Abstract
Initiation of DNA replication involves the ordered assembly of the multi-protein pre-replicative complex (pre-RC) during G1 phase. Previously, DNA topoisomerase II (topo II) was shown to associate with the DNA replication origin located in the lamin B2 gene locus in a cell-cycle-modulated manner. Here we report that activation of both the early-firing lamin B2 and the late-firing hOrs8 human replication origins involves DNA topo II-dependent, transient, site-specific dsDNA-break formation. Topo IIβ in complex with the DNA repair protein Ku associates in vivo and in vitro with the pre-RC region, introducing dsDNA breaks in a biphasic manner, during early and mid-G1 phase. Inhibition of topo II activity interferes with the pre-RC assembly resulting in prolonged G1 phase. The data mechanistically link DNA topo IIβ-dependent dsDNA breaks and the components of the DNA repair machinery with the initiation of DNA replication and suggest an important role for DNA topology in origin activation.
INTRODUCTION
Eukaryotic DNA replication is a stringently regulated and remarkably precise multi-step process, ensuring the duplication of all chromosomes only once per cell cycle (1). Replication is initiated at multiple origins scattered along each chromosome, which are marked by the binding of the evolutionarily conserved origin recognition complex (ORC1-6). ORC acts as a cell-cycle-regulated landing dock for the assembly of the pre-replicative complex (pre-RC), which consists of ORC, Cdc6, Cdt1 and the minichromosome maintenance protein complex (MCM2–7), and its targeting onto chromatin is sufficient to create a functional artificial mammalian replication origin (2). However, unlike the Saccharomyces cerevisiae ORC (ScORC), which recognizes the 11-bp ARS consensus sequence, metazoan ORC exhibits low or no preference for specific DNA sequences (3–5).
One of the basic features of DNA is the intertwining of the DNA strands, which results in a global underwinding of DNA (negatively supercoiled) in all species (6). This underwinding facilitates sister DNA strand separation and affects accessibility of trans-acting factors to DNA (7). The topological state of the DNA in the cell is being managed by the DNA topoisomerases (8), which accomplish this by creating either a single-strand (type I subfamily) or double-strand (type II subfamily) break in DNA, thus permitting the rotation of one or two DNA strands, respectively, around the DNA axis and relaxation of the torsional stress. A role for topoisomerases in the initiation step of DNA replication has been proposed, as they are required for the activation of the simian virus 40 (SV40) (9–12), bovine papilloma virus (BPV) (13) and Epstein-Barr virus (EBV) replication origins (14). Furthermore, it was shown that Drosophila melanogaster ORC (DmORC) exhibits mild sequence specificity, but strong preference for negatively supercoiled DNA, suggesting that the topological state of the DNA is a critical factor for origin specification in that organism (15). Recently, topoisomerases I and II (topo I and II) were also found to interact specifically with the human lamin B2 replication origin in a cell-cycle-regulated manner, indicating that the role of DNA topology during pre-RC assembly may be applicable in mammalian genomes as well (16,17).
The origin of DNA replication located at the 3′ end of the human gene for lamin B2 has been previously mapped to the initiating nucleotide (18,19). It is an early-activated origin, firing in the first minute of S phase (20), and is active in a variety of cell types, including cells of myeloid, epithelial, neuronal, fibroblast origin and primary peripheral blood lymphocytes (21). Similarly, Ors8 has been shown by electron microscopic studies as well as in situ chromosomal and episomal replication assays to be a replication origin in monkey and human cells (22–24). However, unlike its monkey counterpart (mOrs8) which is known to be early activated in monkey kidney (CV-1) cells (25,26), the human homologue (hOrs8) replicates late in S phase (23). There is 90% homology between the mOrs8 and hOrs8.
In this study, we describe the functional synergy of the DNA topology and DNA repair machineries during pre-RC assembly. We present evidence that Ku and topo IIβ directly interact with replication origins in vivo and in vitro, and that their targeting onto chromatin during G1 phase leads to topologic changes in the chromosomal regions that correspond to the human replication origins lamin B2 and hOrs8. The generation of two consecutive topo II-dependent dsDNA breaks in proximity to or within the area covered by the pre-RC complex, during early- and mid-G1 phase, is concurrent with the pre-RC formation. Pharmacological inhibition of topo II results in the inhibition of pre-RC and the downstream pre-Initiation Complex (pre-IC) assembly. The data suggest that dynamic changes in the DNA structure occur during pre-RC assembly and indicate that DNA topology is a critical parameter affecting the initiation of DNA replication.
MATERIALS AND METHODS
Cell culture, cell-cycle analysis and drug treatments
HeLa cells were cultured in αMEM supplemented with 10% fetal bovine serum, penicillin (100 μg/ml), streptomycin (100 μg/ml) and l-glutamine (1 mmol/l) at 37°C and 5% CO2. For M phase synchronization, cells were treated with Nocodazole (100 ng/ml) at 37°C for 24 h and subjected to mitotic shake-off. For G1 phase synchronization, cells were re-seeded and released from M-phase synchronization into the cell cycle by the addition of pre-warmed fresh complete medium. Treatments with the topo II inhibitor merbarone (100 μM) (Calbiochem) and the DNA-PK inhibitor NU-7026 (10 μM) (Calbiochem) were performed for 1 h at 37°C. Cell-cycle progression was monitored by FACS analysis using a Beckman flow cytometer.
Extract preparation, immunoblot analysis and quantification
Nuclear cell extracts (NEs) were prepared as previously described (27) and the protein concentration of each extract preparation was determined using the Bradford Protein Assay. Western blot analysis was carried out according to standard protocols (28) using the antibodies indicated in Supplementary Data.
Electrophoretic mobility-shift assay (EMSA)
Nuclear cell extracts (10 μg) or 100 ng of recombinant Ku (TREVIGEN), Topo IIβ (generous gift from Dr Neil Osheroff, Vanderbilt University Medical Center) and 14-3-3 proteins, were incubated with 0.4 fmol of 32P-labeled PCR products corresponding to the 186-bp core element of the hOrs8 origin of DNA replication, for 1 h on ice. Binding to the oligonucleotide was performed in the presence of high excess poly-dI·dC (Amersham-Pharmacia), used as non-specific competitor, and in a final volume of 20 μl including binding buffer (10 mM Tris–HCl, pH 7.5, 80 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol, 0.1% Triton X-100, 4% glycerol). The amount of poly-dI·dC was optimized to allow detection of specific origin DNA-binding complexes in the presence of excess free probe. The mixtures were subjected to electrophoresis on a native 5% PAGE at 160 V in 0.5× TBE and the gels were then dried and subjected to autoradiography. For electrophoretic mobility-shift competition assays, increasing molar excess amounts of cold probe were included in the reactions as indicated in the figures. For the interference experiments the nuclear extracts were pre-incubated with 1 μg of anti-Ku80 (M-20), anti-TopoIIβ (C-19), anti-ORC2 (H300) (Santa Cruz Biotechnology, Santa Cruz, CA) or normal rabbit serum in binding buffer for 1 h at 4°C followed by the standard-binding reaction.
