Abstract
Unusual clusters of YY1 binding sites are located within several differentially methylated regions (DMRs), including Xist, Nespas and Peg3, which all become methylated during oogenesis. In this study, we performed conditional YY1 knockdown (KD) to investigate YY1's roles in DNA methylation of these DMRs. Reduced levels of YY1 during spermatogenesis did not cause any major change in these DMRs although the same YY1 KD caused hypermethylation in these DMRs among a subset of aged mice. However, YY1 KD during oogenesis resulted in the loss of DNA methylation on Peg3 and Xist, but there were no changes on Nespas and H19. Continued YY1 KD from oogenesis to the blastocyst stage caused further loss in DNA methylation on Peg3. Consequently, high incidents of lethality were observed among embryos that had experienced the reduced levels of YY1 protein. Overall, the current study suggests that YY1 likely plays a role in the de novo DNA methylation of the DMRs of Peg3 and Xist during oogenesis and also in the maintenance of unmethylation status of these DMRs during spermatogenesis.
INTRODUCTION
Genomic imprinting is an unusual mechanism in which one allele is repressed based on its parental origin. Imprinted genes are usually associated with CpG islands at their 5′-ends, and some of these CpG islands show differential DNA methylation patterns between the two alleles (differentially methylated regions, DMR). Imprinted genes tend to be clustered within chromosomal regions, and some of these DMRs are known to control the imprinting of a given domain as imprinting control regions (ICRs) (1,2). As compared to other DMRs, ICRs obtain parental imprinting marks during gametogenesis in the form of DNA methylation (or histone modifications). However, this DNA methylation on ICRs in the germline is regarded as a very unusual event since the promoters of most autosomal genes are protected from DNA methylation during gametogenesis. ICRs also maintain their allele-specific DNA methylation patterns in somatic cells throughout the lifetime of an organism, particularly during early embryogenesis when mammalian genomes go through the global resetting process of DNA methylation. This suggests that ICRs may be subject to special mechanisms for de novo DNA methylation during gametogenesis and for protection from DNA demethylation during early embryogenesis (3,4). These unique aspects of ICRs have been further highlighted by several recent observations. The testis-specific factor CTCFL may interact with PRMT7, a histone-modifying enzyme, to derive de novo DNA methylation on the H19-ICR in the germline (5). In addition, mutant mice lacking Pgc7/Stella and Zfp57 showed target-specific loss of DNA methylation on several ICRs during early embryogenesis, confirming the presence of ICR-specific protection mechanisms against genome-wide DNA demethylation (6–8).
So far, two transcription factors are known to bind to DMRs: CTCF binds to the DMRs of H19, Kcnq1, Rasgrf1, Grb10, Dlk1/Gtl2, Tsix and Xist, and YY1 binds to the DMRs of Nespas, Xist, Tsix and Peg3 (9–19). According to a series of studies, CTCF is required for maintaining the allele-specific methylation pattern on the H19-ICR/DMR during early embryogenesis. However, CTCF is not required to establish DNA methylation during spermatogenesis (20–23). On the other hand, recent siRNA-based YY1 knockdown (KD) experiments derived a somewhat similar conclusion: YY1 has roles in the DNA methylation of Xist, Nespas and Peg3. Both in vitro and in vivo experiments have shown that reducing the cellular levels of YY1 protein caused target-specific changes in the methylation levels of Xist, Nespas and Peg3 (24,25). Another study using one mouse knockout line of YY1 also confirmed a significant role for YY1 in the X chromosomal inactivation process (26). However, these earlier studies did not clarify when and where the in vivo levels of YY1 protein are critical for DNA methylation on these DMRs. In the current study, therefore, we performed a series of YY1 KD experiments focusing on potential roles for YY1 during gametogenesis and early embryogenesis. Our results demonstrate that the proper level of YY1 protein is required for de novo DNA methylation of these DMRs during oogenesis.
MATERIALS AND METHODS
Transgenic mice
To generate conditional YY1 KD transgenic mice, SpeI-linearized DNA of the pSico-YY1 shRNA vector was microinjected into the pronucleus of fertilized eggs of FVB/NJ backgrounds by The Darwin Transgenic Mouse Core Facility (Baylor College of Medicine, Houston, TX, USA) (27). Eight pSico-YY1 (two males and six females) founder transgenic lines were initially generated, and confirmed by the expression of EGFP under a halogen lamp, PCR-based genotyping with tail DNA, and Southern blot using the DNA fragment containing the U6 promoter as a probe. One F0 transgenic mouse successfully passed the transgene through its germ line, and also displayed consistent expression of EGFP. Thus, this line was used for the current study. Conditional YY1 KD was performed by crossing this pSico-YY1 line with two germ cell-specific Cre mice: the Protamine-Cre mice [129-Tg(Pro-cre)58Og/J, The Jackson Lab] and the Zp3-Cre mice [C57BL/6-Tg(Zp3-cre)93Knw/J, The Jackson Lab]. All the experiments involving mice were performed in accordance with National Institutes of Health guidelines for care and use of animals.
