionts and obligate intracellular pathogens (20–23), but also in a free-living photosynthetic marine bacterium, Prochlorococcus marinus (24). The genetic variation of symbiont genomes in comparison to free-living bacteria illustrate a large scale of gene loss that includes genes for regulation of transcription, translation and replication as well as genes for intermediate metabolism and amino acid biosynthesis.
In association with adaptation to a new niche, unnecessary genes without significant functions may accumulate mutations and may be deleted from the genomes. One explanation for genome reduction was derived from a comparative genome analysis between Buchnera aphidicola and Escherichia coli that showed large gene deletions by chromosome rearrangements and multiple events of disintegration dispersed over the whole genome (25,26). However, the mechanism associated with genome reduction or shrinkage is still controversial, with regard to whether a wide deletion may occur as a single event or as an accumulation of smaller deletions. A previous chlamydial genome analysis predicted the replication terminal region within the plasticity zone was involved in genome reduction according to enrichment of species specific, hypothetical and truncated genes (23).
The complete genomic DNA sequences of Acetobacter pasteurianus, which has been used for traditional vinegar productions, was herein determined to characterize the hyper-mutability of AAB, using a multi-phenotype cell complex formed by 21 years maintenance without crucial stresses. The strain that adapted to environments above the growth-limiting temperature was also subjected to genome analyses.
MATERIALS AND METHODS
Strain and culturing
IFO records (maintained in NBRC to date) show that Acetobacter spp. NBRC 3283, formerly assigned as Acetobacter aceti IFO 3283, was originally isolated from a pellicle (a kind of biofilm) on the surface of vinegar fermentation. The 16S rRNA sequences of the Acetobacter spp. NBRC 3283 in this work are all identical to one of A. pasteurianus but not to A. aceti or any other Acetobacter strains. Therefore, the nomenclature of this strain was taxonomically reassigned as A. pasteurianus in this article. Different lots of A. pasteurianus NBRC 3283 were distributed and maintained as a typical Acetobacter strains for research. An NBRC 3283 stock sample for genome analyses was the closest to an original IFO 3283 but contained several substrains. To avoid any nomenclature confusion, the substrains isolated from the IFO 3283 (NBRC 3283) were designated, such as A. pasteurianus IFO 3283-01.
The A. pasteurianus IFO 3283 (NBRC 3283) and Gluconacetobacter xylinus NBRC 3288 were obtained from National Biological Resource Center of Japan (NBRC) and grown routinely at 30°C on YPG medium 1.0% yeast extract, 1.0% polypeptone and 2.0% glycerol. A rich medium, YPGD, contains 1.0% yeast extract, 1.0% polypeptone, 2.0% glycerol and 0.5% glucose, and a minimum medium, NCG, contains 0.5% yeast nitrogen base (BD Difco, Franklin Lakes, NJ, USA), 0.5% casamino acid (BD Difco) and 2.0% glycerol. Potato solid medium was prepared with potato extract from 200 g of boiled potato per liter, 3.0% yeast extract (BD Difco), 0.5% meat extract (Wako Pure Chemical Industries, Osaka, Japan), 1.0% thioglycolate medium (Wako), 0.5% glucose, 1.5% glycerol, 1.5% CaCO3 and 1.5% agar (pH 7.0). Other media and supplementations, such as ethanol and acetic acid, are described in each figure legend. Short-term continuous cultivation was performed by a dilution method, i.e. when the optical density (OD600) of a culture reached ∼3.0, the culture was diluted to OD600 = 0.1 with 100 ml of fresh medium in a 300-ml flask and continuously incubated at 30°C with aeration by rotation at 120 r.p.m.
Between 1954 and 1974, A. pasteurianus IFO 3283 (NBRC 3283) was maintained by a serial passage every 3 months at the Institute for Fermentation, Osaka (IFO). In each passage, a loaf of cells picked from a former slant tube stored at 5°C was spread onto the potato solid medium in a new tube and incubated at 28°C for 4 days to allow the cells to grow enough for the next passage, followed by storage at 5°C for 3 months. Reconstitution of the A. pasteurianus maintenance herein was performed with the potato medium within slant tubes of 18 mm in diameter in a manner similar to that carried out between 1954 and 1974 in IFO.
Escherichia coli JM109 was grown routinely at 37°C on LB medium containing 1.0% NaCl, 1.0% bacto tryptone and 0.5% yeast extract.
DNA sequencing, assembling gap closing and variation detection
Three DNA shotgun libraries of A. pasteurianus IFO 3283 (NBRC 3283) complex were constructed with inserts of 1.5 and 5.0 kb in pUC118 vector (Takara, Otsu, Japan) and 35 kb in the pCC1FOS (Epicentre, Madison, WI, USA) fosmid vector. The plasmid clone DNAs were directly amplified from E. coli colonies and fosmid clone DNAs were extracted from E. coli transformants using the Montage BAC96 MiniPrep Kit (Millipore, Billerica, MA, USA), respectively, and DNAs were end-sequenced using dye-terminator chemistry on an ABI PRISM3730XL sequencer (ABI, Foster, CA, USA), as described previously (27). The 42 048 raw sequence reads corresponding to ∼9.3-fold coverage were assembled using the PHRED/PHRAP/CONSED software package (http://www.phrap.org) (28–30). A number of repeat regions detected in the assembling were masked and then the sequences in most of the masked regions were determined by primer walking to assemble the sequences into supercontigs. Seven supercontigs corresponding to a chromosome and six plasmids were constructed with remains of 30 gaps and 84 low-quality regions under 40 in Phrap score. DNA fragments were then produced to fill all the gaps, except for one locus, by PCR based on the supercontig. The one locus with a few similar transposons was independently sequenced using a fosmid covering the region and Template Generation System II Kit (Finnzymes, Espoo, Finland). All regions with sequences of lower quality than 25 in Phred value were resequenced by primer walking. Assembling of genome DNA was validated by three methods, i.e. Pulse-field gel electrophoresis (Bio-Rad, Hercules, CA, USA) after genomic DNA digestion by NotI, SfiI and FseI, fosmid clone digestion by several restriction enzymes and an Optical Mapping system (OpGen, Madison, WI, USA). When any inconsistency in assembling and coexistence of different nucleotides were detected, the sequence variations were confirmed by resequencing all 32 isolated clones.
The draft genomic DNA sequence of G. xylinus NBRC 3288 was obtained by similar methods as described above, and the DNA sequence of the largest plasmid pGXY010 was analyzed in this work.
For the mutation mapping of A. pasteurianus IFO 3283-01-42C, which is bred from IFO 3283-01 by adaptation to 42°C, DNA sequencing was performed with the Genome Analyzer (Illumina, San Diego, CA, USA). After the text editing of ∼1 billion of raw sequence data, mutation mapping was performed using the MAQ software (31). All candidates of mutation sites were directly resequenced using PCR products as templates by dye-terminator chemistry on an ABI PRISM3730XL sequencer (Invitrogen ABI, Carlsbad, CA, USA).
Genome DNA sequence analysis and annotation
Gene identification was performed with a method described previously (23). rRNA and tRNA genes were predicted using the BLASTN (32) and tRNAscan-SE (33) programs, respectively. Open reading frames (ORFs) longer than 90 bp were first predicted using the combination of three programs, GenomeGambler (34), GeneHacker plus (35) and Glimmer 3.0 (36), and at last protein-coding genes manually selected out of the ORFs. Annotation for each gene was carried out by using the BLASTP (37) and FASTA3 (38) programs against the nonredundant protein database. Transmembrane protein genes were predicted using SOSUI (39). The genes and the annotations of IFO 3283-01 are listed in Supplementary Table S2.
HTRs (or short tandem repeats) were extracted using the JSTRING software, (40), on genomic DNA sequences from A. pasteurianus IFO 3283 (NBRC 3283) substrains, G. oxydans 621H genome (11), Gluconacetobacter diazotrophicus PAL5 (GENBANK: NC-010123, NC-010124, NC-010125), Acidiphilium cryptum JF-5 (GENBANK: from NC-009467 to NC-009474, and NC-009484) and G. bethesdensis CGDNIH1 (12) and a G. xylinus plasmid, pGXY010 (BABN01000001 in this work).
Accession numbers of A. pasteurianus IFO 3283 (NBRC 3283) substrains
Thirty-two isolates from the A. pasteurianus IFO 3283 (NBRC 3283) cell complex stored in 1974 were classified into seven major groups. Seven substrains, one from each group, i.e. IFO 3283-01, IFO 3283-03, IFO 3283-07, IFO 3283-12, IFO 3283-22, IFO 3283-26 and IFO 3283-32, were re-deposited and are available from National Biological Resource Center of Japan (NBRC; http://www.nbrc.nite.go.jp/e/index.html; Table 1). The complete genomic DNA sequences of the seven strains including six plasmids are registered at GenBank/EMBL/DDBJ. The accession numbers are listed in Tables 1 and 2 and in Supplementary Table S1 in detail. The accession number for the largest plasmid, pGXY010, of G. xylinus NBRC 3288 is BABN01000001. The adapted strain, IFO 3283-01-42C, was re-deposited to NBRC and are available as NBRC 105185. The accession numbers of the genomic DNA sequence for IFO 3283-01-42C are listed in Supplementary Table S1.
Table 1.