Chromatin immunoprecipitation assay (ChIP)
ChIP experiments were performed as previously described (29). Briefly, sheared chromatin lysates from 2 × 107 cells were pre-cleared by incubation with Protein G or Protein A agarose, and were subsequently incubated with 20 μg of anti-Ku80, anti-TopoIIβ, anti-PARP-1, anti-ORC2, anti-ORC4, anti-Cdt1, anti-Cdc45 or pre-immune serum. Co-precipitated DNA was washed successively with Lysis Buffer [50 mM HEPES–KOH, pH 7.5, 140 mM NaCl, 2 mM EDTA, 1% Triton X-100, complete protease inhibitors tablet (Roche Molecular Biochemicals)], WB1 (50 mM Tris–HCl, pH 7.5, 500 mM NaCl, 0.1% NP40, 0.05% sodium deoxycholate, complete protease inhibitors), WB2 (50 mM Tris–HCl, pH 7.5, 0.1% NP40, 0.05% sodium deoxycholate, complete protease inhibitors) and TE, isolated and purified.
DNA-break labelling and ChIP
Detection of transient DNA strand break formation was preformed as previously described (30), with slight modifications. Briefly, cells were fixed with Streck Tissue Fixative (STF, Streck Laboratories), which does not cause any DNA damage (31), harvested, washed twice with ice-cold PBS, once with buffer A [0.25% Triton X-100, 10 mM EDTA, 10 mM HEPES (pH 6.5)] and once with buffer B [200 mM NaCl, 1 mM EDTA, 10 mM HEPES (pH 6.5)], and permeabilized with buffer C [100 mM Tris–HCl (pH 7.4), 50 mM EDTA, 1% Triton X-100). Subsequently, the nuclei were sequentially washed with ice-cold PBS, deionized water and 1× terminal deoxynucleotide transferase (TdT; Promega) reaction Buffer. The DNA breaks were labeled with biotin-16-deoxyuridine triphosphate (biotin-16-dUTP) using 50 U of TdT for 1 h at 37°C and residual biotin-dUTP was washed away with Buffer D [100 mM Tris–Cl (pH 7.4), 150 mM NaCl]. Nuclei were refixed and ChIP was performed using an anti-biotin antibody (A1559; Sigma), as described above.
Real-time PCR quantification of DNA
Immunoprecipitated DNA was quantified using the LightCycler FastStart DNA Master SYBR Green I kit (Roche Molecular Biochemicals) according to the manufacturer's instructions, as previously described (29). The sequences and amplification conditions for all primer sets used are listed in Supplementary Table S1, while the amplification, melting and standard curves are shown in Supplementary Figure S2.
Sequence analyses
DNA sequences of characterized replication origins (20,23,32–37) were scanned for the presence of the 12-bp mammalian topo II consensus sequence (38), using the EMBOSS alignment tool (http://www.ebi.ac.uk/Tools/emboss/align/index.html) of the European Bioinformatics Institute (EBI), allowing no gaps.
RESULTS
Ku and topo IIβ bind onto replication origins
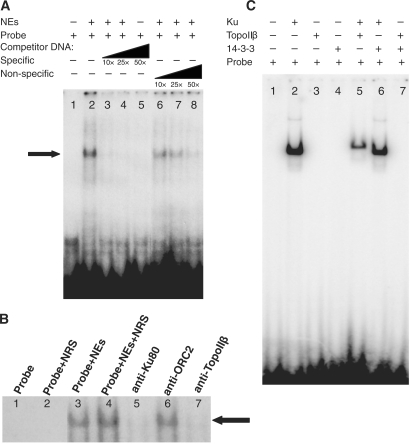
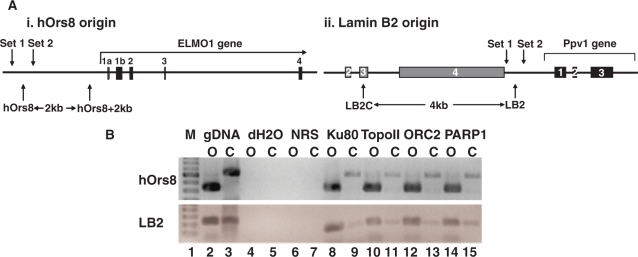
Topo I and II were recently shown to bind onto the lamin B2 replication origin in a cell-cycle-modulated manner (16). Unlike topo I, however, although purified topo II bound lamin B2 in a sequence-specific manner in vivo, it had no sequence preference in vitro, suggesting that it is targeted to specific sites by another protein rather than by direct enzyme/sequence recognition. A human origin binding activity, containing the Ku protein and topo II, was previously purified through its ability to interact specifically with the 186 bp minimal functional sequence of the monkey replication origin Ors8 and support its replication, prompting us to examine whether Ku might target topo II onto replication origins. For this, we performed EMSAs, using antisense oligonucleotides corresponding to the 186-bp core of the human homologue of the monkey Ors8 origin (hOrs8) (23). In the presence of large excess of poly-dI·dC, which blocks non-specific and end-DNA binding, a predominant protein–DNA complex was detected onto the replication origin (Figure 1A, lane 2; indicated by an arrow). Oligonucleotide competition experiments demonstrated greater inhibition of complex formation in the presence of increasing amounts of cold-specific (origin) oligonucleotide (Figure 1A, lanes 3–5) compared to a non-specific oligo of the same size and GC-content (Figure 1A, lanes 6–8), indicating a specific interaction with the origin. The kinetics of the competition reaction suggests that the affinity of the complex for the origin region is 6-fold higher by comparison to the non-specific probe. We next examined whether Ku and topo IIβ are members of the protein–DNA complex formed onto the origin, using antibodies against the 80-kDa subunit of Ku (Ku80) and topo IIβ. As can be seen in Figure 1B, use of the anti-Ku80 (lane 5) and anti-topo IIβ (lane 7) antibodies resulted in the inhibition of the origin-specific complex, indicating the presence of these proteins in it, whereas use of pre-immune rabbit serum (lanes 2 and 4) or an antibody targeted against ORC2 (lane 6) had no effect on complex formation. Use of recombinant proteins showed that topo IIβ is targeted onto origin DNA in vitro by the Ku heterodimer (Ku70/Ku80), reproducibly forming a complex of reduced electrophoretic mobility (Figure 1C, lane 5) by comparison to that formed in the presence of Ku alone (Figure 1C, lane 2). This was not found to be the case for the 14-3-3 protein, which has been shown to bind to cruciform structures at replication origins in vivo (39–41) (Ku does not target 14-3-3 onto hOrs8, since the addition of 14-3-3 does not affect the mobility of the Ku/DNA complex—compare lanes 2 and 6—nor does 14-3-3 target topo IIβ onto it; Figure 1C, lanes 6 and 7, respectively), or a non-origin DNA region (data not shown), suggesting that this targeting is both sequence- and protein-specific. The in vivo association of Ku and topo IIβ with the hOrs8 and lamin B2 replication origins (origin maps are shown in Figure 2A) was analyzed by ChIP assays, using anti-Ku and anti-topo IIβ antibodies (Figure 2B). Both proteins were found to bind in vivo onto both of these origins (Figure 2B, lanes 8 and 10), with some background binding detected at the non-origin-containing chromosomal regions located 4 kb and 2 kb away from the hOrs8 and lamin B2 origins, respectively (Figure 2B, lanes 9 and 11). ORC2, which binds to both origins (16,42), was used as a positive control (Figure 2B, lane 12).
