Abstract
Regulation of mRNA translation is an important mechanism determining the level of expression of proteins in eukaryotic cells. Translation is most commonly initiated by cap-dependent scanning, but many eukaryotic mRNAs contain internal ribosome entry segments (IRESs), providing an alternative means of initiation capable of independent regulation. Here, we show by using dicistronic luciferase reporter vectors that the 5′-UTR of the mRNA encoding human insulin receptor (hIR) contains a functional IRES. RNAi-mediated knockdown showed that the protein PTB was required for maximum IRES activity. Electrophoretic mobility shift assays confirmed that PTB1, PTB2 and nPTB, but not unr or PTB4, bound to hIR mRNA, and deletion mapping implicated a CCU motif 448 nt upstream of the initiator AUG in PTB binding. The IR-IRES was functional in a number of cell lines, and most active in cells of neuronal origin, as assessed by luciferase reporter assays. The IRES was more active in confluent than sub-confluent cells, but activity did not change during differentiation of 3T3-L1 fibroblasts to adipocytes. IRES activity was stimulated by insulin in sub-confluent cells. The IRES may function to maintain expression of IR protein in tissues such as the brain where mRNA translation by cap-dependent scanning is less effective.
INTRODUCTION
Regulation of mRNA translation is an important mechanism determining the level of expression of many proteins in eukaryotic cells. At least two distinct mechanisms are used to bring ribosomes to the mRNA. In the most commonly used process, termed cap-dependent scanning, a protein complex eIF4F (comprised of eIF4E, the cap-binding protein eIF4G, a large scaffold protein and eIF4A, a dead box helicase) interacts initially with the 5′-end of the mRNA and, via the interaction of eIF3 with eIF4G, recruits the 40S ribosomal subunit (1). In the alternative process (internal ribosome entry), the 5′-untranslated region (5′-UTR) of the mRNA is capable, in the presence of trans-acting factors, of adopting a highly complex structure (internal ribosome entry segment, IRES) that allows the ribosome to be acquired to a site that can be a considerable distance from the 5′-end of the mRNA (2,3). The proteins required for this process have yet to be fully identified although it has been found that many cellular and viral IRESs require polypryimidine tract binding protein (PTB) for activity (4,5). Cellular IRESs are generally used under patho-physiological conditions when cap-dependent scanning is compromised, for example during polio virus infection (6), apoptosis (7), mitosis (8), genotoxic stress (9) and heat shock (10). Overall, around 10% of mRNAs have potential for their translation to be initiated by internal ribosome entry (11). However, the subsets of mRNAs that are still translated under different conditions are distinct, suggesting that in each condition there is co-ordinated translational regulation dependent on specific elements. Certain families of highly related mRNAs have also been shown to be translated by internal ribosome entry, for example the Myc family of genes (12–15). The IRESs found in the c-, L- and N-myc 5′-UTRs require a common set of trans-acting factors (ITAFs) for their activity suggesting that they may be regulated in a similar manner (16). IRES-mediated translational regulation is also prevalent amongst growth factors and growth factor receptors. Translation initiation on a number of growth factor mRNAs (IGFII, PDGF2, TGF-β, FGF-2 and VEGF) is specifically regulated during differentiation, growth and stress (17). The mRNAs encoding mammalian insulin like growth factor-1 receptor (IGF-1R) (18), angiotensin type I receptor (19), vasopressin v1b receptor (20) and TrkB neurotrophin receptor (21) all contain functional IRESs. The Drosophila insulin-like receptor mRNA contains an IRES that coordinates physiological signals responsible for monitoring nutrient and cell growth conditions (22).
The level of insulin receptor (IR) expression is an important determinant of cellular insulin sensitivity, but mechanisms regulating IR expression are poorly understood. The IR is widely distributed in almost all mammalian tissues but levels of expression vary widely in different cell types, being highest in classical insulin target tissues such as liver and adipose tissue (23). The IR gene promoter was characterized almost 20 years ago and shown to contain elements typical of housekeeping genes as well as elements responsive to glucocorticoids and insulin (24,25). More recently, up-regulation of IR mRNA induced by serum deprivation was shown to involve the transcription factor FOXO1 (26). It has long been known that prolonged exposure of cells to insulin induces IR down-regulation via internalization and enhanced protein degradation (27). However, the regulation of mammalian IR expression at the level of mRNA translation has not previously been reported. Here, we show that the mRNA encoding human IR contains an IRES which is dependent on PTB and nPTB for activity in vitro and in vivo. The activity of this IRES differs between cell types and it is stimulated by insulin.