Summary of phenotypes and mutation loci in the IFO 3283 (NBRC 3283) multi-phenotype complex
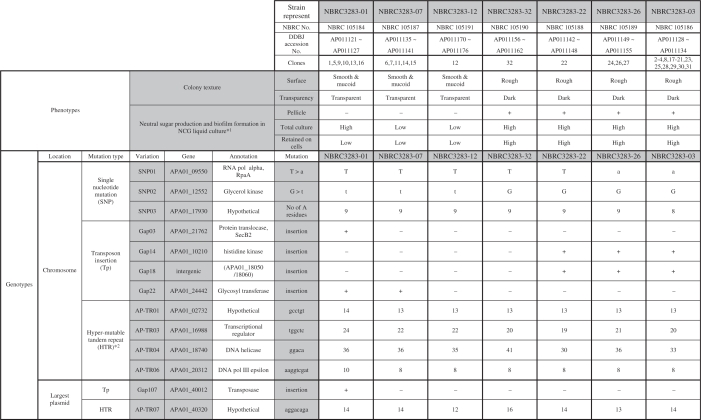 |
*1. High and low sugar production were determined based on the data of Figure 3b.
*2. Repetitive numbers of the representative substrains are indicated.
Table 2.
General features of A. pasteurianus IFO 3283-01 chromosome and plasmids
| IFO 3283-01 | Name | DDBJ accession No. | Size | Copy *1 | GC(%) | rRNA | tRNA | ORFs |
|---|---|---|---|---|---|---|---|---|
| Chromosome | APA01 | AP011121 | 2 907 495 | 1 | 50.7 | 5 operons | 57 | 2628 |
| Plasmid | pAPA01-011 | AP011122 | 191 799 | 0.8 | 59.3 | 0 | 0 | 178 |
| pAPA01-020 | AP011123 | 182 940 | 0.9 | 53.5 | 0 | 0 | 174 | |
| pAPA01-030 | AP011124 | 49 961 | 0.8 | 53.9 | 0 | 0 | 63 | |
| pAPA01-040 | AP011125 | 3204 | 7.4 | 53.7 | 0 | 0 | 3 | |
| pAPA01-050 | AP011126 | 3035 | 19.1 | 55.8 | 0 | 0 | 2 | |
| pAPA01-060 | AP011127 | 1815 | 29.1 | 58.6 | 0 | 0 | 2 |
*1, IFO 3283-01-42C cultured at 30°C.
High-throughput measurement of the total amount of neutral sugar
An amount of total neutral sugar as the polysaccharide components were measured based on a phenol-H2SO4 method (41). By this method, ribose and deoxyribose are detectable as well as glucose, mannose, galactose and fucose, thus the amounts of nucleic acid were compensated. For a liquid culture condition, A. pasteurianus was grown to ∼OD595 = 1 in the NCG medium and the culture was diluted with 0.85% NaCl solution to OD595 = 0.1. For plate culture conditions, A. pasteurianus was grown for 3 days on the YPG plate medium and the cells were harvested and diluted with 0.85% NaCl solution to OD595 = 0.1. The amounts of soluble and insoluble (retained with cells) neutral sugars were measured using the supernatants and cell pellets after centrifugation of the cell suspensions at 10 000 g for 5 min at 4°C. Briefly, 20-µl of samples in a 96-well titer plate was mixed with 20 µl of 5% of phenol and then 100 µl of H2SO4 was added to the mixture. The absorbance of the mixture was measured at 485 nm using a micro-titer plate reader (Beckman, Fullerton, CA, USA). A glucose solution was used for a concentration standard. The amount of nucleic acid in the sample was determined by measurement of the absorbance at 260 nm after mixing 126 µl of samples with 14 µl of 5% SDS solution in a well of a UV transparent multi-titer plate (Corning, New York, NY, USA). Double-strand DNA extracted from A. pasteurianus using a Genomic DNA extraction kit (Qiagen, Germantown, MD, USA) was used as a concentration standard. The amount of protein in the sample was measured using a Protein assay kit (Bio-Rad, Hercules, CA, USA) by absorbance at 595 nm after mixing 40 µl of the diluted cell suspension, 40 µl of 1% SDS solution and 20 µl of Protein assay solution in a well of a multi-titer plate. Bovine serum albumin was used as a concentration standard. As DNA in the sample affects the measurements of total sugar and protein concentration, the amounts of DNA were estimated and subtracted from the total sugar and protein measurements, and then the sugar production per protein was calculated.
High-temperature adaptation and mutation locus analysis
To attempt to breed a thermo-resistant A. pasteurianus strain, A. pasteurianus IFO 3283-01 cells, which exhibit less aggregation and produce sugar in liquid culture, were incubated at 40°C and the culture at approximately OD600 = 3 was inoculated into 200 ml of fresh YPGD liquid medium pre-warmed at 40°C to be OD600 = 0.1. After four passages, the culturing temperature was shifted from 40°C to 42°C. After a temperature tolerant strain was produced, serial cultures at 42°C were still continued 50 times to confirm the viability at that temperature.
Genomic DNA was prepared from clones [40C(3)1 and 40C(5)1] out of the third and fifth cultures at 40°C, respectively, and from clones [42C(1)9, 42C(11)1, 42C(21)1 and 42C(30)1] the first successful culture at 42°C cultivation and 11, 21, 30 succeeding cultures at 42°C, respectively. The genome DNAs were analysed by 1% agarose gel electrophoresis to check small size plasmids and HTR sequences, and by pulse-field gel electrophoresis to study macro scale genome structures.
Five micrograms of genomic DNAs from the IFO 3283-01-42C, which is a clone from the first successful culture at 42°C, were sequenced using a Genome Analyzer (Illumina inc. San Diego CA, USA) following the instruction of Post Genome Ins. (Tokyo, Japan). Two databases were constructed using the 10.0 million sequence reads obtained from the sequencing process and 2.3 million sequence reads with high qualities (at least 20 bases with Solexa score 25 or more in a read). These sequence read sets were separately mapped onto genome sequences of nonparental clones, such as A. pasteurianus IFO 3283-02, to evaluate whether the known mutation loci were identified, thus indicating that those data sets contained sufficient quality and quantity to extract mutation candidates. After extraction of mutation candidates using the genomic DNA sequence of the parental clone, IFO 3283-01, mutation sites were confirmed by PCR and DNA sequencing.
RESULTS AND DISCUSSION
Multi-phenotype cell complex
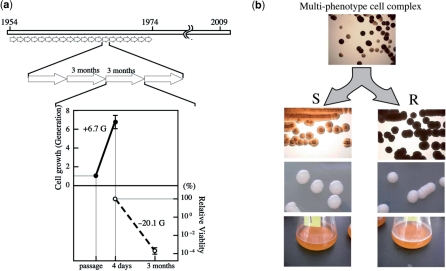
A. pasteurianus IFO 3283 (NBRC 3283) was maintained by a series passages every 3 months since the deposit of the pure culture at Institute for Fermentation, Osaka (IFO) in 1954 until the establishment of the freeze-dry preservation method in 1974 (Figure 1a). Since no colony isolation was performed in the maintenance to avoid the loss of the useful features of the strain, a multi-phenotype cell complex was formed characterized by different textures of rough (R) and smooth (S) colony surfaces, which may be based on a polysaccharide consisting of neutral sugars and a mucoid slime layer, respectively (42) (Figure 1b). The proportions of the rough and smooth colonies were 57% and 43%, respectively.
Figure 1.
Mutation accumulation within maintenance of A. pasteurianus IFO 3283 (NBRC 3283) for 21 years. (a) A schematic illustration of the maintenance and generations from 1954 to 1974. (Bottom) Indicates cell multiplication for the first 5 days up to 6.7 generations (a solid line) and decreased cell viability during the slants storage at 5°C for 85 days (a broken line) in a passage of every 3 months. Gs in the panel mean generation numbers. (b) Phenotypic variations in the IFO 3283 (NBRC 3283) cell complex stored in 1974. The top photo shows at least two types of colonies from multi-phenotype cell complex. Two independent clones, (R; rough colony and S; smooth) are shown in the two sets of bottom photos, the transparency (upper) and texture of the surface (middle) of colonies. (Bottom) Shows that R type clones produce a pellicle (biofilm) on the surface of liquid culture, but not S clones.
To estimate the generations in the 21 years of maintenance, the 3-month slant passage was reconstituted in this work using the potato medium. Within the first 4-day culturing in slant tubes, the cell number increased 101.2-fold, which is 6.7 generations (SD = 0.71). However, cell viability decreased to 0.88 × 10–6 (SD = 4.1) after 85 days storage in 5°C and each survival cell need to increased 0.88 × 106 fold (×220.1) to recover the original amount of cells. Together, cells needed to multiply ×220.1 and ×26.7, corresponding to ×226.7 or 26.7 generations, in each passage (Figure 1a). Consequently, it was estimated that 26.8 generations per passage every 3 months and thus 2.2 × 103 generations occurred within the 21 years, accompanying the formation of the multi-phenotype cell complex.
A. pasteurianus genome DNA sequence and its variations
Shotgun DNA sequencing of the A. pasteurianus IFO 3283 (NBRC 3283) genome was carried out using mixed genomic DNA extracted from the cell complex stored at IFO in 1974. After the assembly of the sequence reads with 9.3-fold coverage, an approximate 2.9-Mbp supercontig and six short contigs were constructed corresponding to the chromosome and six plasmids, respectively. Thirty-eight assembling gaps and sequence discrepancies were identified as candidate loci for genome variations, i.e. 30 gaps by transposon insertions, five possible sites of single nucleotide polymorphisms (SNPs) and three regions with HTRs.