Figure 1.
Ku and topo IIβ interact on the hOrs8 replication origin in vitro. (A) Representative EMSA showing the formation of a protein–DNA complex (indicated by an arrow) formed on hOrs8 (lane 2) in the presence of high concentration of poly-di·dC to block non-specific binding. Addition of increasing amounts of origin probe (lanes 3–5) potently inhibits complex formation even at 10×-molar excess, unlike a non-specific oligo (lanes 6–8) of the same size and GC content, which is less effective. (B) Representative interference experiment showing the presence of Ku and topo IIβ in the complex, using anti-Ku80 (lane 5) and anti-topo IIβ (lane 7) antibodies. An anti-ORC2 antibody (lane 6) and normal rabbit serum (lane 2) were used as negative controls. (C) Representative EMSA using hOrs8 and recombinant Ku protein (lane 2), recombinant Ku and topo IIβ proteins (lane 5), or recombinant Ku and 14-3-3 proteins (lane 6). Additional controls, where Ku has been omitted from the reaction, but in the presence of recombinant topo IIβ (lane 3), 14-3-3 (lane 4), or both topo IIβ and 14-3-3 (lane 7), are also shown. All experiments were performed in triplicate giving rise to reproducible results.
Figure 2.
Ku, topo IIβ and PARP-1 bind onto replication origins in vivo. (A) Maps of the hOrs8 (i) and lamin B2 (ii) origin loci and the primers corresponding to the origin-containing (hOrs8 and LB2, respectively), origin-flanking (Sets 1 and 2) and origin-lacking (hOrs8 + 2 kb and LB2C, respectively) amplicons. The location of the amplicons relative to the gene exons as well as their distance in kb is indicated. (B) ChIP assay using anti-Ku80, -topo IIβ and -PARP-1 antibodies. The amount of DNA immunoprecipitated corresponding to the hOrs8 and lamin B2 origin-containing (O) and their respective origin-lacking control (C) regions, is shown. Immunoprecipitation with normal rabbit serum (NRS) and an anti-NF-κB antibody were used as negative controls, while ORC2, previously known to bind to both origins, was used as a positive control. A sample with water as template (dH2O) was used as a negative control for the real-time PCR reaction, whereas a sample with genomic DNA as template (gDNA) was used as positive control.
In light of the recent finding that topo IIβ, in complex with Ku and PARP-1, participate in the recruitment of the transcription machinery and activation of the pS2 gene, by modifying the chromatin architecture of its promoter (30), we examined whether PARP-1 is also present in the origin-specific protein complex. Indeed, PARP-1 was found to associate with the lamin B2 and hOrs8 origins (Figure 2B, lane 14), similarly to ORC2, Ku and topo IIβ.
The origin association of these proteins was also analyzed as a function of the cell cycle (Figure 3). For this, cells were synchronized in M phase, using mitotic shake-off, and then released into G1 phase by the addition of complete media. Upon this treatment, cells started entering G1 phase after 2 h and reached S phase at 10 h, as monitored by FACS analysis (Figure 3A) and the levels of the G2/M, G1 and S phase Cyclins B, E and A, respectively (Figure 3B). All three proteins (Ku, topo IIβ and PARP-1) bound to the lamin B2 and hOrs8 origins during early and mid-G1 phase, while they dissociated upon cell entry into S phase, following the same pattern (Figure 3C). The expression levels of these proteins during G1 phase were constant (Figure 3B), suggesting that their origin-association pattern is solely due to their chromatin recruitment. Furthermore, all proteins co-precipitated in pull-down experiments (Figure 3D), indicating that they either associate with replication origins in close proximity to one another or they function as a holocomplex, similar to their role in transcription activation (30).
Figure 3.
Origin association of the Ku, topo IIβ and PARP-1 complex during G1 phase. (A) Cell-cycle progression following release from M phase. Cell-cycle distribution of asynchronous, M phase-synchronized and M phase-released cells was monitored by FACS analysis. Synchronization of cells in M phase was done using nocodazole treatment and mitotic shake and hourly samples were taken for 11 h after their release from M phase. (B) Representative western blot analysis, showing the levels of the M phase-specific Cyclin B, G1 phase-specific Cyclin E and S phase-specific Cyclin A following release from M phase. The protein levels of Ku80, topo IIβ and PARP-1 showing constant expression during G1 phase are also shown. ORC2, known to be stably expressed throughout the cell cycle, was used as the loading control. (C) ChIP assay showing an analysis of the association of Ku80, topo IIβ and PARP-1 with hOrs8 (i) and lamin B2 (ii) as a function of the cell cycle. Abundance of the origin-containing or origin-lacking regions in the Ku80, topo IIβ and PARP-1 immunoprecipitates at the indicated timepoints following release from M phase. Values are expressed as molecules of immunoprecipitated DNA per 2 × 107 cells and represent the average of three experiments and 1 SD. The horizontal bar represents background DNA immunoprecipitated by NRS. (D) Reciprocal pull-down experiment using anti-Ku80, -topo IIβ and -PARP-1, -ORC2 antibodies and NRS (negative control), and immunostaining with the same antibodies. Nuclear extracts (NEs) corresponding to 10% of the input protein used in the co-immunoprecipitation reaction, were used as positive control in the western blot analysis (lane 2).