MATERIALS AND METHODS
Cloning and sequencing of HIR 5′-UTR
Primers were designed to amplify human insulin receptor (HIR) 5′-UTR based on previously identified transcription start sites (28) and using RNA extracted from human epithelial carcinoma (HeLa) cells. A single product of the expected size (∼540 nt) was obtained and sequenced. The sequence was identical to that in the human genome database (Homo sapiens chromosome 19 genomic contig, reference assembly; version NT_011255.14, GI:37552371), which differs slightly from the published sequence (28). Following digestion with EcoR1 and Nco1 the fragment was inserted into the dicistronic reporter vector pRF (10) and into the vector pRBRF, a dicistronic promoterless construct described previously (29).
Cell culture and transient transfections
3T3-L1 cells were cultured and differentiated as described previously (30). Other cell lines N1E-115 (mouse neuroblastoma), HeLa, N2A (mouse neuroblastoma), NB2a (neuroblastoma originating from mouse cortex), MCF7 (derived from human breast adenocarcinoma) and B50 (neuroblastoma derived from rat hippocampal neurons) were cultured using conditions described on the ATCC web page (see www.atcc.org). Cells were transfected using FuGene 6 (Roche) following the supplier's instructions, or with calcium phosphate (31).
The activities of firefly and Renilla luciferases in in vitro translation reactions or lysates prepared from transfected cells were measured using a Dual-Luciferase reporter assay system (Promega) and light emission was measured over 10 s using an OPTOCOMP I luminometer. The activity of β-galactosidase in lysates prepared from cells transfected with pβ-Gal was measured using a Galactolight plus assay system (Tropix). IRES activity was calculated as the average of (IRES-driven firefly luciferase expression/β-Gal expression). Errors were calculated as the standard deviation of the three calculated IRES activities, and expressed as a percentage of the average activity.
Quantitative RT–PCR
Total cellular RNA was isolated from parallel transfections using the RNAqueous kit (Ambion) according to the manufacturer's protocol and treated with Turbo DNase (Ambion) to ensure that any contaminating genomic or plasmid DNA was removed. Total cDNA for quantiative PCR assay was prepared from total RNA with the Improm II Reverse Transcription System (Promega) using random primers. The Power SYBR Green PCR Kit (Applied Biosystems) was used to detect Renilla and firefly luciferase mRNA, using the primer pairs Ren1 (forward primer, TTTACATGGTAACGCGGCCT; reverse primer, GGTCTGGTATAATACACCGCGC; amplifying bases 146–210 of the Renilla ORF), Ren2 (forward primer, GTTTATTGAATCGGACCCAGGA; reverse primer, CCCATTTCATCAGGTGCATCTT; amplifying bases 758–865 of the Renilla ORF) and Fire1 (forward primer, GAAGAGATACGCCCTGGTTCCT; reverse primer, TGTCCACCTCGATATGTGCATC; amplifying bases 90–130 of the firefly ORF) which were designed using the Primer Express Program v2.0 (Applied Biosystems). Amplification reactions were carried out and analysed using a Stratagene Mx3000P Real-Time PCR System, with a standard curve of pRF plasmid DNA diluted over five orders of magnitude used to quantify the amount of starting template in each sample. All assays were carried out in triplicate on three independent occasions.
Reduction of PTB expression by RNAi
On day one, HeLa cells were seeded at a density of 105 cells per well of a 24-well plate and grown overnight in DMEM 10% FCS. On day two, 6 µl of a 20 µM stock of siRNAs PTB P1 (AAC UUC CAU CAU UCC AGA GAA) and C2 (AAG GUC CGG CUC CCC CAA AUG) (32) were mixed with 44 µl Optimem-1 medium (Invitrogen), and then combined with a mixture of 3 µl Lipofectamine 2000 (Invitrogen) and 12 µl Optimem-1 and incubated at room temperature for 25 min. A further 38 µl of Optimem-1 was added, and 100 µl of the mixture transferred to the cells. On day three, the cells were washed with PBS, detached from the plate by addition of 120 µl of trypsin/EDTA and incubation at 37°C for 2–3 min. DMEM (380 µl) was added and the cells then split equally between three wells of a 24-well plate, each containing 0.5 ml of DMEM/FCS. On day four, the cells were transfected with siRNA as on day two, except that one-third of the quantities were used per well. On day four 0.3 µg of dicistronic reporter per well was transfected using FuGene6 (Roche) according to the manufacturer's instructions. The cells were harvested on day seven and luciferase activity was determined.