The DNA sequences of the mutation candidates were completely determined using PCR products amplified from genomic DNAs of the 32 isolates (serially named from IFO 3283-01 to IFO 3283-32) from the IFO 3283 (NBRC 3283) cell complex, except one gap in the largest plasmid was sequenced by serial terminal deletions after cloning of the corresponding region from the IFO 3283-01. The results showed that four transposon insertions (numbered as Gap03, Gap14, Gap18 and Gap22), three SNPs (SNP01, SNP02 and SNP03) and three HTRs (AP-TR01, AP-TR03 and AP-TR04) were identified as chromosome variations and a transposon insertion (Gap107) and an HTR (AP-TR07) in the largest plasmid were found as a plasmid variation (Table 1). In the other 25 loci of the gaps, sequencing using long genome fragments shows that there are a few similar transposons but no genome variations in each locus. The complexity of transposons might prevent the program to assemble the loci with shotgun sequence data. Neither nucleotide substitutions nor insertion/deletion were found in the other two SNP loci out of the five candidates.
The mutation rate of non-mutator E. coli (chromosome size: 4.64 Mb) in the exponential phase was previously estimated as 2–8 × 10–4 per generation (43–45). When the rate is adapted to the A. pasteurianus (chromosome size: 2.9 Mb), its mutation rate may be 1.25–5.0 × 10–4 per generation. Therefore, the mutation amount are predicted by multiplication of the mutation rate and the generation numbers, resulting in 0.3–1.1 mutations within the 2.2 × 103 generations in the 21 years. Since the actual chromosome mutation number is eight (three SNPs and five transposon insertions) and A. pasteurianus seems to accumulate mutations faster than the non-mutator E. coli, the ‘experienced’ hyper-mutability of A. pasteurianus might be explained by the high rate of mutation (6). It is also reported that the mutation rate of E. coli in the starvation or ‘stationary phase’ increases up to 10–100 times higher than that in the exponential phase (46). However, A. pasteurianus cells were allowed to grow on a rich medium during the maintenance, 85 days storage in 5°C remarkably decreased the cell viability to 0.88 x 10−6. This suggests that A. pasteurianus cells might be sensitive to the nutrition depletion and that increases the mutation rate.
General features of A. pasteurianus genome
The genome constituents and its basic structures are the same in the 32 strains of A. pasteurianus IFO 3283 (NBRC 3283) cell complex, except the 13 variable loci. The IFO 3283-01 genome consists of a circular chromosome, APA01, of 2 907 495 bp in length with an average G+C content of 50.7% and six plasmids (Table 2). A saturated shotgun sequencing analysis of IFO 3283-01-42C derived from IFO 3283-01 showed that the copy numbers of the three largest plasmids are similar that of the one chromosome and the copy numbers of other plasmids are from 7.4 to 29.1-fold of one of chromosome (Table 2).
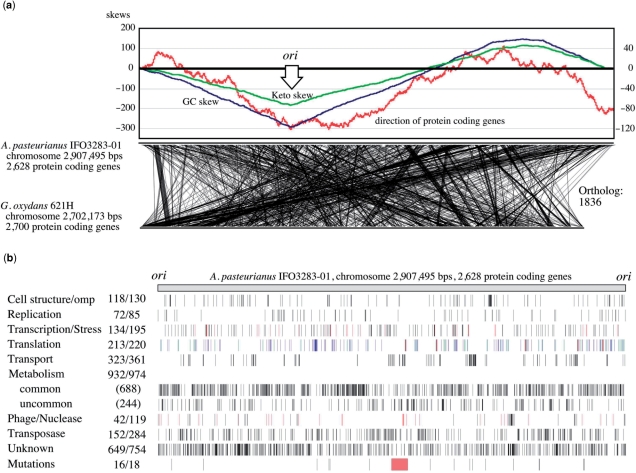
The putative replication origin (ori) of the chromosome was predicted based on the GC skew (47) and the coding direction of genes (48) (Figure 2a). There is a relatively strong correlation between the directions of transcription and replication. The replication terminal was not specifically determined, but may be located in the region (ter region) between 1.25 and 1.55 Mbp from the ori. The chromosome of IFO 3283-01 contains 2628 protein-coding genes, 57 tRNA genes and five copies of ribosomal RNA operons and plasmids encode 422 protein-coding genes (Table 2, Figure 2b and details in Supplementary Table S2).
Figure 2.
Genome map and structure of A. pasteurianus IFO 3283-01 chromosome. (a) The origin of replication was determined based on three values from cumulus of the GC (blue) and keto (green) skews, and transcription directions (red), and shown as ori (47). The locations of orthologs are shown by linking between A. pasteurianus and G. oxydans (11). (b) Genes in the chromosome are mapped after gene categorization into nine groups such as cell structure, replication and so on. Transcription and stress are indicated in black and red, respectively. Genes in translation are separated into protein coding in blue, and 52 tRNA in green and 15 rRNA in red. Numbers next to the categories are the number of genes in the chromosome and whole genome on the left and right, respectively. Genes in ‘common’ of metabolism means that its orthologs are found in both genomes of Gluconobacter oxydans (11) and Granulibacter bethesdensis (12). The mutation loci identified in this work are also mapped. Genome variation by three SNPs in red, four transposon insertions in blue and five HTRs in green, and terminal deletion in red of the 42°C evolved strain.
Specific functions were assigned to 75.1% (2291 genes) of the total 3050 protein-coding genes, and 24.9% were hypothetical genes. A remarkable number of genes, 285 (9.0%), was found to encode transposases (Figure 2b and Supplementary Table S3). A total 1836 orthologous genes between chromosomes of A. pasteurianus IFO 3283-01 and Gluconobacter oxydans 621H (11) are linked but a number of fragmented short regions show synteny between these species (Figure 2a). Locations of genes categorized into several groups are shown in Figure 2b. Notable biases of gene density were found in the groups of transport and phage related genes.
Metabolism
Acetobacter aceti IFO3281 utilizes glycerol as the best carbon source for growth (49), and glycerol was the best carbon substrate for A. pasteurianus as well under our experimental conditions (data not shown). Metabolic cascades of glycerol utilization was not unveiled yet by experimental methods, but it may be predicted by the results from Acetobacter xylinum (renamed to G. xylinus) (50,51) and the gene contents of A. pasteurianus in this work. The predicted metabolic pathways were illustrated in the Supplementary Figure S1. There are generally two pathways to produce dihyroxyacetone (DHA) phosphate from glycerol, one is via DHA catalyzed by glycerol dehydrogenase and the other is via glycerol 3-phosphate by glycerol kinase. The DHA phosphate may be converted to D-glyceraldehyde 3-phosphate by triosephosphate isomerase and thus enter into the glycolysis and gluconeogenesis pathway. However, the A. pasteurianus genome contains no genes encoding membrane-bound glycerol dehydrogenase which is well characterized in Gluconobacter (52), the capability to produce both DHA and glycerol 3-phosphate by G. xylinus (50,51) were reported and A. pasteurianus genome includes the glycerol kinase gene (described later) and several gens for alcohol dehydrogenase genes, such as sorbitol dehydrogenase. Therefore, A. pasteurianus might proceed glycerol utilization in both pathways. Acetobacter pasteurianus genome does not carry two genes encoding 6-phosphofructokinase (EC:2.7.1.11) and pyruvate ferredoxin oxidoreductase (EC:1.2.7.1) in the glycolysis pathway. The metabolism of sugars and glycolysis may be associated through the pentose phosphate pathway and acetate metabolism, respectively (11). There are no genes encoding succinyl-CoA synthetase (EC:6.2.1.4 and 6.2.1.5) in the citrate cycle shown previously and the precess might be substituted with the acetic acid resistance gene product, AarC (53). KEGG-based metabolic maps may be postulated in our web site, http://web.cc.yamaguchi-u.ac.jp/∼yazuma/Acetic_acid_bacteia_files/Metabolic%20map%20index.html.
Linage of genome variation
Unlike chromosome variations in diploid eukaryotes, mutations in a bacterial chromosome may conserve the order, in which the mutations occur in chromosomes. Chromosome variations observed in A. pasteurianus genomes, three SNPs and four transposon insertions were analyzed to classify a generation order of variations and a strain lineage (Table 1 and Figure 3a). HTRs were not considered for this purpose because the repetitive numbers of HTRs were modified even in the process of PCR-directed DNA sequencing.
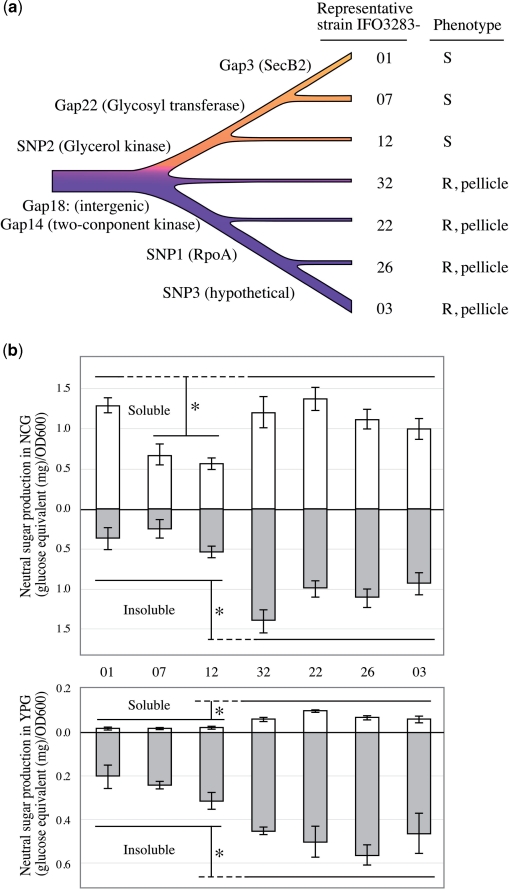
Figure 3.