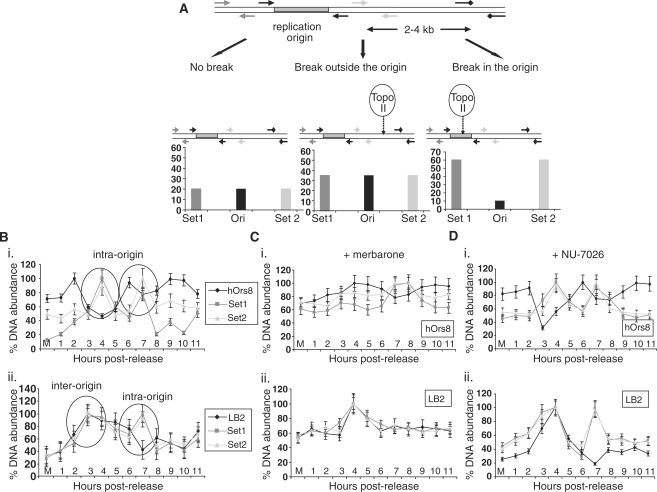
Topo-II-mediated, transient dsDNA breaks during G1 phase
We next examined whether the enzymatic activity of topo IIβ might alter the DNA topology at replication origins during pre-RC assembly. DNA-break formation was screened, using a recently developed protocol that detects DNA-break formation at specific chromosomal loci by combining biotin-dUTP labelling by TdT and subsequent ChIP analysis, using an anti-biotin antibody (see ‘Materials and Methods’ section, Figure 4A). In HeLa cells traversing through G1 phase, putative DNA breaks were detected in both the lamin B2 and hOrs8 replication origins (Figure 4B), but no enrichment of biotin incorporation was observed at the non-origin containing chromosomal regions located 4 kb and 2 kb away, respectively, as evidenced by the background levels of the corresponding amplicons (data not shown). Time-course DNA-break labelling indicated that the origin-specific DNA breaks occurred in a biphasic and dynamic fashion. An initial break was detected early in G1 phase (within 4 h upon release) followed by a second one during mid-G1 phase (within 7 h post-release). Generation of both breaks was transient, as they started reverting to their initial status within 1 h. Interestingly, both breaks at the hOrs8 origin occurred within the origin core (Figure 4Bi), whereas in the lamin B2 origin only one break was created within the area covered by the pre-RC complex (Figure 4Bii). Consistent with this finding is the presence of a topo II site upstream of the pre-RC area at the lamin B2 origin, which might correspond to a scaffold attachment region (16). To ascertain whether the origin DNA cleavages are indeed topo-II mediated, the time-course DNA-break labelling assay was repeated in the presence of the topo II inhibitor, merbarone (5-N-phenylcarboxamido-2-thiobarbituric acid), which inhibits the enzyme's cleaving activity without damaging the DNA or stabilizing intermediate DNA-topo II cleavable complexes (as per manufacturer's specifications). The advantage of topo II pharmacological inhibition over an RNAi approach is the ability to add the inhibitor after cell exit from G2/M, when topo II is known to participate in chromosome condensation and segregation, thus focusing on its role in G1 phase. Treatment with merbarone prevented the DNA breaks at the hOrs8 (Figure 4Ci), and lamin B2 (Figure 4Cii) origins, indicating that they represent transient double-stranded DNA (dsDNA) breaks triggered by DNA topo II.
Figure 4.
DNA-break formation during G1 phase. (A) Schematic diagram explaining the three different scenarios of the DNA-break labelling assay. No break formation (left) should lead to background levels of DNA immunoprecipitated by the anti-biotin antibody. In the case of a break in proximity to the origin, but outside of the core region (middle), increased amounts of the PCR products upstream (Set 1), downstream (Set 2) and flanking (Ori) the origin region should be produced, while an intra-origin break (right) should result in increased Set 1 and Set 2, but decreased Ori products. (B) Generation of putative DNA breaks in hOrs8 (i) and lamin B2 (ii) during G1 phase. Encircled are the two consecutive DNA breaks formed at the two chromosomal loci, two intra-origin breaks in hOrs8, and one inter-origin and one intra-origin in lamin B2. Values are expressed as ratios of the individual DNA abundances at each timepoint to the maximal abundance observed during the course of the assay, following normalization with the non-origin containing regions. Error bars represent 1 SD of three separate experiments. (C) Effect of merbarone on DNA-break formation at the hOrs8 (i) and lamin B2 (ii) replication origins. Error bars represent 1 SD of three separate experiments. (D) DNA-break labelling assay at the hOrs8 (i) and lamin B2 (ii) replication origins following NU-7026 treatment. Error bars represent 1 SD of three separate experiments.
In light of the presence of Ku, the binding subunit of DNA-PK, at the origin area, we also examined the effect of the inhibition of the DNA-PK enzymatic activity, using the DNA-PK inhibitor NU-7026. In contrast to merbarone, NU-7026 had no effect on DNA-break formation at either of the two orgins (Figure 4D), suggesting either that Ku only functions in tethering topo IIβ onto them, or that the DNA-PK activity is only activated under stress conditions.