Electrophoretic mobility shift assays
Electrophoretic mobility shift assays (EMSAs) were preformed as described previously (33) using ∼23 000 c.p.m. of labelled transcript which was incubated with unr (0.2 µg) and/or PTB isoforms (0.1 µg) at room temperature for 10 min. Loading buffer was added and samples loaded directly onto 5% or 10% acrylamide TBE gels that were then electrophoresed at 150 V for 1 h. Gels were dried and exposed on a PhosphorImager.
In vitro transcription reactions
Vector DNA was linearized by restriction digestion using a site downstream of the region of interest (HpaI for dicistronic, NcoI for monocistronic); transcripts were synthesized in a reaction mixture containing 1× transcription buffer [40 mM HEPES–KOH (pH 7.9), 6 mM MgCl2, 2 mM spermidine, 10 mM dithiothreitol (DTT), 10 mM NaCl], 40 U RNAguard or RNasin, 1 mM ATP, 1 mM UTP, 1 mM CTP, 0.5 mM GTP, 1 mM m7G(5′)ppp(5′)G, 1 µg DNA template and 20 U T7 or T3 RNA polymerase to a final volume of 50 µl. For radiolabelled RNAs, CTP was replaced with 50 µCi [a-32P]CTP. For biotinylated RNA, the transcription reactions were supplemented with 0.1 mM biotin UTP. The reaction mixture was incubated at 37°C for 1.5 h and the RNA purified.
Protein expression
Unr with a C-terminal His-tag was purified from cultures of Sf9 cells that had been infected with a recombinant baculovirus expressing unr-His, according to the supplier's instructions (Qiagen). Cells were harvested, lysed in phosphate-buffered saline containing 0.1% Triton, and the tagged protein purified on a Nickel affinity column. His-tagged PTB1, PTB2, nPTB and hnRNPK were over-expressed in Escherichia coli from PET28a vector by addition of IPTG to the growth medium and proteins were again purified using a Nickel affinity column (29).
In vitro translation reactions
The rabbit reticulocyte flexi-lysate in vitro translation system (Promega) was primed with 5 ng/ml RNA and used according to the manufacturer's instructions. The reaction was performed in a final volume of 12.5 µl and 0.5–2 µg of each protein was added where appropriate. Luciferase activities were assayed as described above, and the firefly and Renilla values expressed relative to the control plasmid pRF, which was assigned a value of 1. All experiments were performed in triplicate on at least three independent occasions.
SDS–PAGE and western blotting
Cell extracts were prepared as described previously (34), subjected to electrophoresis on SDS–polyacrylamide gels and the proteins transferred to polyvinylidene difluoride (PVDF) membrane (Millipore). Blots were probed with anti-PTB monoclonal antibody (derived from the BB7 hybridoma), or with anti-actin antibody (Sigma) as loading control, and reactions visualized using chemiluminescent reagents.
Secondary structure prediction
Chemical structure probing was performed exactly as described previously (29). Secondary structure predictions were generated using the web implementation of the Mfold algorithm incorporating version 3.0 of the Turner rules.