Isolation of sperm, egg and blastocyst-stage embryos
Sperm was isolated from the epididymis of male mice according to a previously established protocol (28). In brief, the retrieved epididymis was incubated at room temperature for 1 h with gentle rocking in the buffer containing 130 mM NaCl, 20 mM Tris, 2 mM EDTA pH 7.4. After 4 min spin at 2000 r.p.m. on a tabletop centrifuge (Cat. Centrifuge 5415D, Eppendorf), the precipitated sperm was washed two more rounds with the same buffer. The isolated sperm was examined under a microscope to measure the total number and purity of each sperm pool (Supplementary Data 2). The sperm was subsequently used for isolating DNA.
Mature eggs in MII phase were isolated through a widely used superovulation protocol (28,29). In brief, female mice were first injected subcutaneously with 5 IU of Pregnant Mare's Serum (PMS) (Cat. G4877, Sigma), and the same mice were injected with 5 IU of the human Chorionic Gonadotropin (hCG) hormone (Cat. C1063, Sigma) 48 h after the initial injection. These mice were sacrificed 12 h after the second injection, and mature eggs were isolated from the swollen ampullae of the oviducts. The isolated eggs were incubated in hyaluronidase solution (Cat. H3506, Sigma) for several minutes to separate them from the cumulus cells, and subsequently washed three additional times to remove potential somatic tissue contamination. Finally, the eggs were used to isolate DNA.
For the isolation of blastocyst-stage embryos, we followed the same ovulation protocol as the above. After the second injection, however, we put these female mice together with male littermates. We sacrificed the female mice 3 days after the breeding setup, and flushed the embryos from their uterus (30). The isolated blastocysts were further examined under the microscope to estimate of their developmental stages and purity. Each blastocyst was individually used to isolate DNA for genotyping.
Bisulfite conversion and combined bisulfite restriction analysis (COBRA)
The DNAs isolated from sperm and egg were individually treated with the bisulfite conversion reaction according to the manufacturer's protocol (EZ DNA methylation kit, Zymo Research). The converted DNAs were used as templates for the PCR reaction using specific primers designed to amplify each ICR. The oligonucleotide sequences for this study are provided as Supplementary Data 1. PCR was performed with the Maxime PCR premix kit (Intron Biotech). To determine the DNA methylation levels of target regions, we used the following two approaches: (i) the restriction enzyme digestion-based COBRA (31) and (ii) subcloning and sequencing of each PCR product. For the COBRA analysis, each PCR product was digested with the two different types of restriction enzymes. The first type of enzymes (HpaI, TaqI and HhaI) recognizes and digests CpGs and thus the digestion by these enzymes indicates the methylated status of CpGs in the original DNA. In contrast, the second type of enzymes (HphI) recognizes and digests TpGs and thus the digestion in this case indicates the unmethylated status of CpGs. The methylation levels of a given genomic DNA can be inferred from the relative ratios of the undigested and digested DNA amounts. Some of the PCR products were also individually subcloned into the pGEM T-Easy vector (Promega), and the purified DNAs from 10 to 20 clones were sequenced to survey DNA methylation levels of a given PCR product.
RESULTS
Overall conditional YY1 KD strategy
We have previously established one transgenic mouse line containing the pSico-YY1 construct, the expression of which can be activated on by the Cre recombinase (25; Figure 1A). This previous study has also demonstrated the feasibility of this YY1 KD scheme using the EIIa-Cre allele. In the current study, we used the same transgenic line for conditional KD of YY1 during gametogenesis. We crossed heterozygous pSico-YY1 mice with two germ cell-specific, homozygous Cre mice: either Protamine-Cre to target spermatogenesis or Zp3-Cre to target oogenesis (Figure 1B). The subsequent F1 mice that were double positive for pSico-YY1 and Pro-Cre (or Zp3-Cre) were used for the following two experiments. First, we isolated germ cells from these F1 mice to survey potential changes in the DNA methylation levels of DMRs. Second, we bred these F1 mice to isolate blastocyst-stage embryos and also to test the developmental potential of germ cells with the YY1 KD. The proper YY1 KD by this scheme was also confirmed through independent western blot experiments (Supplementary Data 3). Since the two germ cell-specific Cre mice have different genetic backgrounds than the pSico-YY1 mice (Pro-Cre is from 129; Zp3-Cre is from C57BL; pSico-YY1 is from FVB), we also used the F1 littermates lacking the pSico-YY1 allele as negative controls for the above two sets of experiments.
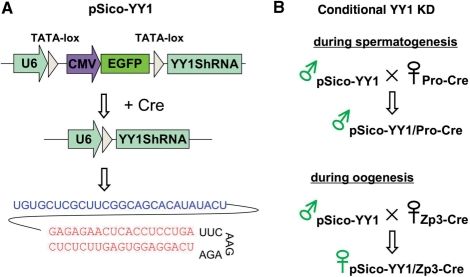
Figure 1.