Functional significance of DNA-break formation
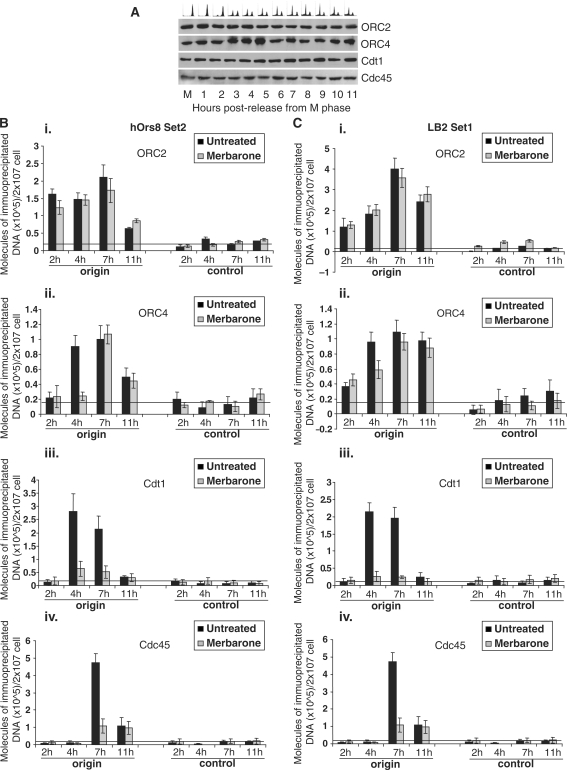
In view of the presence of topo IIβ in close proximity to the DNA replication start site, which suggested a role in the initiation step of DNA replication, we examined the effect of topo II inhibition on pre-RC assembly. Using ChIP assays, we analyzed the kinetics of recruitment of the pre-RC members at the hOrs8 and lamin B2 replication origins during G1 phase. Pre-RC is known to assemble onto replication origins in a stepwise manner (Sclafani and Holzen); origins are initially marked by the ORC hexamer, which is recognized and bound by Cdc6 and Cdt1 during G1-phase. Binding of the latter two proteins permits the subsequent loading of the putative DNA helicase, the MCM2–7, which becomes active by forming a complex with Cdc45 and GINS (43). Consistent with this, the hOrs8 (Figure 5B; black bars) and lamin B2 (Figure 5C; black bars) origins were initially bound by ORC2 (2 h upon release from M phase; Figure 5Bi and Ci, black bars), followed by ORC4 (4 h upon release from M phase; Figure 5Bii and Cii, black bars), Cdt1 (4 h upon release from M phase; Figure 5Biii and Ciii, black bars) and finally Cdc45 (7 h upon release from M phase; Figure 5Biv and Civ, black bars). The expression levels of these proteins during G1 phase were constant (Figure 5A), suggesting that their origin-association pattern is due to their stepwise recruitment. Treatment with merbarone blocked recruitment of ORC4 (4 h upon release from M phase; Figure 5Bii, compare corresponding grey and black bar), Cdt1 (4 h and 7 h upon release from M phase; Figure 5Biii, compare corresponding grey and black bars) and Cdc45 (7 h upon release from M phase; Figure 5Biv, compare corresponding grey and black bar) at the hOrs8 replication origin, whereas ORC2 binding was not affected (Figure 5Bi; grey bars). Similarly, inhibition of dsDNA-break formation led to impaired recruitment of ORC4, Cdt1 and Cdc45, but not of ORC2, onto the lamin B2 origin (Figure 5Cii, iii, iv and i, respectively; grey bars). This is in agreement with previous studies, showing that Ku participates in the stepwise pre-RC assembly at a stage downstream of ORC2 (42,44). Furthermore, merbarone treatment resulted in prolonged G1 phase (Supplementary Figure S1) supporting the notion that pre-RC assembly was affected by topo II activity. Overall, these results suggest that dynamic changes in the DNA topology of origin-containing chromosomal regions naturally occur during pre-RC assembly and that topo II activity is required for replication initiation.
Figure 5.
Origin association of replication initiator proteins during G1 phase. (A) Immunostaining showing the nuclear expression of ORC2, ORC4, Cdt1 and Cdc45 upon release from M phase. ORC2, known to be stably expressed throughout the cell cycle, represents the loading control. (B–C) ChIP assay showing the association of ORC2 (i), ORC4 (ii), Cdt1 (iii) and Cdc45 (iv) with the hOrs8 (B) and lamin B2 (C) origins with (grey bars) or without (black bars) merbarone treatment. Abundance of immunoprecipitated origin DNA is expressed in molecules per 2 × 107 cells and the error bars represent three experiments and 1 SD. The horizontal bars represent background DNA immunoprecipitated by NRS.
DISCUSSION
High order chromatin organization is believed to play an important role in metazoan DNA replication origin specification and selection (5,45). Previous studies implicated chromatin topology in the initiation step of replication of viral and D. melanogaster genomes (9,11–15,46). In this study, we examined whether this role is extended to the human genome as well. Our findings indicate that activation of the human lamin B2 and hOrs8 replication origins involves the generation of topo-II-dependent transient dsDNA breaks, which occur in a biphasic manner, during early- and mid-G1 phase. However, although both breaks at the hOrs8 replication origin occurred within the origin core, in the lamin B2 origin only one break was created within the area covered by the pre-RC complex and the other one occurred in close proximity to, but outside the origin region. It will be interesting to determine whether this is associated with the differential activation timing of the two origins, lamin B2 being early-firing (20) and hOrs8 being late-firing (23). Mammalian cells possess two isoforms of topo II, α and β, with high sequence homology (68% identity and 86% similarity) (47,48). Topo IIα levels fluctuate during the cell-cycle peaking in G2/M (49,50) and thus appears to be required for chromosome condensation and segregation, whereas topo IIβ is less well understood (51). Recently, it was reported that the topo IIβ isoform is implicated in the initiation of DNA replication of KSHV, which utilizes the host molecular machinery in order to proliferate (52). In agreement, we found that topo IIβ binds onto human replication origins, suggesting a DNA replication-specific role for this isoform.
Targeting of topo IIβ onto chromatin is dependent on complex formation with the Ku protein, the DNA-binding subunit of DNA-PK, which has been previously linked to the initiation step of DNA replication (29,42,44,53). PARP-1, a multi-functional protein involved in chromatin regulation, transcription regulation, DNA repair and cell death (54), was also found to be a member of this complex. These proteins were previously shown to form an activation complex on the promoter of the pS2 gene participating in local changes of chromatin architecture and recruitment of the transcription machinery during estradiol-dependent transcriptional activation (30,55). Similarly, a topo IIβ/Ku/PARP-1 holoenzyme seems to act in origin firing, binding onto replication origins during G1 phase and regulating the chromatin topology of the surrounding chromosomal region.
Although the best characterized DNA-binding activity of Ku is its DNA-end-binding activity (56,57), a number of studies, using closed DNA minicircles or plasmids that lack free ends, have shown that Ku also exhibits sequence-specific DNA binding, binding to regulatory DNA elements and affecting gene transcription and DNA replication (58,59). In vivo studies have also supported the binding of Ku to specific regulatory elements (29,30,60–63). In this study, excess amounts of the non-specific competitor poly-dI·dC were used, as a source of free DNA-ends in order to compete out Ku's affinity for DNA-ends and reveal origin DNA-specific complex formation. Although resolution of the crystal structure of the Ku heterodimer on DNA showed that it has a preformed ring that encircles duplex DNA making no contacts with DNA bases (64), a number of mechanisms can be speculated for Ku's recognition of specific internal sequences, including the dimerization of Ku upon recognition of specific sequences, as well as the extrusion of specific internal sequences to hairpins or cruciforms, which are then recognized by Ku. In support of these scenarios, the SAP domain of the Ku70 carboxyterminus has been proposed to be able to recognize specific DNA sequences (64,65), whereas the Ku heterodimer has been shown to have affinity for DNA secondary structures (66).