Mutation accumulation within maintenance of A. pasteurianus IFO 3283 (NBRC 3283) for 21 years. (a) Genotype lineage (chromosome) and phenotypes of 32 substrains of the IFO 3283 multi-phenotype complex is summarized as an illustration based on Table 1. S and R indicate smooth and rough in the colony textures. (b) Profiles of neutral sugar content of the representing substrains measured in liquid and plate media. (Top and bottom) Show the amounts of neutral sugar in liquid and plate culture, respectively. White and gray bars indicate the amounts of soluble and insoluble neutral sugar, respectively. Vertical error bars show 1 SD calculated from at least four independent experiments and asterisk means significantly different in sugar production (P < 0.02).
SNP01 at 746 nt of rpoA gene (APA01-09550) shows alterations of T or A among the 32 isolates, which resulted in amino acid alterations of leucine or glutamine, respectively. Multiple alignment with other alpha-proteobacteria homologues suggested that leucine is the ancestral amino acid and thus T746 must be mutated to A. Similarly, G144 at SNP02 in glycerol kinase gene (APA01-12552) might be an ancestral one and mutated to T. Alteration of nine and eight adenine at SNP03 generates different coding frames to encode 604 and 90 aa in downstream of the SNP03, respectively. However, no significantly similar proteins to both 604 and 90 aa were found in nonredundant database, the frame with 604 aa is more likely an ancient type because no overlapping genes were observed on the coding region and the coding sequence does not contain a significant amount of minor codons, such as AGG and CTA.
When the corresponding genes (alleles) at chromosome variations associated with transposon insertions (Gap03, Gap14, Gap22 and Gap18) were compared among the 32 strains, all transposons were inserted in a same site in each allele, and no insertion or deletion were observed in alleles without transposons. This suggests that each transposon insertion happened in a certain strain and inherited, and no removing occurred.
More assumptions were added for the lineage analysis of the chromosome variation: (i) no reversible mutation occurred, (ii) no chromosome recombination happened between independent strains and (iii) when a wider range of strains with a certain mutation incorporates a smaller one with a different mutation, the former mutation may happen earlier than the latter one. For example, Gap22 is observed in IFO3238-01, -05, -09, -10, -13, -16, -07, -06, -11, -14 and -15, but Gap03 in only IFO3238-01, -05, -09, -10, -13 and -16. In this case, Gap22 was thought to happen earlier than Gap03 (Table 1).
Based on these assumptions, the 32 isolates were classified into seven groups in the order shown in Figure 3a. One transposon insertion (Gap107) in the largest plasmid was excluded from the categorization of the genome variations, because plasmids are possibly transmittable among the independent cells. However, no transmission of the plasmids was observed between different genome groups in this work. One representative clone each was chosen from the seven groups, i.e. IFO 3283-01, IFO 3283-03, IFO 3283-07, IFO 3283-12, IFO 3283-22, IFO 3283-26 and IFO 3283-32 (Table 1). IFO 3283-32 is the only strain without mutations, which may retain the original genotype, and may also show the original phenotype of A. pasteurianus NBRC 3283. The substrains carrying no (or a few) mutations were minor in the IFO 3283 cell complex.
Relationship between phenotypes and genotypes
The remarkable phenotypic differences in the 32 clones are the colony textures on plate culture and pellicle (a liquid/air interface biofilm) formation in liquid culture (Table 1, Figure 1b and Figure 3a). Strains forming rough surface colonies produce more polysaccharide consisting of neutral sugars than the other (42,54). Polysaccharide productivities of the 32 clones were measured. Since the clones in each group showed a similar production of neutral sugar, the averages of the sugar production in each group were calculated and are shown in Figure 3b.
Groups represented by IFO 3283-32, -22, -26 and -03 showed significantly high production of neutral sugar on plate and in liquid culture. On the other hand, IFO 3283-07 and -12 groups produced less under both conditions. Interestingly, the IFO 3283-01 group produced soluble sugar in liquid culture as high as IFO3283-32 etc., but did less in the other conditions like the IFO 3283-07 and -12 groups (Figure 3b). The constituents of the slime layer produced by smooth colony strains are possibly polysaccharides but chemically undetectable by the Phenol-H2SO4 method used in this study, and have never been determined.
Based on the genome lineage analysis, the single nucleotide nonsynonimous mutation in the glycerol kinase gene (SNP02) seems to relate to sugar production on plate and liquid media. However, at a terminal on the lineage, the high productivity of soluble polysaccharide of the IFO 3283-01 in liquid culture seems to be accompanied with disruption of secB2 gene (Gap03). Colony textures and pellicle formation of all 32 isolates in media containing 0.5% glucose were same as without glucose (Supplementary Figure S2), thus suggesting that the mutations, SNP02 and Gap03, affect polysaccharide production and the structural modification rather than glucose metabolism.
Glycerol kinase catalyzes the rate-limiting reaction in the pathway of glycerol utilization and the mutation is located in a beta sheet used to form the inter-homodimer Zn(II) binding site (55). Therefore, the SNP02 mutation in glycerol kinase gene might directly affect glucose control of glycerol utilization (56). However, since the strain with SNP02 can grow on glycerol as a sole carbons source at the similar speed as the strain without the mutation, the glycerol utilization by glycerol kinase may be similar to each other strain. SecB is a molecular chaperone in the general secretion system (57). The A. pasteurianus genome as well as G. oxydans (11) and a zoonosis pathogen Francisella tularensis (58), contain a well-conserved secB1 (APA01-07990) and a divergent secB2 (APA01-21762). The precise function of SecB2 and the relationship to polysaccharide synthesis are unknown. There are two genes related to glycerol metabolism (APA01-12570 and -12560) downstream of the glycerol kinase gene, and six genes (from APA01-21770 to -21820) in a predicted operon with secB2 (Supplementary Table S4). It is possible that these gene products affect the polysaccharide and pellicle production as well.
Fixation of mutant strains in population
Acetobacter pasteurianus IFO 3283 (NBRC 3283) was originally isolated from a pellicle. However, the strains of IFO 3283-07, -12 and -01 groups, which lost the ability to produce a pellicle, were fixed in this population and became a major portion of the cell complex (total 12 clones out of 32). It is a fundamental but difficult question to answer how mutant strains could be prominent during cell maintenance without special selective pressures.
The strains in the IFO 3283-07, -12 and -01 groups showed longer lag phase in the YPG medium containing 2.5% ethanol than the others; however, in the absence of ethanol the former strains demonstrated a slightly, but significantly higher density in the stationary phase than in the latter one (data not shown). All strains showed no significant difference in viability in 5°C storage for 3 months (data not shown). The strains of IFO 3283-07, -12 and -01 groups form more smooth colonies to spread easily on the surface of solid medium. This could explain the fixation in the population.
No phenotypic differences were observed among the groups of IFO 3283-03, -22, -26 and -32, but the strains in the groups of IFO 3283-32 and IFO 3283-22 turned to minor groups (1 each out of 32). Instead, strains in IFO 3283-03 and -26 turned to a major one (18/32) in the complex with two more mutations, SNP01 and SNP03. The SNP01 mutation in rpoA gene changes Leu to Gln at 149 aa, which locates at a hinge region between an N-terminal domain for initiation of subunit assembly and a C-terminal domain for transcriptional factor binding and DNA association (59). It is possible that the mutation affects the affinity of the alpha subunit to other transcriptional factors, and subsequently the transcriptional level of its regulon(s) modifies fitting ability to the given environment. A gene (APA01-17930) at SNP03 is hypothetical and probably monocistronic, thus the significance of SNP03 is unpredictable. The annotation of the genes related to SNP01 and other mutation sites are listed in Supplementary Table S4.
Transposons and plasmids
Acetobacter pasteurianus genome contains more than 280 transposons, ∼9% of total genes in the genome. Phylogenetic analysis revealed that the transposases in those transposons construct several paralogous clusters and the transposase in IS1380, which was previously reported as one of A. pasteurianus (9), is most abundant and the total copy number is 74. The phylogenetic tree was shown in Supplementary Figure 3, and a list of transposases and the result of paralogous clustering were shown in Supplementary Table S3. Thirty-two genes assigned for specific functions are truncated by the transposons (Supplementary Figure S3) and the gene truncation may affect the metabolism or response to environment stimuli. For instance, a gene cluster similar to the nitrate assimilation gene cluster of G. bethesdensis CGDNIH1 (12) was identified in A. pasteurianus genome, but the assimilatory nitrate reductase catalytic subunit (APA01_42100) was truncated by a transposon while the genes encoding a assimilatory nitrate reductase electron-transfer subunit (APA01_42120) and a nitrate transporter (APA01_42150) contain a frameshift in each coding region. It suggests A. pasteurianus is unable to utilize nitrate as a source of nitrogen by the reduction of nitrate via nitrite to ammonium.