Overall Strategy of conditional YY1 KD. (A) Schematic representation of the pSico-YY1 shRNA vector. The U6 promoter is separated from the downstream YY1-shRNA sequence by the EGFP expression cassette. Cre recombinase, available as a transgenic allele, recognizes the two TATA-loxP sites and joins these two split regions, resulting in the production of the YY1 shRNA sequence, which leads to KD. The sequence in blue is the leader sequence of the U6 RNA gene, which has been added to increase the stability of the YY1 shRNA, and the sequence in red is the sequence of the YY1 shRNA. (B) Conditional KD scheme for gametogenesis. The pSico-YY1 transgenic line was crossed with one of two germ cell-specific Cre mice: Protamine-Cre for spermatogenesis and Zp3-Cre for oogenesis. The resulting F1 mice were used for testing potential effects of YY1 KD during either spermatogenesis or oogenesis.
YY1 KD during spermatogenesis
To study YY1 KD during spermatogenesis, we prepared 20 male F1 mice (pSico-YY1, Pro-Cre). These mice were sacrificed at three different ages, 1, 3 and 6 months old, for the isolation of sperm DNA. Five F1 mice per time point were used to isolate sperm from their epididymis. The remaining five mice were used for breeding experiments, which will be described later (Figure 5). Genomic DNA was purified from the isolated sperm and used to analyze DNA methylation via bisulfite conversion. The bisulfite-converted DNAs were subsequently amplified with PCR using specific primer sets designed for each DMR (Supplementary Data 1). The amplified PCR product from each DMR was analyzed with restriction-digestion based COBRA (31) and/or subcloning and sequencing (Figure 2).
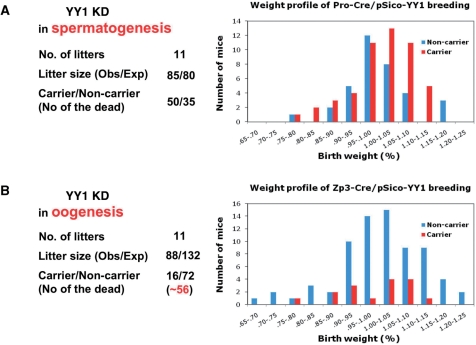
Figure 5.
Developmental potential of the gametes with YY1 KD. The germ cells with YY1 KD were also tested through breeding the male F1 mice (pScio-YY1, Pro-Cre) (A) and the female F1 mice (pSico-YY1, Zp3-Cre) (B) with their littermates. These two breeding experiments were summarized as shown on the left. The two graphs on the right represent the birth weight profiles derived from the 1-day-old neonates of these two breeding experiments. The neonates were analyzed in terms of their birth weights. Each mouse was first categorized according to its genotype [carrier (red) versus noncarrier (blue)]. The relative weight percentile for each mouse was calculated through dividing its weight by the averaged value for its litter. This relative weight value was used to further classify each mouse into different categories ranging from 65% to 125% as shown on the x-axis. The total number of mice in each weight category is plotted as a value on the y-axis.
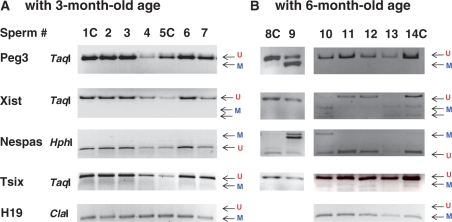
Figure 2.
YY1 KD effects during spermatogenesis. Sperm DNA was isolated individually from the F1 mice (pSico-YY1, Pro-Cre) and the littermate controls (+, Pro-Cre). The individual F1 mice are indicated by different numbers, while the control littermates are indicated by numbers followed by C (1C, 5C, 8C and 14C). (A) Individual DNA samples from the 3-month-old set were treated with the bisulfite conversion method, followed by the restriction-enzyme-based method—COBRA. Digestion and lack of digestion by TaqI and ClaI indicate the methylated (M) and unmethylated (U) status, respectively, of a given CpG site in the original DNA as shown in the digestion of Peg3, Xist, Tsix and H19. In contrast, this is opposite for the HphI digestion, which recognizes TpG as shown in the digestion of Nespas. Digestion by this enzyme indicates the unmethylated status of a CpG site, whereas lack of digestion indicates methylation. (B) A subset of the 6-month-old mice is also shown in the same manner as the above. The results for H19 (8#C and 9) are not available due to the unsuccessful PCR amplification.
Sperm from the 3-month-old F1 mice with YY1 KD did not show any change in the DNA methylation levels of the YY1-associated DMRs, including Peg3, Xist, and Nespas (Figure 2A). Five different samples of sperm DNA (#2, 3, 4, 6 and 7) were all unmethylated in the three DMRs, as were the two control samples (#1C and 5C). We also observed a similar pattern from the group of 1-month-old F1 mice (data not shown). However, a subset of the 6-month-old F1 mice (3 out of 10 F1) showed target-specific changes in the methylation levels of the YY1-associated DMRs (#9, 10 and 13; Figure 2B). Although the littermate controls still showed typical unmethylated DNA patterns on Peg3, Nespas and Xist (#8C, 11, 12 and 13C), these F1 mice showed hypermethylation on these DMRs: #9 for Peg3, #10 and 13 for Xist and #9 and 10 for Nespas. We also tested other DMRs, including Tsix and H19, but the methylation levels on these DMRs were not affected in these F1 mice, indicating a target-specific outcome. This hypermethylation was initially noticed due to the sudden sterility observed from one of the five F1 male mice (#9) that had been used for our breeding experiments (Figure 5). Since this change was mainly observed in older mice, this outcome may not be solely caused by YY1 KD during spermatogenesis. In sum, the above experiments suggest that reduced levels of YY1 protein during spermatogenesis may not have any major impact on the DNA methylation levels of the DMRs.