Pharmacological inhibition of the topo II activity by the specific inhibitor merbarone, resulted in suppression of DNA-break formation and blocked the pre-RC assembly as well as the downstream initiator protein Cdc45. In contrast, DNA-PK inhibition had no effect on DNA-break formation suggesting that Ku functions in tethering topo IIβ onto replication origins. This would increase the sequence specificity of topo IIβ cleavage, in a manner similar to that shown for RAG recombinases, which have similar enzymatic properties to DNA topoisomerases (67). Alternatively, DNA-PK activity may be involved in dsDNA-break repair under conditions of topo II malfunction, acting as a backup mechanism to ensure chromosomal stability. Interestingly, pre-RC formation was blocked at two stages, upstream of ORC4 and Cdt1 recruitment, respectively. In contrast, origin-binding by ORC2 remained unaffected, suggesting that topo II participates in pre-RC assembly rather than origin recognition. This is in agreement with previous studies showing that Ku participates in the stepwise pre-RC assembly at a stage downstream of ORC2 (42,44). ORC2 targeting may instead be attributed to other accessory factors with intrinsic DNA sequence specificity, such as AIF-C (68), Trf2 (69), EBNA-1 (70), HMGA1a (71) or topo I (16).
Overall, the data support the notion that topo IIβ activity is required for the initiation of DNA replication, at least at the lamin B2 and hOrs8 replication origins studied here. Sequence analysis of eight well-characterized human replication origins identified the presence of the mammalian topo II consensus sequence (Table 1), suggesting that the mechanism shown here may be more generalized and applicable genome-wide, as well as underscoring the importance of DNA topology as an important parameter that is crucial for mammalian origin activation.
Table 1.
Number of topo II sites identified in eight well-characterized human replication origins. Their similarities to the mammalian topo II consensus sequence, as well as the references where the origins were initially described are also indicated
| Replication origin | Number of sites | Similarity | Reference |
|---|---|---|---|
| Human c-myc | 1 | 12/12 | (46) |
| Human Ors8 | 4 | 10/12 (three copies), 11/12 | (19) |
| Human lamin B2 | 2 | 11/12 | (47) |
| Human β-globin | 2 | 10/12, 12/12 | (48) |
| Human hsp70 | 1 | 11/12 | (49) |
| Human Top1 | 2 | 10/12 | (50) |
| Human DNMT1 (C3) | 1 | 11/12 | (51) |
| Human rDNA | 5 | 11/12 (four copies), 12/12 | (52) |
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institute of Health Research; Canadian Cancer Society/National Institute of Cancer of Canada (to M.Z.H.); McGill Faculty of Medicine internal studentship award in the past and Roland and Marcel Gosselin Graduate Studentship and the Victor K.S. Lui Fellowship at present (to E.R.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Neil Osheroff (Vanderbilt University Medical Center, Nashville, TN) and Dr Zoi Lygerou (University of Patras, Greece) for generously providing recombinant topo IIβ and anti-Cdt1 antibody, respectively.
REFERENCES
- 1.Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu. Rev. Genet. 2007;41:237–280. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeda DY, Shibata Y, Parvin JD, Dutta A. Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev. 2005;19:2827–2836. doi: 10.1101/gad.1369805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert DM. In search of the holy replicator. Nat. Rev. Mol. Cell Biol. 2004;5:848–855. doi: 10.1038/nrm1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cvetic C, Walter JC. Eukaryotic origins of DNA replication: could you please be more specific? Sem. Cell Dev. Biol. 2005;16:343–353. doi: 10.1016/j.semcdb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Rampakakis E, Arvanitis DN, Di Paola D, Zannis-Hadjopoulos M. Metazoan origins of DNA replication: regulation through dynamic chromatin structure. J. Cell Biochem. 2009;106:512–520. doi: 10.1002/jcb.22070. [DOI] [PubMed] [Google Scholar]
- 6.Schvartzman JB, Stasiak A. A topological view of the replicon. EMBO Rep. 2004;5:256–261. doi: 10.1038/sj.embor.7400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiasa H, Marians KJ. Topoisomerase IV can support oriC DNA replication in vitro. J. Biol. Chem. 1994;269:16371–16375. [PubMed] [Google Scholar]
- 8.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 9.Tsurimoto T, Melendy T, Stillman B. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature. 1990;346:534–539. doi: 10.1038/346534a0. [DOI] [PubMed] [Google Scholar]
- 10.Halmer L, Vestner B, Gruss C. Involvement of topoisomerases in the initiation of simian virus 40 minichromosome replication. J. Biol. Chem. 1998;273:34792–34798. doi: 10.1074/jbc.273.52.34792. [DOI] [PubMed] [Google Scholar]
- 11.Simmons DT, Gai D, Parsons R, Debes A, Roy R. Assembly of the replication initiation complex on SV40 origin DNA. Nucleic Acids Res. 2004;32:1103–1112. doi: 10.1093/nar/gkh236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trowbridge PW, Roy R, Simmons DT. Human topoisomerase I promotes initiation of simian virus 40 DNA replication in vitro. Mol. Cell Biol. 1999;19:1686–1694. doi: 10.1128/mcb.19.3.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, Clower RV, Melendy T. Cellular topoisomerase I modulates origin binding by bovine papillomavirus type 1 E1. J. Virol. 2006;80:4363–4371. doi: 10.1128/JVI.80.9.4363-4371.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawanishi M. Topoisomerase I and II activities are required for Epstein-Barr virus replication. J. Gen. Virol. 1993;74(Pt 10):2263–2268. doi: 10.1099/0022-1317-74-10-2263. [DOI] [PubMed] [Google Scholar]
- 15.Remus D, Beall EL, Botchan MR. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 2004;23:897–907. doi: 10.1038/sj.emboj.7600077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdurashidova G, Radulescu S, Sandoval O, Zahariev S, Danailov MB, Demidovich A, Santamaria L, Biamonti G, Riva S, Falaschi A. Functional interactions of DNA topoisomerases with a human replication origin. EMBO J. 2007;26:998–1009. doi: 10.1038/sj.emboj.7601578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falaschi A, Abdurashidova G, Sandoval O, Radulescu S, Biamonti G, Riva S. Molecular and structural transactions at human DNA replication origins. Cell Cycle. 2007;6:1705–1712. doi: 10.4161/cc.6.14.4495. [DOI] [PubMed] [Google Scholar]