RESULTS
The IR 5′-UTR contains an IRES
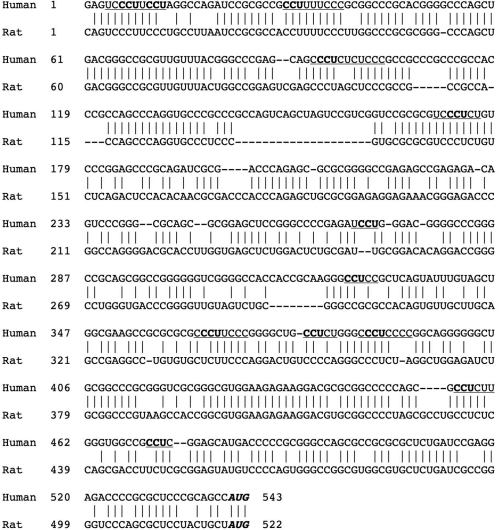
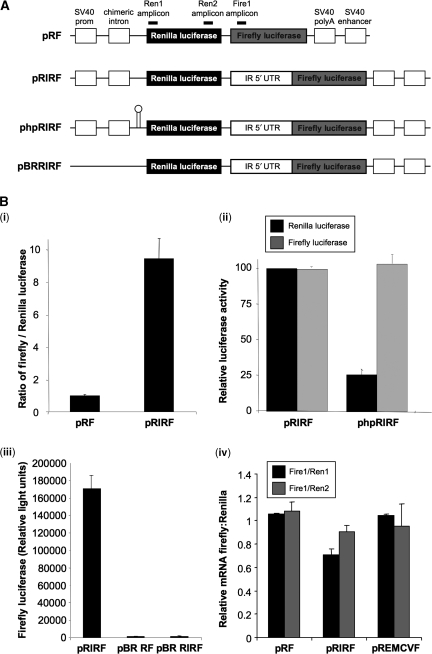
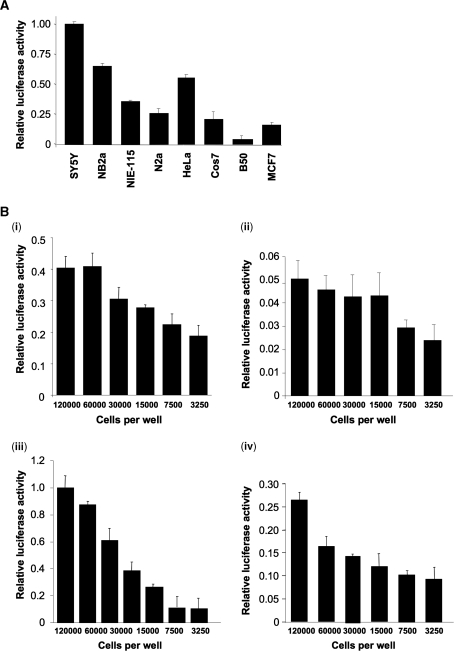
Transcription of the human IR gene is initiated at several sites around 540 nt upstream of the translation start site (28). The 5′-UTR of the human IR mRNA is long and GC-rich and is highly conserved between species, the mouse and rat mRNAs showing ∼70% identity to the human sequence consistent with functional significance of this region (Figure 1). The IR 5′-UTR is similar in this regard to that of the highly related type I IGF-1R which has been shown to contain an IRES (18). To test whether the IR 5′-UTR also contains an IRES the corresponding DNA was obtained by RT–PCR and inserted into the dicistronic reporter vector pRF (Figure 2A). This vector contains the Renilla and firefly luciferases as the upstream and downstream cistrons respectively and an increase in firefly luciferase activity is indicative of an IRES in the preceding sequence (10). The resulting plasmid (pRIRF) was transfected into HeLa cells and luciferase activity was determined. The presence of the IR 5′-UTR stimulated the translation of the firefly luciferase approximately nine times over that attributable to read-through (Figure 2Bi). To rule out apparent IRES activity resulting from enhanced read-through the IR 5′-UTR was inserted into the vector phpRF that contains a stable hairpin upstream of the Renilla cistron. The presence of the hairpin reduced the activity from the Renilla cistron by 70% but had no effect on the amount of firefly luciferase produced (Figure 2Bii). To ensure that the IRES activity was not due to the presence of cryptic promoters in the 5′-UTR of IR the DNA corresponding to this region was inserted into the vector pBRRF which does not contain a promoter. This vector was again transfected into HeLa cells and luciferase activity was determined. No luciferase activity was detected showing that there was no promoter present in this region (Figure 2Biii). To determine whether increases in firefly luciferase expression were due to an increase in monocistronic mRNAs being produced by alternative splicing events that removed the Renilla ORF or by cryptic promoter activity, a quantitative real-time PCR assay was carried out to determine the amount of reporter mRNA (Figure 2Biv). Two amplicons were chosen within the Renilla ORF (Ren1 and Ren2) with the firefly amplicon (Fire1) selected close to the AUG initiation codon. A ratio of 1 : 1 of firefly : Renilla mRNA would be expected in an intact dicistronic message and is indeed observed in mRNA from the control vector pRF, and also in a vector containing the IRES from EMCV. Insertion of the insulin receptor 5′-UTR between the cistrons of the pRF vector results in a change in the ratio between the cistrons. However, as the resulting ratios are less than 1, this suggests that there is no cryptic promoter activity in the IR 5′-UTR, and there is no evidence for a cryptic splicing event that results in loss of the Renilla cistron (both events which would lead to the production of functional, capped, monocistronic firefly luciferase mRNAs that would be translated by the cap-dependent scanning mechanism). Taken together these data strongly suggest that IR 5′-UTR contains a functional IRES.