YY1 KD during oogenesis
To study YY1 KD during oogenesis, we prepared 50 female F1 mice (pSico-YY1, Zp3-Cre) along with 50 littermate controls (+, Zp3-Cre). We performed three different rounds of egg isolation with these 8-week-old F1 mice. In each round, 10 F1 and 10-littermate control mice were superovulated and sacrificed. The remaining 20 F1 and 20 littermates were used for other breeding experiments. Each set of the 10 female mice yielded ∼400 mature oocytes. We did not find any gross difference between the F1 mice with YY1 KD and the littermate control set in terms of morphology of mature eggs, although the number of the isolated egg was slightly smaller in the mice with YY1 KD (Figure 3C). This suggests that our YY1 KD scheme probably did not cause any hindrance to the oogenesis process itself.
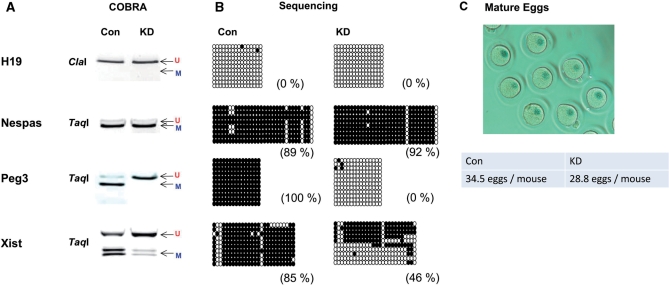
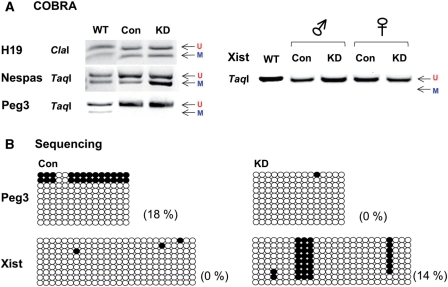
Figure 3.
YY1 KD effects during oogenesis. About 400 eggs at MII phase were isolated from each of the following two types of female mice: the KD mice (pScio-YY1, Zp3-Cre) and the control (Con) littermates (+, Zp3-Cre). Genomic DNA from these two types of mice was first treated by bisulfite conversion, followed by restriction enzyme digestion (A) and sequencing (B). The PCR product from the H19-ICR was digested by ClaI, while the remaining products from the DMRs of Nespas, Peg3 and Xist were digested by TaqI. The results derived from sequencing of individual PCR products were summarized in the following manner. Each circle represents one CpG site: closed and open ones indicate methylated and unmethylated states, respectively, of the analyzed CpG sites. Each row represents one clone derived from the original PCR product. The methylation levels (%) of each DMR were calculated through dividing the number of methylated CpGs by the total number of surveyed CpG sites. We monitored and compared the purity, number and morphology of the isolated egg samples, and a representative image of the isolated egg is also shown (C).
Due to the limited amount of egg DNA, we targeted only five DMRs for this analysis, including H19, Nespas, Xist, Tsix and Peg3. We successfully amplified most of these DMRs except for Tsix. The DNA amplified from the eggs was subsequently used to analyze the DNA methylation levels of the DMRs (Figure 3). According to the results from COBRA and individual sequencing, YY1 KD during oogenesis did affect the DNA methylation levels of the two YY1-associated DMRs, Peg3-DMR and Xist-DMR. As expected, the control set showed complete DNA methylation on these two DMRs. However, the YY1 KD set displayed almost no methylation (100–0%) on the Peg3-DMR and much lower levels (85–46%) of DNA methylation on the Xist-DMR. In contrast, the DNA methylation levels on the DMRs of Nespas and H19 were similar between the YY1 KD and control sets. These results confirmed that reducing YY1 levels during oogenesis has target-specific effects on the methylation levels of the DMRs of Peg3 and Xist. This further suggests that the proper level of YY1 protein is likely required for de novo DNA methylation of these two DMRs during oogenesis.
YY1 KD during the preimplantation stage
Since YY1 KD during oogenesis had a major impact on the DNA methylation levels of the DMRs of Peg3 and Xist, we further followed up the progression of these changes by allowing eggs with YY1 KD to fertilize and develop. Female F1 mice (pSico-YY1, Zp3-Cre) were time-mated with male littermates, and used for harvesting blastocyst-stage embryos. DNA was individually purified from each blastocyst for genotyping. PCR-based genotyping determined the sex and presence of the pSico-YY1 allele in each blastocyst. This allowed us to group all the retrieved blastocysts from each litter of 7–25 embryos into four different categories: male or female, with or without the pSico-YY1 allele. The DNA from each category of blastocysts was combined together as a group, and used for DNA methylation analyses as shown in Figure 4.