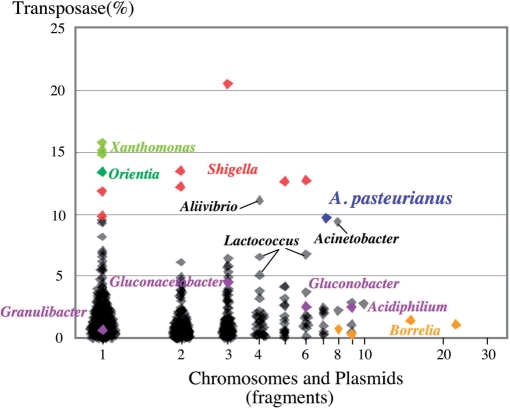
Acetobacter pasteurianus genome harbors 6 plasmids besides a number of transposons, which is 9% of total genes in the genome. Spiral bacteria Borrelia genomes harbor remarkable amount of plasmids (60), but transposons in Borrelia genomes account for only a few percentages of the total genes. Reversely, Shigera (61), Xanthomonas (62) and Orientia (63) contain a significant proportion of transposon but carry few plasmids in the genomes. The plot analysis of the proportion of transposase genes in the total genes and the fragment number including chromosomes and plasmids using genome sequences of 777 bacterial species in the public database, suggests that the combination of relatively large amounts of plasmids and transposons is one of the characteristics leading to the hyper-mutability of A. pasteurianus genome (Figure 4).
Figure 4.
Features of A. pasteurianus hyper-mutability. Proportion of transposase genes in the total number of genes (vertical) and DNA fragment number of the genome including chromosomes and plasmids (horizontal) are plotted using each genome of 777 bacterial species with gene sequences in the public database (ftp://ftp.ncbi.nlm.nih.gov/genbank/genomes/Bacteria). Blue and purple spots indicate the proportion for A. pasteurianus IFO 3283-01 and other AAB, respectively. Remarkable bacterial in this plotting, such as Borrelia (60), Shigera (61), Xanthomonas (62) and Orientia (63), are indicated in orange, red, light green and green. Profiles for other bacteria are shown in black, and those close to A. pasteurianus were indicated by bars, such as Aliivibrio, Acinetobacter and Lactococcus.
HTR
Three HTRs that cause genome hyper-variation were detected through the genome DNA sequencing and its assembling. Tandem repeats (TRs) were extracted using the JSTRING software (40) to determine all of the HTRs from the genome DNA sequence of A. pasteurianus. Seven TRs (named as AP-TR01–07) in A. pasteurianus were found to have a score over 200 (64), including the three HTRs (AP-TR01, 04 and 07; Table 1). Six TRs, AP-TR01 and AP-TR03-07, are built from homogeneous repetitive sequences, and AP-TR02 is built from a heterogeneous one. All TRs are tracked into individual protein coding regions. Out of the seven TRs, AP-TR01, 03, 04, 06 and 07 all showed hyper-mutability by PCR-directed DNA sequencing and therefore were designated as A. pasteurianus HTRs herein.
The repetitive sequence units and numbers of HTRs and genes including HTRs are listed in Table 1. For instance, a sequence unit and repetitive number of AP-TR01 in the IFO 3283-01 are gcctgt and 14, respectively. In sequencing to determine the repetitive numbers of HTRs, ∼5% of one repeat shorter and longer sequences were observed as backgrounds (data not shown). It seems that HTRs could be expanded and contracted by repetitive units during DNA multiplication from a single cell to PCR-directed colony sequencing, corresponding to several dozen generations (Supplementary Figure S2).
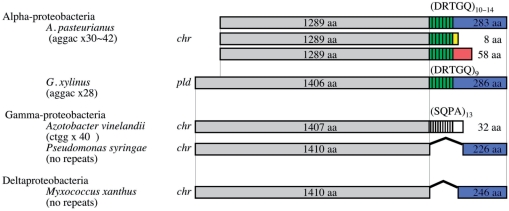
The repetitive sequence units of AP-TR04 and AP-TR07 are penta (aggac) and octa (aggacaga) nucleotides, respectively. Therefore, the repetitive numbers alter the coding frames following the HTRs. When the repetitive number of AP-TR04 is multiple of three, the gene, APA01-18740, encodes a 1289-aa sequence consisting of three domains, a DNA helicase domain in the N-terminal, DRTGQ repeats at the TR and 283 aa in the C-terminal In the other two cases of the repetitive numbers, the gene encodes 8 aa and 58 aa in the C-termini (Figure 5). An interesting sequence was found in a draft DNA sequence of G. xylinus NBRC 3288 genome (unpublished data). The largest plasmid of G. xylinus, pGXY010, contains homogeneous TR (aggac)28 in a DNA helicase gene similar to APA01-18740. The DNA helicase gene with the TR may be horizontally transferred between two independent genera by a plasmid and this horizontal gene/plasmid transfer might be another mechanism for genome instability of AAB.
Figure 5.
A schematic illustration of the structures of gene products with hyper-mutable tandem repeat sequences. Products of the genes with AP-TR04 are shown on top. Similar DNA helicases in the largest plasmid (BABN01000001) of Gluconacetobacter xylinus NBRC 3288, Azotobacter vinelandii DJ (65), Pseudomonas syringae pv. tomato str. DC3000 (NC-004578) and Myxococcus xanthus DK 1622 (NC-008095) are shown below. N-terminal regions in gray indicate a conserved DNA helicase domain. Tandem repeat regions are shown in green with strips for A. pasteurianus and G. xylinus and in white stripes for an A. vinelandii homolog. Amino acid sequences in C-terminal regions indicated in blue, yellow and red are encoded by same DNA sequences but different frame based on repetitive number of the AP-TR04 in A. pasteurianus. Amino acid sequences in C-terminal region in blue from four species are similar to each other. Gene locations, chromosome and plasmid, are indicated as chr and pld, respectively.
Genome DNA sequence of Azotobacter vinelandii DJ (65) was recently published and we found a high similarity between the two genes, A. pasteurianus APA01-18740 and A. vinelandii Avin_22890, which the authors assigned as a pseudogene (65). However, A. pasteurianus and A. vinelandii belong to different bacterial groups, alpha- and gamma-proteobacteria, respectively, the APA01-18740 and Avin_22890 contain TRs at a very similar site in each gene. Interestingly the repetitive sequence, (agcc)40, in the Avin_22890 is different from one in the APA01-18740. However the primary habitats of present A. vinelandii and A. pasteurianus are different, both A. vinelandii and A. pasteurianus are an obligate aerobic bacteria and found in soil worldwide, and it is possible that A. pasteurianus and A. vinelandii have shared its habitation and exchanged their genome information in their evolutional processes. It remains debatable whether the two genes are related to each other in their history or the existences of the two TRs in similar sites are just coincidental. Nevertheless, it remains difficult to explain how the different repetitive sequence units were generated.
The most favorable explanation responsible for the mechanism of TR instability attributing to hereditary neurological diseased such as polyglutamine expansion is a DNA strand slippage model (66,67), in which either the template or newly synthesized DNA strand forms partially stable secondary structures with repetitive sequences and overcoming the complex replication results in expansion or deletion of TRs. Several cellular mechanisms may underlay the process, such as general replication machinery including DNA polymerase III, helicases, recombination factors and DNA repair system (68–70). Where expansion and deletion of tandem repeats are thought to be enhanced by mutations that reduce the fidelity of replication (68), two genes with HTRs encoding a DNA helicase and DNA a polymerase III exonuclease epsilon subunit may be involved in the hyper-mutability of A. pasteurianus.
It is still unclear whether the involvement of translesion synthesis (TLS) or TLS polymerase is significant in the mechanism of expansions or deletions of TRs (69,71). Genome of A. pasteurianus contains genes encoding RecA, RecFOR, SSB, and DNA polymerase I, III and IV, but not RecBCD, or DNA polymerase II or V. Therefore, it is possible that TLS is involved in A. pasteurianus hyper-mutability, if so, DNA polymerase IV, of which a mutant was shown to increase instability of TRs in E. coli, is the only error-prone DNA polymerase in A. pasteurianus (69).
Hyper-mutable TRs may play critical roles in the phenotypic modification of bacteria, such as phase variation and genetic diversity in pathogens. A TR is located in the promoter sequence of the Neisseria meningitides nadA gene coding a pathogenic adhesion protein and the number of repeats influences the binding status of transcriptional regulators to the nadA promoter and the nadA gene expression (72). High-frequency phase variation of LPS structures, characteristic of Haemophilus influenzae requires licA gene regulation. Changes of the TR repeat number in the 5′ region of licA result in a translational switch for the licA product involved in LPS biosynthesis (73,74). A. pasteurianus HTRs locate in the middle of protein coding region, thus the meaning of HTRs may be different from the cases for transcriptional and translational regulation.
Four other AAB genomes were analyzed using the JSTRING software, (40) to determine the prevalence of TRs in AAB. The G. oxydans 621H genome (11) also contains two homogeneous repeats. Genomes of other AAB, G. bethesdensis CGDNIH1 (12), G. diazotrophicus PAL5 and A. cryptum JF-5, include many heterogeneous repeats, such as a unique long repeating sequence (cccgtggcg)205, but rarely a homogeneous one. As a result, TRs are prevalent in AAB, but sequences of the repeat units, repetitive numbers and genes with the TRs are varied. The TRs might be significant, but their roles are largely unknown.