Figure 1.
5′-UTRs of human and rat IR mRNA. The 5′-UTR of hIR mRNA, as previously defined (28) was amplified from HeLa cells. The sequence determined was identical to that in the human genome database (Homo sapiens chromosome 19 genomic contig, reference assembly; version NT_011255.14, GI:37552371). This sequence differs slightly from that published when the transcription start sites were determined (28). The 5′-UTR of rat IR mRNA has not been formally defined, and the rat sequence shown is from rat genome database (Rattus norvegicus chromosome 12 genomic contig, reference assembly, Version NW_047365.1, GI:34870143). Polypyrimidine tracts in the human sequence are underlined, and CCU motifs indicated in bold. The translation start codon is indicated in italics.
Figure 2.
The 5′-UTR of hIR mRNA contains an IRES. (A) Schematic representation of the constructs pRF, pRIRF, pHPRIRF and pBRRIRF. (B) IRES activity was demonstrated using bicistronic vectors and controls. Data shown are mean ± SD of three independent experiments (each involving triplicate assays). (i) The plasmids pRF and pRIRF were transfected into HeLa cells and luciferase activity was determined. The value shown is the ratio firefly activity/Renilla activity relative to the transfection control β-galactosidase. (ii) The IR 5′-UTR was subcloned into the vector phpRF (10) which contains a stable hairpin upstream of the Renilla luciferase cistron to inhibit ribosome scanning. This vector was transfected into HeLa cells and luciferase activities determined and the values expressed relative to those of the transfection control β-galactosidase and relative to pRIRF which was set at 100%. (iii) IR 5′-UTR was subcloned into the dicistronic promoterless vector pBRRF, creating pBRRIRF, to test whether apparent IRES activity might instead reflect a cryptic promoter. Lucifersae activities were determined as in (ii). (iv) Quantitative real-time PCR assays were carried out to determine the amount of reporter mRNA and to measure whether increases in firefly luciferase expression were due to an increase in monocistronic mRNAs being produced by alternative splicing events that removed the Renilla ORF or by cryptic promoter activity. Two amplicons were chosen within the Renilla ORF (Ren1 and Ren2 (shown in A) with the firefly amplicon (Fire1) selected close to the AUG initiation codon. Amounts of mRNA from each amplicon were assayed and compared to a standard curve generated from pRF plasmid DNA, from which equal amounts of firefly and Renilla cDNA are amplified. In the transfected samples, a similar ratio of firefly : Renilla mRNA would be expected in an intact dicistronic message. Results from a dicistronic vector containing the viral IRES from EMCV are shown for comparison.
The IR-IRES requires PTB for activity in vitro and in cultured cells
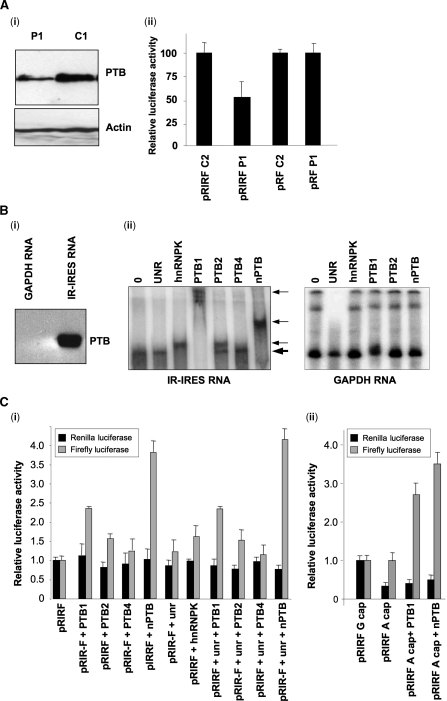
We have shown previously that PTB is a general IRES-trans-acting factor binding to CCUn in polypyrimidine rich stretches of RNA (4). To determine whether IR-IRES activity was sensitive to a reduction in the levels of PTB in cultured cells RNAi was used to reduce expression of this protein (Figure 3Ai). The dicistronic vector that contains the IR-IRES was transfected into these cells and luciferase activity was determined (Figure 3Aii). IR-IRES activity was reduced to 50% of the untreated values by RNAi suggesting strongly that PTB is an ITAF for IR-IRES cells grown in culture (Figure 3Aii).