Figure 4.
YY1 KD effects during early embryogenesis. Blastocysts were isolated through crossing the female F1 mice (pSico-YY1, Zp3-Cre) with littermate breeders. The isolated blastocysts were categorized based on their genotypes and genders. The DNA from each category of blastocysts was first treated with the bisulfite conversion, followed by the restriction digestion-based COBRA (A) and sequencing (B). The digested PCR products for H19, Nespas and Peg3 are derived from male blastocysts, while Xist is shown in four categories due to the fact that X chromosome number varies depending on gender. The PCR products were digested by the following restriction enzymes: H19 was digested by ClaI, while Nespas, Peg3 and Xist were digested by TaqI. The sequencing results for Xist are from female blastocysts (Con and KD).
According to the results from the COBRA analyses, about half of DNA in the H19-DMR was methylated and this pattern was observed among all of the different categories of blastocysts. This confirmed that the continued YY1 KD from oogenesis to the blastocyst stage did not have any major effect on the methylation levels of the H19-DMR. A similar conclusion has also been drawn from the analyses on the Nespas-DMR although the YY1 KD set showed slightly more methylation levels than the control (Con) set. In contrast, the DMRs of Xist and Peg3 displayed complete loss of DNA methylation on both the YY1 KD and control sets. A similar outcome was observed in both genders. Complete loss of DNA methylation on the Peg3-DMR was predicted since the majority of eggs had already lost their DNA methylation during oogenesis (Figure 3). It is important to note that the actual effects of our conditional KD scheme are likely manifested in a manner similar to those of maternal-effect mutations (32). All the eggs should have similar YY1 KD effects, regardless of whether they are carriers or noncarriers of the pSico-YY1 allele. Indeed, this was well reflected in the results—complete loss of DNA methylation even in the control set. To further confirm this possibility, we derived another set of blastocyst-stage embryos from an independent mating between 10 F1 females (+, Zp3-Cre) and male littermates. As shown in Figure 4A (marked as WT), the Peg3-DMR displayed half-methylation pattern similar to those of the two other DMRs, H19 and Nespas. In the case of Xist-DMR, however, the WT sample also showed complete loss in DNA methylation as seen in both the YY1 KD and control sets. This suggests that the observed complete loss of DNA methylation on the Xist-DMR is likely a normal pattern in blastocyst-stage embryos, but not an outcome of YY1 KD. Therefore, the analysis on the Xist-DMR was inconclusive. Overall, the above experiments confirmed that continuing YY1 KD from oogenesis to the blastocyst stage had a consistent outcome on DNA methylation as seen from the YY1 KD during oogenesis: complete loss for the Peg3-DMR.
Developmental potential of the germ cells with YY1 KD
The developmental potential of the germ cells with the reduced levels of YY1 was also tested through individually breeding each of the two following types of F1 mice: male F1 (pSico-YY1, Pro-Cre) and female F1 (pSico-YY1, Zp3-Cre) (Figure 5). First, we crossed the five F1 male mice (pSico-YY1, Pro-Cre) with their littermates, yielding over 20 litters. Out of these 20 litters, 11 were analyzed in terms of their genotypes, sexes and birth weights (Figure 5A). We also performed another parallel breeding experiment with control mice lacking the pSico-YY1 allele (+, Pro-Cre). The overall litter sizes were similar between the test breeding (85/11) and the control breeding (80/11), indicating no lethality during the gestation period. Moreover, the ratio between the carriers and noncarriers of the pSico-YY1 allele (50 : 35) was slightly higher than the predicted ratio (1 : 1) based on Mendelian inheritance. Although the carriers somewhat outnumbered the noncarriers, these results appear to be in a good agreement with the previous results revealing no major impact due to the YY1 KD during spermatogenesis (Figure 2). We also measured the body weight of each neonate as an indicator of its health status. Each weight value was divided by the average weight of its litter to yield a percentile score (%). The subsequent birth weight profiles were compared between carrier and noncarrier groups within the test breeding set. This comparison revealed that carriers were slightly healthier (or heavier) than the noncarriers. This is also consistent with the fact that the carriers outnumbered the noncarriers (50 : 35). This suggests that YY1 KD during spermatogenesis might have been somehow beneficial to embryo development, although this unexpected conclusion warrants further investigation.