- 18.Giacca M, Zentilin L, Norio P, Diviacco S, Dimitrova D, Contreas G, Biamonti G, Perini G, Weighardt F, Riva S, et al. Fine mapping of a replication origin of human DNA. Proc. Natl Acad. Sci. USA. 1994;91:7119–7123. doi: 10.1073/pnas.91.15.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdurashidova G, Deganuto M, Klima R, Riva S, Biamonti G, Giacca M, Falaschi A. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science. 2000;287:2023–2026. doi: 10.1126/science.287.5460.2023. [DOI] [PubMed] [Google Scholar]
- 20.Biamonti G, Giacca M, Perini G, Contreas G, Zentilin L, Weighardt F, Guerra M, Della Valle G, Saccone S, Riva S, et al. The gene for a novel human lamin maps at a highly transcribed locus of chromosome 19 which replicates at the onset of S-phase. Mol. Cell Biol. 1992;12:3499–3506. doi: 10.1128/mcb.12.8.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S, Giacca M, Norio G, Biamonti G, Riva S, Falaschi A. Utilization of the same DNA replication origin by human cells of different derivation. Nucleic Acids Res. 1996;24:3289–3294. doi: 10.1093/nar/24.17.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Todd A, Landry S, Pearson CE, Khoury V, Zannis-Hadjopoulos M. Deletion analysis of minimal sequence requirements for autonomous replication of ors8, a monkey early-replicating DNA sequence. J. Cell Biochem. 1995;57:280–289. doi: 10.1002/jcb.240570212. [DOI] [PubMed] [Google Scholar]
- 23.Callejo M, Sibani S, Di Paola D, Price GG, Zannis-Hadjopoulos M. Identification and functional analysis of a human homologue of the monkey replication origin ors8. J. Cell Biochem. 2006;99:1606–1615. doi: 10.1002/jcb.20868. [DOI] [PubMed] [Google Scholar]
- 24.Pearson C, Shihab-El-Deen A, Price G, Zannis-Hadjopoulos M. Electron microscopic analysis of in vitro replication products of ors 8, a mammalian origin enriched sequence. Somat. Cell Mol. Genet. 1994;20:147–152. doi: 10.1007/BF02254755. [DOI] [PubMed] [Google Scholar]
- 25.Kaufmann G, Zannis-Hadjopoulos M, Martin RG. Cloning of nascent monkey DNA synthesized early in the cell cycle. Mol. Cell Biol. 1985;5:721–727. doi: 10.1128/mcb.5.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zannis-Hadjopoulos M, Frappier L, Khoury M, Price GB. Effect of anti-cruciform DNA monoclonal antibodies on DNA replication. EMBO J. 1988;7:1837–1844. doi: 10.1002/j.1460-2075.1988.tb03016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. 2nd. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sibani S, Price GB, Zannis-Hadjopoulos M. Decreased origin usage and initiation of DNA replication in haploinsufficient HCT116 Ku80+/− cells. J. Cell Sci. 2005;118:3247–3261. doi: 10.1242/jcs.02427. [DOI] [PubMed] [Google Scholar]
- 30.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 31.Kodym R, Horth E. Determination of radiation-induced DNA strand breaks in individual cells by non-radioactive labelling of 3′ OH ends. Int. J. Radiation Biol. 1995;68:133–139. doi: 10.1080/09553009514551031. [DOI] [PubMed] [Google Scholar]
- 32.Iguchi-Ariga SM, Okazaki T, Itani T, Ogata M, Sato Y, Ariga H. An initiation site of DNA replication with transcriptional enhancer activity present upstream of the c-myc gene. EMBO J. 1988;7:3135–3142. doi: 10.1002/j.1460-2075.1988.tb03180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Lin CM, Brooks S, Cimbora D, Groudine M, Aladjem MI. The human beta-globin replication initiation region consists of two modular independent replicators. Mol. Cell Biol. 2004;24:3373–3386. doi: 10.1128/MCB.24.8.3373-3386.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taira T, Iguchiariga S, Ariga H. A novel DNA replication origin identified in the human heat shock 70 gene promoter. Mol. Cell Biol. 1994;14:6386–6397. doi: 10.1128/mcb.14.9.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keller C, Ladenburger EM, Kremer M, Knippers R. The origin recognition complex marks a replication origin in the human TOP1 gene promoter. J. Biol. Chem. 2002;277:31430–31440. doi: 10.1074/jbc.M202165200. [DOI] [PubMed] [Google Scholar]
- 36.Araujo FD, Knox JD, Ramchandani S, Pelletier R, Bigey P, Price G, Szyf M, Zannis-Hadjopoulos M. Identification of initiation sites for DNA replication in the human dnmt1 (DNA-methyltransferase) locus. J. Biol. Chem. 1999;274:9335–9341. doi: 10.1074/jbc.274.14.9335. [DOI] [PubMed] [Google Scholar]
- 37.Coffman FD, He M, Diaz ML, Cohen S. DNA replication initiates at different sites in early and late S phase within human ribosomal RNA genes. Cell Cycle. 2005;4:1223–1226. doi: 10.4161/cc.4.9.1961. [DOI] [PubMed] [Google Scholar]
- 38.Fosse P, Rene B, Le Bret M, Paoletti C, Saucier JM. Sequence requirements for mammalian topoisomerase II mediated DNA cleavage stimulated by an ellipticine derivative. Nucleic Acids Res. 1991;19:2861–2868. doi: 10.1093/nar/19.11.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pearson CE, Zannis-Hadjopoulos M, Price GB, Zorbas H. A novel type of interaction between cruciform DNA and a cruciform binding protein from HeLa cells. EMBO J. 1995;14:1571–1580. doi: 10.1002/j.1460-2075.1995.tb07143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Todd A, Cossons N, Aitken A, Price GB, Zannis-Hadjopoulos M. Human cruciform binding protein belongs to the 14-3-3 family. Biochemistry. 1998;37:14317–14325. doi: 10.1021/bi980768k. [DOI] [PubMed] [Google Scholar]
- 41.Novac O, Alvarez D, Pearson CE, Price GB, Zannis-Hadjopoulos M. The human cruciform binding protein, CBP, is involved in DNA replication and associates in vivo with mammalian replication origins. J. Biol. Chem. 2002;22:22. doi: 10.1074/jbc.M107902200. [DOI] [PubMed] [Google Scholar]
- 42.Sibani S, Price GB, Zannis-Hadjopoulos M. Ku80 binds to human replication origins prior to the assembly of the ORC complex. Biochemistry. 2005;44:7885–7896. doi: 10.1021/bi047327n. [DOI] [PubMed] [Google Scholar]