Figure 3.
The IR-IRES requires PTB for activity in vivo and in vitro. (A) Cells were transfected with siRNA to reduce the expression of PTB (P1) or scrambled control siRNA (C2) and then transfected with the plasmid harbouring the IR-IRESs (pRIRF) or the vector control (pRF). (i) Cells were harvested and lysed and a proportion of the resulting material was immunoblotted using antibodies against PTB, or against actin as a loading control. (ii) Samples were assayed for firefly and Renilla luciferase activity and the IRES activity was calculated as firefly activity relative to the transfection control β-galactosidase (ii). Data shown are mean ± SD of three independent experiments (each involving triplicate assays). (Bi) Biotinylated IR-IRES or GAPDH (control) RNA was incubated with HeLa cell lysates. Protein/RNA complexes were isolated on strepavadin-agarose beads, eluted and subjected to SDS–PAGE. The gels were transferred to nitrocellulose and probed with anti-PTB antibodies. (Bii) EMSAs were performed using radiolabelled IR-IRES or GAPDH RNA which was incubated with 0.2 µg of unr, hnRNPK, or PTB1-4. Products were separated on a 5% TBE polyacrylamide gel. The thick arrow indicates the position of the unshifted band, thinner arrows indicate the positions of shifted bands. (Ci) In vitro translation reactions were performed in reticulocyte lysates primed with 100 ng of capped pRIRF RNA and 50–200 ng of PTB1-4, hnRNPK and unr, either alone or in combination. IRES activity is expressed relative to the reactions that were carried out in the absence of trans-acting factors and this value was set to 1. All experiments were performed in triplicate on three independent occasions. (Cii) In vitro translation reactions were performed in reticulocyte lysates primed with 100 ng of ‘A’ capped pRIRF RNA and 100 ng of either PTB or nPTB-1. IRES activity is expressed relative to the reactions that were carried out using RNA that contained a ‘G’ cap in the absence of trans-acting factors and this value was set to 1. All experiments were performed in triplicate on three independent occasions.
To determine whether the IR-IRES was able to interact with PTB from cultured cells, biotinylated RNA was transcribed in vitro from cut plasmid DNA which contained the IR-IRES element (pSKIRL) or from a control GAPDH plasmid (pSKGAP) and incubated with HeLa cell cytoplasmic extracts. RNA/protein complexes were purified using magnetic beads which contained strepavidin as described previously (16). After extensive washing, proteins were eluted with buffer containing 300 mM NaCl and the products were separated by SDS–PAGE and immunoblotted for PTB. The data show that PTB interacts preferentially with IR-IRES RNA, relative to the GAPDH control sequence (Figure 3Bi).
In mammalian cells there are three widely distributed isoforms of PTB, namely PTB1, PTB2 and PTB4, and a paralog nPTB that has enhanced expression in neuronal tissue (5). The RNAi used would decrease the levels of PTBs -1, -2 and -4 therefore assays were performed in vitro to identify whether any one of these proteins preferentially interacted with the IR-IRES. EMSAs were carried out using purified PTB isoforms and additional proteins that have been shown to bind other cellular and viral IRESs. Radiolabelled IR-IRES RNA generated by in vitro transcription reactions was incubated with the purified proteins. A complex that migrated more slowly was observed when IR-RNA was incubated with hnRNPK, PTB1, PTB2 and nPTB but not unr or PTB4 (Figure 3Bii) or a number of additional potential ITAFs including PCBP1, PCBP2 or La (data not shown). It has been shown for the Apaf-1 IRES that unr only binds in the presence of PTB1, however no binding of unr to the IR-IRES was observed in the presence of PTB1 (data not shown). To confirm that the interaction with these proteins and the IR-IRES-RNA was specific, EMSAs were performed using GAPDH RNA. In this case, there was no shift in the presence of these proteins (Figure 3Bii). Interestingly, although the molecular weights of PTB1, PTB2 and nPTB are almost identical the IR-IRES RNA migration was delayed to very different extents by these three proteins (Figure 3Bii). This could suggest that the number of PTB molecules interacting with the RNA differs for different isoforms, or that the IR-IRES RNA adopts alternative structures upon binding to these three different yet highly related proteins.