In contrast, the results from the breeding of the five female F1 mice (pSico-YY1, Zp3-Cre) with their littermates yielded a very different outcome. The litter size from the test breeding experiment, in which YY1 was knock-downed, was much smaller than that of the control breeding (88 : 132). Furthermore, the number of the carriers was much smaller than that of the noncarriers (16 : 72) within the test breeding set. This indicated a high incidence of lethality, about 80%, during the gestation period. This also agrees with the previous result that YY1 KD during oogenesis had a major impact on the methylation levels of Peg3 and Xist (Figure 3). There was no obvious difference in terms of gender ratios in both sets: the carrier set (female : male = 8 : 8) and the noncarrier set (female : male = 37 : 35). In addition, the weight profile from this breeding set did not reveal any gender-specific effect within both the carrier and noncarrier sets, which could occur by the disruption of X chromosomal inactivation (Figure 5B). This is different from the result of a previous conditional YY1 KD using the EIIa-Cre allele, which is designed to express the YY1-shRNA during blastocyst stage (25). In that experiment, YY1 KD effects were more pronounced in female mice with much lighter body weight. This might simply reflect the fact that the YY1 KD effect during oogenesis by the Zp3-Cre did not last long enough to be manifested in the surviving pups. In contrast, the YY1 KD effect by EIIa-Cre targeting the blastocyst stage may have been shown in the surviving pups more readily than that by the Zp3-Cre. Overall, the results from the breeding experiments confirmed that proper levels of YY1 protein during oogenesis are crucial for the survival of embryos during the later gestation period.
DISCUSSION
The current study performed a series of conditional YY1 KD experiments to characterize the roles of YY1 during gametogenesis. According to the results, reduced YY1 levels during oogenesis had a major impact on the establishment of proper DNA methylation on the DMRs of Peg3 and Xist. However, a similar KD during spermatogenesis had no effect on the methylation levels of the YY1-associated DMRs. This suggests that YY1 likely plays a role in the de novo DNA methylation of these two DMRs during oogenesis.
So far, two transcription factors, CTCF and YY1, are known to bind to the DMRs of imprinted domains. According to the results from H19-ICR/DMR, CTCF is not required for establishing the paternal-specific DNA methylation, but acts as a protector against DNA methylation on the maternal allele during early embryogenesis (22,23). In contrast, the results from the current study demonstrated that YY1 is likely required for establishing the maternal-specific DNA methylation on Peg3 and Xist during oogenesis (Figure 3). These results are also consistent with the results from the previous studies using in vitro and in vivo systems. In vitro YY1 KD using Neuro2A cell lines resulted in hypomethylation of some of the YY1-associated DMRs, such as Peg3 and Xist (24). In vivo YY1 KD using similar transgenic mouse lines resulted in both hyper and hypomethylation on Xist, Nespas and Peg3 (25). The observed hypermethylation on Peg3 and Xist from the previous study, however, might be an unrelated side effect of YY1 KD experiments since this was observed only at that time. On the other hand, we have consistently observed hypomethylation on Peg3 and Xist throughout this series of YY1 KD experiments. This suggests that the hypomethylation on Peg3 and Xist is most likely a genuine outcome of the reduced levels of YY1. If that is the case, the result presented in this study further suggests a previously unrecognized role for YY1, targeting de novo DNA methylation to the DMRs of Peg3 and Xist. YY1 has never been described before as a targeting protein for de novo DNA methylation, but this is a likely scenario given the large number of YY1-interacting partners, especially histone-modifying enzymes such as PRMT1 and SFMBT2 (33,34). It is plausible to think that some of these histone-modifying partners pre-mark the DMRs of Xist and Peg3 for DNA methylation during oogenesis. Therefore, it will be very interesting to test this predicted role of YY1 in the near future.
The current study suggests the intriguing possibility that YY1 may target de novo DNA methylation to the DMRs of Peg3 and Xist during oogenesis. However, YY1 binding sites are not limited to these DMRs: these sites are found in the promoters of numerous genes, and also in many repetitive regions of the vertebrate genomes. Then, what are the unique features associated with Peg3 and Xist that might signal for de novo methylation during oogenesis? We believe that this specificity might originate from the unusual features associated with the YY1 binding sites of these two DMRs. First, these DMRs have clusters of multiple YY1 binding sites, on average seven sites for the Xist-DMR (500 bp in length) and 12 sites for the Peg3-DMR (2 kb in length) (13,35). Besides, these YY1 binding sites appear to be the strongest binding sites for the YY1 protein (35). The local concentration of the YY1 protein around these DMRs might be much higher than the rest of the genome due to the high densities of multiple YY1 binding sites giving the strongest binding affinity. These unusually high levels of YY1 protein might be a targeting signal for de novo methylation to these DMRs. Second, the YY1 binding sites within these DMRs are all oriented in a same direction. This unusual unidirectionality of the YY1 binding sites might be an indication for some unknown chromatin structures around these DMRs. This could be also a signal for de novo DNA methylation. According to recent findings, the spacing between individual CTCF sites within the H19-ICR/DMR appears to be a critical factor for maintaining the DNA methylation of this DMR (36). This suggests that some unknown features associated with the CTCF binding sites, such as chromatin structure, are also important for the DNA methylation of this DMR. In a similar line, it is important to note that the CTCF binding sites found in the H19-DMR also share similar unique features with the YY1 binding sites of Peg3 and Xist: multiple binding sites with high affinity and unidirectionality (37). These shared unusual features further support the possibility of some unique chromatin structures around the DMRs. Therefore, it will be very interesting to test in the near future if these features are indeed responsible for targeting de novo methylation to the DMRs during gametogenesis.