Adaptation to unviable high-temperatures
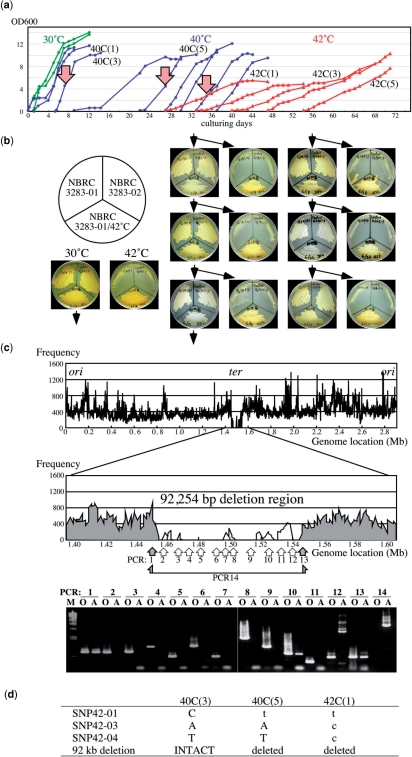
To clarify whether A. pasteurianus has other systems for genome instability and to show the utility of the genome instability to produce useful features, we performed an adaptation to unviable temperatures under conditions accelerating evolution in vitro. Acetobacter pasteurianus IFO 3283-01, which produces soluble polysaccharide in liquid culture and formed less aggregation, was used. Acetobacter pasteurianus can grow at 39°C, but the growth is unstable at 40°C. No growth was observed at 40.5°C or higher (data not shown). Then adaptation at high temperature was initiated by a long-term cultivation at 40°C to allow the cells grow stably at 40°C. Remarkable morphological changes were observed during this cultivation. The cells lengthened from ∼2–3 µm up to 300 µm within the first 3 days, and 10 days later the cell length shortened to ∼5 µm (Supplementary Figure S4). The long cells contained multi-nucleoid bodies. It is not clear yet whether the long cells turned into shorter or that short cells present at beginning occupied a majority in the population.
After 27 days cultivation at 40°C, representing five times continuous dilution-growth culturing passages, the cells in fifth culture at 40°C [termed 40C(5)] could be successfully established to grow at 42°C (Figure 6a). Growth at 42°C was tested for 365 days by serial dilution-growth cultivation (partially shown in Figure 6a). In principle, the acquired resistance may result from physiological or genetic adaptation (75). When the acquired 42°C resistance resulted from a transient physiological adaptation similar to phenomenon of adaptive pH response in Acetobacter (8,76,77), the cells would rapidly lose the 42°C resistance upon cultivation under nonlimiting conditions at 30°C culturing. The first evolved culture, 42C(1), and the parental strain IFO 3283-01 were grown on YPG plates at 30°C and 42°C for 3.5 days. The cells grown at 30°C were successively cultured on YPG plates at 30°C and 42°C twice a week. After 13 serial platings at 30°C, for total for 7 weeks representing 150 generations, the ability of growth at 42°C still remained (Figure 6b), thus suggesting that the acquisition of growth ability at 42°C is based on mutation(s).
Figure 6.
Adaptation to an unviable environment at high-temperatures. (a) Growth profiles of A. pasteurianus IFO 3283-01 in the successful case for adaptation. Green, blue and red curves indicate cell growth at 30, 40 and 42°C, respectively. Three arrows, 40C(3), 40C(5) and 42C(1), indicate the cell cultures analyzed for mutations. (b) A heritability analysis of growth at 42°C. The seed strain (IFO 3283-01), the bred substrain (IFO 3283-01/42°C) and a different cell type strain (IFO32383-02) were inoculated on same plate and tested for growth at 30°C and 42°C. Cells grown on a 30°C plate were then inoculated onto the next two plates for growth at 30°C and 42°C. (c) A mapping analysis of sequencing reads on the chromosome of IFO 3283-01/42°C (top). Vertical and horizontal axes indicate chromosome location and frequency of high-quality sequence reads in 1000 bases. PCR analyses were carried out for the large deletion region and positions of primer pairs for PCR analyses were shown in the middle. DNA was used from the clone IFO 3283-01-42C isolated from the culture 42C(1) indicated by the right arrow in the panel a. Gray zones and gray arrows in the middle panel indicate existence of the region in the genomic DNA, while white areas, no-mapped regions and white arrow show the region deleted from the genome. PCR products analyzed in agarose gel electrophoresis were shown in the bottom. O and A on the top of each lane designate DNA templates for PCR prepared from the original (IFO 3283-01) and the adapted strain (IFO 3283-01-42C), respectively. (d) A summary of the mutations found in the adapted strain (IFO 3283-01-42C). Original and mutated sequences are shown in capital and small letters, respectively.
Genomic DNA prepared from a clone from 42C(30) and the parental strain IFO 3283-01 were compared by pulse field gel electrophoresis after digestion by restriction nucleases to clarify the genome-wide modifications. No remarkable differences were observed except a fragment probably corresponding to the smallest plasmid (data not shown). Plasmid contents were analyzed and it was revealed that proportion of the smallest plasmid in the genomic DNA was remarkably changed at the 11th culture 42C(11) and it was undetectable in the 42C(21) culture (Supplementary Fig. S4). Repetitive numbers of the three HTRs were modified in the cultivation at 42°C (Supplementary Figure S4).
The clone, A. pasteurianus IFO 3283-01-42C, isolated from the first culture at 42°C may contain neither genome-wide rearrangement nor plasmid depletion, and therefore mutation mapping was carried out by saturated genome shotgun sequencing with a Genome Analyzer (Illumina, San Diego, CA, USA). Approximately 10 million reads of 35-bp raw data were obtained. Using the raw data, a high-quality data set with only 1.4 million high-quality reads was prepared, corresponding to 14-fold of A. pasteurianus genome (Supplementary Figure S4). Mapping analysis was performed with the whole and high-quality data sets using the MAQ software (31).
Analyses with the whole and high-quality data sets indicated that there were 13 and 4 SNP candidates in chromosome, respectively, but no SNP candidates in the plasmids. Three SNP candidates were chosen from both analyses from the two data sets, thus DNA sequences at 14 SNP candidate loci were determined using PCR-direct DNA sequencing. Consequently, the three SNP candidates, i.e. SNP42-01, SNP42-03 and SNP42-04, chosen with both data sets were shown as actual SNPs in IFO 3283-01-42C genome. Besides SNPs, sequence mapping indicated that there are clustered regions without mapped sequences at the predicted replication terminal region (Figure 6c). The entire regions were thoroughly sequenced using PCR products, and it revealed that a region 92 kb in length involving APA01-13760 to APA01-14470 was truncated.
To clarify the contributions of the SNPs and the 92-kb deletion region for adaptive evolution at high temperature, clones from 40C(3) and 40C(5) were tested for viability at 42°C and the genome variations shown in IFO 3283-01-42C. The 40C(5) clone was able to multiply at 42°C but not the 40C(3) one. No mutations were observed in the 40C(3) clone, but the SNP42-01 and 92-kb deletion were detected in 40C(5). This indicated that the SNP42-01 and/or 92-kb deletion may be involved in the adaptive evolution (Figure 6d).
The SNP42-01 was located in APA01-42C-00810 coding a two-component hybrid sensor istidine kinase and regulator. The mutation, from C1187 to T, is nonsynonymous, and resulted in amino acid substitution T396I in a conserved domain among histidine kinases. Therefore, the mutation may affect the signal transduction by an unknown environmental stimulation. The 92-kb deleted region includes 72 genes including a rRNA operon, 16 putative pseudogenes, 21 metabolism-related genes, 14 transposons, 13 transporter genes and six genes encoding unknown function proteins (Supplementary Table S2). It is very difficult to evaluate the association of the SNP42-01 gene and genes in the deleted region with the adaptive evolution. While shorter plasmids and mitochondrial genomes show faster replication (78), the 92-kb deletion corresponding to approximate 3% of the whole genome may affect growth speed or cell viability by decreasing the stress for proliferation in this stressful environment.
CONCLUDING REMARKS
Microorganisms utilized in academic and industrial fields must be maintained to conserve identical characteristics wherever and whenever used. Extraordinary consideration for storage and handling should be paid to hyper-mutable microorganisms isolated from nature. In the middle of the 20th century, it was believed that AAB should be maintained by serial passage to avoid phenotypic modification, but neither frozen nor maintained at 4°C, because of the extraordinarily low survival rate and high mutation frequency (79,80). Furthermore, colony isolation during the passage was prohibited to avoid cloning unanticipated mutants. Attempting to clarify the hyper-mutability of A. pasteurianus, known as a hyper-mutable bacterium (6,9,10), therefore required the genome DNA sequencing of cells grown in two different culturing conditions.
First, A. pasteurianus IFO 3283 (NBRC 3283), which formed a multi-phenotype cell complex following a series of slant passages for 21 years, was subjected to ‘mixed’ genome DNA sequencing. The genome analysis revealed that more than 280 transposons, ∼9% of the genes, exist in the genome consisting of a 2.9-Mb chromosome and 6 plasmids. The A. pasteurianus genome may be characterized by a combination of a large number both transposons and plasmids. Actually five variations by transposon insertion, as well as three SNPs, were found in the IFO 3283 (NBRC 3283) complex. A plasmid of G. xylinus carries a gene specifically similar to one in the A. pasteurianus chromosome, and a plasmid of A. pasteurianus was depleted during high-temperature adaptation. These findings may illustrate the involvement of plasmids in the genetic instability of AAB. The existence of HTRs in open reading frames is a characteristic of the A. pasteurianus genome. However, it seems to be involved in the feasibility of DNA maintenance, the physiological meaning is largely unknown.
Second, A. pasteurianus was used for the breeding of a strain proliferative at an unviable high temperature (42°C). Genome analyses of the bred strain by saturated genome shotgun sequencing revealed that a 92-kb deletion (∼3% of the whole genome size) and three single nucleotide mutations occurred in the genome during the high-temperature adaptation. However, the generating mechanism and genetic meaning of the large deletion is unknown, it seems that the smaller genome yields a survival advantage in the highly stressful conditions, possibly because of faster replication or lower heat generation based on less burden in DNA replication.