- 43.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl Acad. Sci. USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rampakakis E, Di Paola D, Zannis-Hadjopoulos M. Ku is involved in cell growth, DNA replication and G1-S transition. J. Cell Sci. 2008;121:590–600. doi: 10.1242/jcs.021352. [DOI] [PubMed] [Google Scholar]
- 45.Aladjem MI. Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat. Rev. 2007;8:588–600. doi: 10.1038/nrg2143. [DOI] [PubMed] [Google Scholar]
- 46.Halmer L, Vestner B, Gruss C. Involvement of topoisomerases in the initiation of simian virus 40 minichromosome replication. J. Biol. Chem. 1998;273:34792–34798. doi: 10.1074/jbc.273.52.34792. [DOI] [PubMed] [Google Scholar]
- 47.Austin CA, Sng JH, Patel S, Fisher LM. Novel HeLa topoisomerase II is the II beta isoform: complete coding sequence and homology with other type II topoisomerases. Biochim. Biophys. Acta. 1993;1172:283–291. doi: 10.1016/0167-4781(93)90215-y. [DOI] [PubMed] [Google Scholar]
- 48.Jenkins JR, Ayton P, Jones T, Davies SL, Simmons DL, Harris AL, Sheer D, Hickson ID. Isolation of cDNA clones encoding the beta isozyme of human DNA topoisomerase II and localisation of the gene to chromosome 3p24. Nucleic Acids Res. 1992;20:5587–5592. doi: 10.1093/nar/20.21.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heck MM, Hittelman WN, Earnshaw WC. Differential expression of DNA topoisomerases I and II during the eukaryotic cell cycle. Proc. Natl Acad. Sci. USA. 1988;85:1086–1090. doi: 10.1073/pnas.85.4.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kimura K, Saijo M, Ui M, Enomoto T. Growth state- and cell cycle-dependent fluctuation in the expression of two forms of DNA topoisomerase II and possible specific modification of the higher molecular weight form in the M phase. J. Biol. Chem. 1994;269:1173–1176. [PubMed] [Google Scholar]
- 51.Austin CA, Marsh KL. Eukaryotic DNA topoisomerase IIβ. BioEssays. 1998;20:215–226. doi: 10.1002/(SICI)1521-1878(199803)20:3<215::AID-BIES5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, Li H, Tang Q, Maul GG, Yuan Y. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: involvement of host cellular factors in the replication. J. Virol. 2008;82:2867–2882. doi: 10.1128/JVI.01319-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burckstummer T, Bennett KL, Preradovic A, Schutze G, Hantschel O, Superti-Furga G, Bauch A. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods. 2006;3:1013–1019. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- 54.Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 55.Lis JT, Kraus WL. Promoter cleavage: a topoIIbeta and PARP-1 collaboration. Cell. 2006;125:1225–1227. doi: 10.1016/j.cell.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 56.Mimori T, Hardin JA. Mechanism of interaction between Ku protein and DNA. J. Biol. Chem. 1986a;261:10375–10379. [PubMed] [Google Scholar]
- 57.Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 58.Giffin W, Torrance H, Rodda DJ, Prefontaine GG, Pope L, Hache RJ. Sequence-specific DNA binding by Ku autoantigen and its effects on transcription. Nature. 1996;380:265–268. doi: 10.1038/380265a0. [DOI] [PubMed] [Google Scholar]
- 59.Ruiz MT, Matheos D, Price GB, Zannis-Hadjopoulos M. OBA/Ku86: DNA binding specificity and involvement in mammalian DNA replication. Mol. Biol. Cell. 1999;10:567–580. doi: 10.1091/mbc.10.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Knuth MW, Gunderson SI, Thompson NE, Strasheim LA, Burgess RR. Purification and characterization of proximal sequence element-binding protein 1, a transcription activating protein related to Ku and TREF that binds the proximal sequence element of the human U1 promoter. J. Biol. Chem. 1990;265:17911–17920. [PubMed] [Google Scholar]
- 61.Camara-Clayette V, Thomas D, Rahuel C, Barbey R, Cartron JP, Bertrand O. The repressor which binds the −75 GATA motif of the GPB promoter contains Ku70 as the DNA binding subunit. Nucleic Acids Res. 1999;27:1656–1663. doi: 10.1093/nar/27.7.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Falzon M, Fewell JW, Kuff EL. EBP-80, a transcription factor closely resembling the human autoantigen Ku, recognizes single- to double-strand transitions in DNA. J. Biol. Chem. 1993;268:10546–10552. [PubMed] [Google Scholar]
- 63.Shi L, Qiu D, Zhao G, Corthesy B, Lees-Miller S, Reeves WH, Kao PN. Dynamic binding of Ku80, Ku70 and NF90 to the IL-2 promoter in vivo in activated T-cells. Nucleic Acids Res. 2007;35:2302–2310. doi: 10.1093/nar/gkm117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 65.Aravind L, Koonin EV. SAP – a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 66.Dynan WS, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sawchuk DJ, Mansilla-Soto J, Alarcon C, Singha NC, Langen H, Bianchi ME, Lees-Miller SP, Nussenzweig MC, Cortes P. Ku70/Ku80 and DNA-dependent protein kinase catalytic subunit modulate RAG-mediated cleavage: implications for the enforcement of the 12/23 rule. J. Biol. Chem. 2004;279:29821–29831. doi: 10.1074/jbc.M403706200. [DOI] [PubMed] [Google Scholar]
- 68.Minami H, Takahashi J, Suto A, Saitoh Y, Tsutsumi K. Binding of AlF-C, an Orc1-binding transcriptional regulator, enhances replicator activity of the rat aldolase B origin. Mol. Cell Biol. 2006;26:8770–8780. doi: 10.1128/MCB.00949-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Atanasiu C, Deng Z, Wiedmer A, Norseen J, Lieberman PM. ORC binding to TRF2 stimulates OriP replication. EMBO Rep. 2006;7:716–721. doi: 10.1038/sj.embor.7400730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schepers A, Ritzi M, Bousset K, Kremmer E, Yates JL, Harwood J, Diffley JF, Hammerschmidt W. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 2001;20:4588–4602. doi: 10.1093/emboj/20.16.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thomae AW, Pich D, Brocher J, Spindler MP, Berens C, Hock R, Hammerschmidt W, Schepers A. Interaction between HMGA1a and the origin recognition complex creates site-specific replication origins. Proc. Natl Acad. Sci. USA. 2008;105:1692–1697. doi: 10.1073/pnas.0707260105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.