In contrast, a similar YY1 KD during spermatogenesis did not cause any major impact on the DNA methylation levels of the tested DMRs (Figure 2). This may reflect the following possibilities. First, YY1 may not be involved in establishing the DNA methylation of these DMRs during spermatogenesis. It is important to note that the majority of YY1-associated DMRs are not usually methylated during spermatogenesis. In this regard, the DMRs of Xist, Nespas and Peg3 are not different from the CpG-rich promoter regions of all the other nonimprinted genes in terms of their DNA methylation status during spermatogenesis—no methylation. This implies that YY1-associated DMRs could be subject to the normal epigenetic modifications of other nonimprinted genes without the involvement of YY1. Second, YY1 may be required only for maintaining the unmethylated status of these DMRs during spermatogenesis. This has been somewhat corroborated by the observation that the YY1-associated DMRs were hypermethylated in the older mice set of the Pro-Cre breeding experiment (Figure 2). About 30% of the 6-month-old mice with the YY1 KD showed a target-specific hypermethylation in Xist, Nespas and Peg3. Since this effect was observed only in aged mice, this may not be a genuine outcome that had been solely caused by the YY1 KD. However, this is also a possible scenario given the fact that a similar effect was also observed in the CTCF case, where the reduced levels of CTCF resulted in hypermethylation in the unmethylated allele of H19 during early embryogenesis (22,23). Moreover, we believe that the detection of this hypermethylation only in the aged mice may be related to the timing of YY1 KD by the Pro-Cre allele. The Pro-Cre allele is manly designed to express the YY1-shRNA at the later stage of spermatogenesis. Thus, it is reasonable to think that the sperm from the aged mice may have experienced the YY1 KD more likely than the sperm from the younger male mice. In this regard, it will be interesting to test a similar YY1 KD using different Cre alleles, such as tissue-nonspecific alkaline phosphatase (TNAP)-Cre, which targets much earlier stages of gametogenesis (38). In sum, the YY1 KD during spermatogenesis appears to have no major effect so far, but we may need to perform more experiments to further confirm this conclusion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH grant (R01GM66225 to J.K.). Funding for open access charge: National Institutes of Health grant (R01GM66225).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Drs Andrea Ventura and Tyler Jacks at MIT for providing the pSico and pSicoR vectors; Isabel Lorenzo and Graeme Mardon at Baylor College of Medicine for excellent transgenic service; Jennifer M. Huang, Chris Faulk, and Hana Kim for critical reading of the manuscript.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr. Opin. Cell Biol. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 2.Ideraabdullah FY, Vigneau S, Bartolomei MS. Genomic imprinting mechanisms in mammals. Mutat. Res. 2008;647:77–85. doi: 10.1016/j.mrfmmm.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 4.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat. Rev. Genet. 2008;9:29–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 5.Jelinic P, Stehle JC, Shaw P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006;4:e355. doi: 10.1371/journal.pbio.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 2006;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- 7.Mackay DJ, Callaway JL, Marks SM, White HE, Acerini CL, Boonen SE, Dayanikli P, Firth HV, Goodship JA, Haemers AP, et al. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat. Genet. 2008;40:949–951. doi: 10.1038/ng.187. [DOI] [PubMed] [Google Scholar]
- 8.Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell. 2008;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 10.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 11.Fitzpatrick GV, Pugacheva EM, Shin JY, Abdullaev Z, Yang Y, Khatod K, Lobanenkov VV, Higgins MJ. Allele-specific binding of CTCF to the multipartite imprinting control region KvDMR1. Mol. Cell Biol. 2007;27:2636–2647. doi: 10.1128/MCB.02036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon B, Herman H, Hu B, Park YJ, Lindroth A, Bell A, West AG, Chang Y, Stablewski A, Piel JC, et al. Rasgrf1 imprinting is regulated by a CTCF-dependent methylation-sensitive enhancer blocker. Mol. Cell Biol. 2005;25:11184–11190. doi: 10.1128/MCB.25.24.11184-11190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hikichi T, Kohda T, Kaneko-Ishino T, Ishino F. Imprinting regulation of the murine Meg1/Grb10 and human GRB10 genes; roles of brain-specific promoters and mouse-specific CTCF-binding sites. Nucleic Acids Res. 2003;31:1398–1406. doi: 10.1093/nar/gkg232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takada S, Paulsen M, Tevendale M, Tsai CE, Kelsey G, Cattanach BM, Ferguson-Smith AC. Epigenetic analysis of the Dlk1-Gtl2 imprinted domain on mouse chromosome 12: implications for imprinting control from comparison with Igf2-H19. Hum. Mol. Genet. 2002;11:77–86. doi: 10.1093/hmg/11.1.77. [DOI] [PubMed] [Google Scholar]