Alpha-proteobacteria, including AAB, contains many intracellular symbionts and parasites such as bacteria in the families, Rhizobiaceae, Rickettsiaceae and Brucellaceae, whose genomes are generally shorter and show increased evolution rates in comparison to those of closely related free-living bacteria (20). However, A. pasteurianus is assumed to be a free-living bacterium, it may have evolved to fit in competitive natural niches of seasonal fruits and flowers accompanied by the genetic flexibility and instability.
ACCESSION NUMBERS
The eight substrains of A. pasteurianus (IFO 3283-01, -03, -07, -12, -22, -26, -32 and IFO 3283-01-42C) reported herein have been re-deposited in the National Biological Resource Center (NBRC) and its genome DNA sequences were registered in DDBJ/EMBL/GenBank database. The accession numbers of NBRC and DDBJ are shown in Tables 1 and 2, and in Supplementary Table S1 in detail (AP011121–AP011176). An accession number for the largest plasmid of G. xylinus NBRC 3288, pGXY010, is BABN01000001.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
A Grant-in-Aid from the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN), Mizkan Group Corporation and Innovation Center of Yamaguchi University. Funding for open access charge: PROBRAIN.
Conflict of interest statement. None declared.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Lambert B, Kersters K, Gossele F, Swings J, De Ley J. Gluconobacters from honey bees. Antonie Van Leeuwenhoek. 1981;47:147–157. doi: 10.1007/BF02342197. [DOI] [PubMed] [Google Scholar]
- 2.Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, Lee DG, Shin SC, Ha EM, Lee WJ. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 3.Yamada Y, Yukphan P. Genera and species in acetic acid bacteria. Int. J. Food Microbiol. 2008;125:15–24. doi: 10.1016/j.ijfoodmicro.2007.11.077. [DOI] [PubMed] [Google Scholar]
- 4.Greenberg DE, Porcella SF, Stock F, Wong A, Conville PS, Murray PR, Holland SM, Zelazny AM. Granulibacter bethesdensis gen. nov., sp. nov., a distinctive pathogenic acetic acid bacterium in the family Acetobacteraceae. Int. J. Syst. Evol. Microbiol. 2006;56:2609–2616. doi: 10.1099/ijs.0.64412-0. [DOI] [PubMed] [Google Scholar]
- 5.Adachi O, Moonmangmee D, Toyama H, Yamada M, Shinagawa E, Matsushita K. New developments in oxidative fermentation. Appl. Microbiol. Biotechnol. 2003;60:643–653. doi: 10.1007/s00253-002-1155-9. [DOI] [PubMed] [Google Scholar]
- 6.Beppu T. Genetic organization of Acetobacter for acetic acid fermentation. Antonie Van Leeuwenhoek. 1993;64:121–135. doi: 10.1007/BF00873022. [DOI] [PubMed] [Google Scholar]
- 7.Sokollek SJ, Hertel C, Hammes WP. Description of Acetobacter oboediens sp. nov. and Acetobacter pomorum sp. nov., two new species isolated from industrial vinegar fermentations. Int. J. Syst. Bacteriol. 1998;48(Pt 3):935–940. doi: 10.1099/00207713-48-3-935. [DOI] [PubMed] [Google Scholar]
- 8.Steiner P, Sauer U. Proteins induced during adaptation of Acetobacter aceti to high acetate concentrations. Appl. Environ. Microbiol. 2001;67:5474–5481. doi: 10.1128/AEM.67.12.5474-5481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takemura H, Horinouchi S, Beppu T. Novel insertion sequence IS1380 from Acetobacter pasteurianus is involved in loss of ethanol-oxidizing ability. J. Bacteriol. 1991;173:7070–7076. doi: 10.1128/jb.173.22.7070-7076.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coucheron DH. An Acetobacter xylinum insertion sequence element associated with inactivation of cellulose production. J. Bacteriol. 1991;173:5723–5731. doi: 10.1128/jb.173.18.5723-5731.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prust C, Hoffmeister M, Liesegang H, Wiezer A, Fricke WF, Ehrenreich A, Gottschalk G, Deppenmeier U. Complete genome sequence of the acetic acid bacterium Gluconobacter oxydans. Nat. Biotechnol. 2005;23:195–200. doi: 10.1038/nbt1062. [DOI] [PubMed] [Google Scholar]
- 12.Greenberg DE, Porcella SF, Zelazny AM, Virtaneva K, Sturdevant DE, Kupko JJ, III, Barbian KD, Babar A, Dorward DW, Holland SM. Genome sequence analysis of the emerging human pathogenic acetic acid bacterium Granulibacter bethesdensis. J. Bacteriol. 2007;189:8727–8736. doi: 10.1128/JB.00793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sallstrom B, Andersson SG. Genome reduction in the alpha-Proteobacteria. Curr. Opin. Microbiol. 2005;8:579–585. doi: 10.1016/j.mib.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Nobusato A, Uchiyama I, Ohashi S, Kobayashi I. Insertion with long target duplication: a mechanism for gene mobility suggested from comparison of two related bacterial genomes. Gene. 2000;259:99–108. doi: 10.1016/s0378-1119(00)00456-x. [DOI] [PubMed] [Google Scholar]
- 15.Finkel SE, Kolter R. Evolution of microbial diversity during prolonged starvation. Proc. Natl Acad. Sci. USA. 1999;96:4023–4027. doi: 10.1073/pnas.96.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bjedov I, Tenaillon O, Gerard B, Souza V, Denamur E, Radman M, Taddei F, Matic I. Stress-induced mutagenesis in bacteria. Science. 2003;300:1404–1409. doi: 10.1126/science.1082240. [DOI] [PubMed] [Google Scholar]
- 17.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nilsson AI, Koskiniemi S, Eriksson S, Kugelberg E, Hinton JC, Andersson DI. Bacterial genome size reduction by experimental evolution. Proc. Natl Acad. Sci. USA. 2005;102:12112–12116. doi: 10.1073/pnas.0503654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim WS, Park JH, Ren J, Su P, Dunn NW. Survival response and rearrangement of plasmid DNA of Lactococcus lactis during long-term starvation. Appl. Environ. Microbiol. 2001;67:4594–4602. doi: 10.1128/AEM.67.10.4594-4602.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersson SG, Kurland CG. Reductive evolution of resident genomes. Trends Microbiol. 1998;6:263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- 21.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 22.Moran NA. Microbial minimalism: genome reduction in bacterial pathogens. Cell. 2002;108:583–586. doi: 10.1016/s0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- 23.Azuma Y, Hirakawa H, Yamashita A, Cai Y, Rahman MA, Suzuki H, Mitaku S, Toh H, Goto S, Murakami T, et al. Genome sequence of the cat pathogen, Chlamydophila felis. DNA Res. 2006;13:15–23. doi: 10.1093/dnares/dsi027. [DOI] [PubMed] [Google Scholar]
- 24.Dufresne A, Garczarek L, Partensky F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005;6:R14. doi: 10.1186/gb-2005-6-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moran NA, Mira A. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-12-research0054. RESEARCH0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silva FJ, Latorre A, Moya A. Genome size reduction through multiple events of gene disintegration in Buchnera APS. Trends Genet. 2001;17:615–618. doi: 10.1016/s0168-9525(01)02483-0. [DOI] [PubMed] [Google Scholar]
- 27.Sekine M, Tanikawa S, Omata S, Saito M, Fujisawa T, Tsukatani N, Tajima T, Sekigawa T, Kosugi H, Matsuo Y, et al. Sequence analysis of three plasmids harboured in Rhodococcus erythropolis strain PR4. Environ. Microbiol. 2006;8:334–346. doi: 10.1111/j.1462-2920.2005.00899.x. [DOI] [PubMed] [Google Scholar]
- 28.Gordon D, Desmarais C, Green P. Automated finishing with autofinish. Genome Res. 2001;11:614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 30.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakiyama T, Takami H, Ogasawara N, Kuhara S, Kozuki T, Doga K, Ohyama A, Horikoshi K. An automated system for genome analysis to support microbial whole-genome shotgun sequencing. Biosci. Biotechnol. Biochem. 2000;64:670–673. doi: 10.1271/bbb.64.670. [DOI] [PubMed] [Google Scholar]
- 35.Yada T, Nakao M, Totoki Y, Nakai K. Modeling and predicting transcriptional units of Escherichia coli genes using hidden Markov models. Bioinformatics. 1999;15:987–993. doi: 10.1093/bioinformatics/15.12.987. [DOI] [PubMed] [Google Scholar]
- 36.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 40.De Fonzo V, Aluffi-Pentini F, Parisi V. JSTRING: a novel Java tandem repeats searcher in genomic sequences with an interactive graphic output. Open Appl. Inform. J. 2008;2:14–17. [Google Scholar]
- 41.Dubois M, Gilles K, Hamilton JK, Rebers PA, Smith F. A colorimetric method for the determination of sugars. Nature. 1951;168:167. doi: 10.1038/168167a0. [DOI] [PubMed] [Google Scholar]
- 42.Deeraksa A, Moonmangmee S, Toyama H, Yamada M, Adachi O, Matsushita K. Characterization and spontaneous mutation of a novel gene, polE, involved in pellicle formation in Acetobacter tropicalis SKU1100. Microbiology. 2005;151:4111–4120. doi: 10.1099/mic.0.28350-0. [DOI] [PubMed] [Google Scholar]