- 15.Chao W, Huynh KD, Spencer RJ, Davidow LS, Lee JT. CTCF, a candidate trans-acting factor for X-inactivation choice. Science. 2002;295:345–347. doi: 10.1126/science.1065982. [DOI] [PubMed] [Google Scholar]
- 16.Navarro P, Page DR, Avner P, Rougeulle C. Tsix-mediated epigenetic switch of a CTCF-flanked region of the Xist promoter determines the Xist transcription program. Genes Dev. 2006;20:2787–2792. doi: 10.1101/gad.389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pugacheva EM, Tiwari VK, Abdullaev Z, Vostrov AA, Flanagan PT, Quitschke WW, Loukinov DI, Ohlsson R, Lobanenkov VV. Familial cases of point mutations in the XIST promoter reveal a correlation between CTCF binding and pre-emptive choices of X chromosome inactivation. Hum. Mol. Genet. 2005;14:953–965. doi: 10.1093/hmg/ddi089. [DOI] [PubMed] [Google Scholar]
- 18.Kim J, Kollhoff A, Bergmann A, Stubbs L. Methylation-sensitive binding of transcription factor YY1 to an insulator sequence within the paternally expressed imprinted gene, Peg3. Hum. Mol. Genet. 2003;12:233–245. doi: 10.1093/hmg/ddg028. [DOI] [PubMed] [Google Scholar]
- 19.Kim JD, Hinz AK, Bergmann A, Huang JM, Ovcharenko I, Stubbs L, Kim J. Identification of clustered YY1 binding sites in imprinting control regions. Genome Res. 2006;16:901–911. doi: 10.1101/gr.5091406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoenherr CJ, Levorse JM, Tilghman SM. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 2003;33:66–69. doi: 10.1038/ng1057. [DOI] [PubMed] [Google Scholar]
- 21.Engel N, West AG, Felsenfeld G, Bartolomei MS. Antagonism between DNA hypermethylation and enhancer-blocking activity at the H19 DMD is uncovered by CpG mutations. Nat. Genet. 2004;36:883–888. doi: 10.1038/ng1399. [DOI] [PubMed] [Google Scholar]
- 22.Fedoriw AM, Stein P, Svoboda P, Schultz RM, Bartolomei MS. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science. 2004;303:238–240. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- 23.Engel N, Thorvaldsen JL, Bartolomei MS. CTCF binding sites promote transcription initiation and prevent DNA methylation on the maternal allele at the imprinted H19/Igf2 locus. Hum. Mol. Genet. 2006;15:2945–2954. doi: 10.1093/hmg/ddl237. [DOI] [PubMed] [Google Scholar]
- 24.Kim JD, Hinz AK, Choo JH, Stubbs L, Kim J. YY1 as a controlling factor for the Peg3 and Gnas imprinted domains. Genomics. 2007;89:262–269. doi: 10.1016/j.ygeno.2006.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J, Kim JD. In vivo YY1-knockdown effects on genomic imprinting. Hum. Mol. Genet. 2008;17:391–401. doi: 10.1093/hmg/ddm316. [DOI] [PubMed] [Google Scholar]
- 26.Donohoe ME, Zhang LF, Xu N, Shi Y, Lee JT. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol. Cell. 2007;25:43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 27.Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl Acad. Sci. USA. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bunch PO, Saling PM. Generation of a mouse membrane fraction with zona receptor activity. Biol. Reprod. 1991;44:672–680. doi: 10.1095/biolreprod44.4.672. [DOI] [PubMed] [Google Scholar]
- 29.Eppig JJ, Telfer E. Isolation and culture of oocytes. Methods Enzymol. 1993;225:77–84. doi: 10.1016/0076-6879(93)25008-p. [DOI] [PubMed] [Google Scholar]
- 30.Hogan B, Beddington R, Constantini F, Lacy E, editors. In Manipulating the Mouse Embryo, 2nd ed. New York: Cold Spring Harbor Laboratory Press; 1994. pp. 296–298. [Google Scholar]
- 31.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howell C, Bestor T, Ding F, Latham K, Mertineit C, Trasler J, Chaillet J. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- 33.Rezai-Zadeh N, Zhang X, Namour F, Fejer G, Wen YD, Yao YL, Gyory I, Wright K, Seto E. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev. 2003;17:1019–1029. doi: 10.1101/gad.1068003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klymenko T, Papp B, Fischle W, Köcher T, Schelder M, Fritsch C, Wild B, Wilm M, Müller J. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JD, Kim J. YY1’s longer DNA binding motifs. Genomics. 2009;93:152–158. doi: 10.1016/j.ygeno.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott RH, Douglas J, Baskcomb L, Huxter N, Barker K, Hanks S, Craft A, Gerrard M, Kohler JA, Levitt GA, et al. Constitutional 11p15 abnormalities, including heritable imprinting center mutations, cause nonsyndromic Wilms tumor. Nat. Genet. 2008;40:1329–1334. doi: 10.1038/ng.243. [DOI] [PubMed] [Google Scholar]
- 37.Kim J. Multiple YY1 and CTCF binding sites in imprinting control regions. Epigenetics. 2008;3:115–118. doi: 10.4161/epi.3.3.6176. [DOI] [PubMed] [Google Scholar]
- 38.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.