- 43.Kibota TT, Lynch M. Estimate of the genomic mutation rate deleterious to overall fitness in E. coli. Nature. 1996;381:694–696. doi: 10.1038/381694a0. [DOI] [PubMed] [Google Scholar]
- 44.Boe L, Danielsen M, Knudsen S, Petersen JB, Maymann J, Jensen PR. The frequency of mutators in populations of Escherichia coli. Mutat. Res. 2000;448:47–55. doi: 10.1016/s0027-5107(99)00239-0. [DOI] [PubMed] [Google Scholar]
- 45.Drake JW. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl Acad. Sci. USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loewe L, Textor V, Scherer S. High deleterious genomic mutation rate in stationary phase of Escherichia coli. Science. 2003;302:1558–1560. doi: 10.1126/science.1087911. [DOI] [PubMed] [Google Scholar]
- 47.Grigoriev A. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 1998;26:2286–2290. doi: 10.1093/nar/26.10.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nomura M, Morgan EA. Genetics of bacterial ribosomes. Annu. Rev. Genet. 1977;11:297–347. doi: 10.1146/annurev.ge.11.120177.001501. [DOI] [PubMed] [Google Scholar]
- 49.Kylma AK, Granstrom T, Leisola M. Growth characteristics and oxidative capacity of Acetobacter aceti IFO 3281: implications for L-ribulose production. Appl. Microbiol. Biotechnol. 2004;63:584–591. doi: 10.1007/s00253-003-1406-4. [DOI] [PubMed] [Google Scholar]
- 50.Nabe K, Izuo N, Yamada S, Chibata I. Conversion of glycerol to dihydroxyacetone by immobilized whole cells of Acetobacter xylinum. Appl. Environ. Microbiol. 1979;38:1056–1060. doi: 10.1128/aem.38.6.1056-1060.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinhouse H, Benziman M. Phosphorylation of glycerol and dihydroxyacetone in Acetobacter xylinum and its possible regulatory role. J. Bacteriol. 1976;127:747–754. doi: 10.1128/jb.127.2.747-754.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adachi O, Ano Y, Shinagawa E, Matsushita K. Purification and properties of two different dihydroxyacetone reductases in Gluconobacter suboxydans grown on glycerol. Biosci. Biotechnol. Biochem. 2008;72:2124–2132. doi: 10.1271/bbb.80199. [DOI] [PubMed] [Google Scholar]
- 53.Mullins EA, Francois JA, Kappock TJ. A specialized citric acid cycle requiring succinyl-coenzyme A (CoA):acetate CoA-transferase (AarC) confers acetic acid resistance on the acidophile Acetobacter aceti. J. Bacteriol. 2008;190:4933–4940. doi: 10.1128/JB.00405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moonmangmee S, Kawabata K, Tanaka S, Toyama H, Adachi O, Matsushita K. A novel polysaccharide involved in the pellicle formation of Acetobacter aceti. J. Biosci. Bioeng. 2002;93:192–200. doi: 10.1263/jbb.93.192. [DOI] [PubMed] [Google Scholar]
- 55.Pettigrew DW, Liu WZ, Holmes C, Meadow ND, Roseman S. A single amino acid change in Escherichia coli glycerol kinase abolishes glucose control of glycerol utilization in vivo. J. Bacteriol. 1996;178:2846–2852. doi: 10.1128/jb.178.10.2846-2852.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chagneau C, Heyde M, Alonso S, Portalier R, Laloi P. External-pH-dependent expression of the maltose regulon and ompF gene in Escherichia coli is affected by the level of glycerol kinase, encoded by glpK. J. Bacteriol. 2001;183:5675–5683. doi: 10.1128/JB.183.19.5675-5683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van der Sluis EO, Driessen AJ. Stepwise evolution of the Sec machinery in Proteobacteria. Trends Microbiol. 2006;14:105–108. doi: 10.1016/j.tim.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 58.Larsson P, Oyston PC, Chain P, Chu MC, Duffield M, Fuxelius HH, Garcia E, Halltorp G, Johansson D, Isherwood KE, et al. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 2005;37:153–159. doi: 10.1038/ng1499. [DOI] [PubMed] [Google Scholar]
- 59.Ishihama A. Role of the RNA polymerase alpha subunit in transcription activation. Mol. Microbiol. 1992;6:3283–3288. doi: 10.1111/j.1365-2958.1992.tb02196.x. [DOI] [PubMed] [Google Scholar]
- 60.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 61.Jin Q, Yuan Z, Xu J, Wang Y, Shen Y, Lu W, Wang J, Liu H, Yang J, Yang F, et al. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 2002;30:4432–4441. doi: 10.1093/nar/gkf566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.da Silva AC, Ferro JA, Reinach FC, Farah CS, Furlan LR, Quaggio RB, Monteiro-Vitorello CB, Van Sluys MA, Almeida NF, Alves LM, et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature. 2002;417:459–463. doi: 10.1038/417459a. [DOI] [PubMed] [Google Scholar]
- 63.Cho NH, Kim HR, Lee JH, Kim SY, Kim J, Cha S, Kim SY, Darby AC, Fuxelius HH, Yin J, et al. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc. Natl Acad. Sci. USA. 2007;104:7981–7986. doi: 10.1073/pnas.0611553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parisi V, De Fonzo V, Aluffi-Pentini F. STRING: finding tandem repeats in DNA sequences. Bioinformatics. 2003;19:1733–1738. doi: 10.1093/bioinformatics/btg268. [DOI] [PubMed] [Google Scholar]
- 65.Setubal JC, Dos Santos P, Goldman BS, Ertesvag H, Espin G, Rubio LM, Valla S, Almeida NF, Balasubramanian D, Cromes L, et al. The genome sequence of Azotobacter vinelandii, an obligate aerobe specialized to support diverse anaerobic metabolic processes. J. Bacteriol. 2009;191:4534–4545. doi: 10.1128/JB.00504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wells RD, Parniewski P, Pluciennik A, Bacolla A, Gellibolian R, Jaworski A. Small slipped register genetic instabilities in Escherichia coli in triplet repeat sequences associated with hereditary neurological diseases. J. Biol. Chem. 1998;273:19532–19541. doi: 10.1074/jbc.273.31.19532. [DOI] [PubMed] [Google Scholar]
- 67.Lechner RL, Engler MJ, Richardson CC. Characterization of strand displacement synthesis catalyzed by bacteriophage T7 DNA polymerase. J. Biol. Chem. 1983;258:11174–11184. [PubMed] [Google Scholar]
- 68.Iyer RR, Pluciennik A, Rosche WA, Sinden RR, Wells RD. DNA polymerase III proofreading mutants enhance the expansion and deletion of triplet repeat sequences in Escherichia coli. J. Biol. Chem. 2000;275:2174–2184. doi: 10.1074/jbc.275.3.2174. [DOI] [PubMed] [Google Scholar]
- 69.Jacob KD, Eckert KA. Escherichia coli DNA polymerase IV contributes to spontaneous mutagenesis at coding sequences but not microsatellite alleles. Mutat. Res. 2007;619:93–103. doi: 10.1016/j.mrfmmm.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Delagoutte E, Goellner GM, Guo J, Baldacci G, McMurray CT. Single-stranded DNA-binding protein in vitro eliminates the orientation-dependent impediment to polymerase passage on CAG/CTG repeats. J. Biol. Chem. 2008;283:13341–13356. doi: 10.1074/jbc.M800153200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bichara M, Wagner J, Lambert IB. Mechanisms of tandem repeat instability in bacteria. Mutat. Res. 2006;598:144–163. doi: 10.1016/j.mrfmmm.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 72.Martin P, Makepeace K, Hill SA, Hood DW, Moxon ER. Microsatellite instability regulates transcription factor binding and gene expression. Proc. Natl Acad. Sci. USA. 2005;102:3800–3804. doi: 10.1073/pnas.0406805102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weiser JN, Shchepetov M, Chong ST. Decoration of lipopolysaccharide with phosphorylcholine: a phase-variable characteristic of Haemophilus influenzae. Infect. Immun. 1997;65:943–950. doi: 10.1128/iai.65.3.943-950.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weiser JN, Love JM, Moxon ER. The molecular mechanism of phase variation of H. influenzae lipopolysaccharide. Cell. 1989;59:657–665. doi: 10.1016/0092-8674(89)90011-1. [DOI] [PubMed] [Google Scholar]
- 75.Sauer U. Evolutionary engineering of industrially important microbial phenotypes. Adv. Biochem. Eng. Biotechnol. 2001;73:129–169. doi: 10.1007/3-540-45300-8_7. [DOI] [PubMed] [Google Scholar]
- 76.Bearson S, Bearson B, Foster JW. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 1997;147:173–180. doi: 10.1111/j.1574-6968.1997.tb10238.x. [DOI] [PubMed] [Google Scholar]
- 77.Steiner P, Sauer U. Long-term continuous evolution of acetate resistant Acetobacter aceti. Biotechnol. Bioeng. 2003;84:40–44. doi: 10.1002/bit.10741. [DOI] [PubMed] [Google Scholar]
- 78.Casane D, Dennebouy N, de Rochambeau H, Mounolou JC, Monnerot M. Genetic analysis of systematic mitochondrial heteroplasmy in rabbits. Genetics. 1994;138:471–480. doi: 10.1093/genetics/138.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lopez A, Johnson LW, Wood CB. Observations on a laboratory method for submerged acetic fermentation. Appl. Microbiol. 1961;9:425–433. doi: 10.1128/am.9.5.425-433.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sourek J, Kulhanek M. Long-term storage of acetic acid bacteria by means of lyophilization. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg. 1969;123:580–585. [PubMed] [Google Scholar]