Most cellular IRESs are inactive in vitro suggesting an absolute requirement for ITAFs that are not present in these systems (35). To test whether these proteins were also able to stimulate IRES activity in vitro dicistronic RNA was derived from pRIRF and pRF and was used to prime reticulocyte lysates which were supplemented with the proteins indicated (Figure 3C). There was stimulation of IR-IRES activity in the presence of PTB1 and nPTB by 2- and 3.5-fold, respectively and a smaller but significant stimulation was observed with hnRNPK and PTB2. There was no additive effect on IRES activity by combinations of PTB and unr (Figure 3C) or hnRNPK with PTB isoforms (data not shown). To confirm that the stimulation of protein synthesis from the firefly cistron was due to enhanced IRES activity, RNA transcripts were derived which contain an ‘A’ cap rather than a ‘G’ cap and these were used to prime rabbit reticulocyte lysates. The data show that, as expected, there is a reduction in the synthesis of Renilla luciferase from these transcripts but no effect on IRES-mediated firefly luciferase expression (Figure 3Cii). Moreover, addition of PTB or nPTB to the in vitro translation reaction that contained the ‘A’ cap mRNAs caused the same degree of stimulation of IRES-activity as before (Figure 3Cii).
Identification of the minimal active element
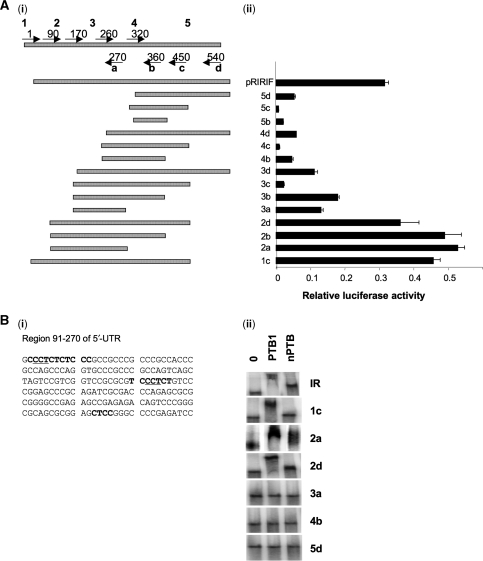
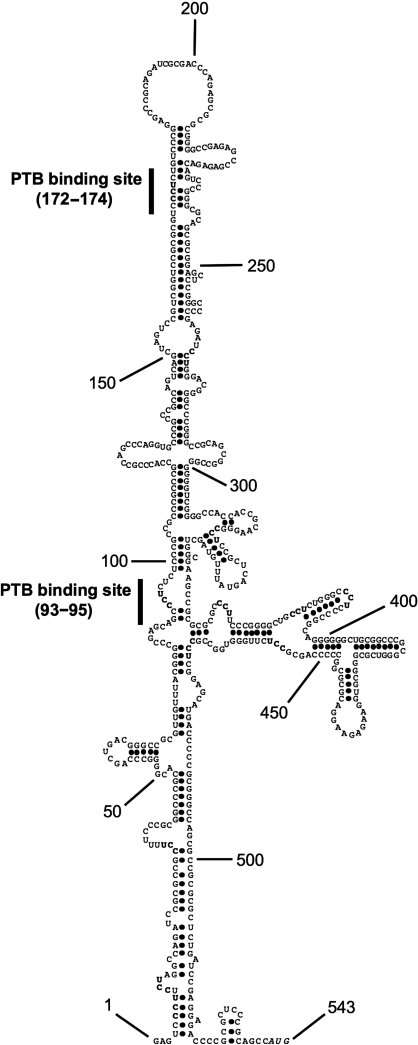
It has been shown previously that PTB interacts with cellular IRES RNA in polypyrimidine rich regions of RNA that contain one or more repeats of (CCU)n (4). Deletion mapping was performed to identify the minimal active element of the IR-IRES to locate any such motifs. Primers were designed to various regions of the IR 5′-UTR and a series of fragments was generated. For example, using primer 1 as the forward primer and primer a, as the reverse primer, the fragment 1a was generated (Figure 4). These fragments were then sub-cloned into the dicistronic vector pRF, and the corresponding set of constructs were transfected into HeLa cells and luciferase activity was determined (Figure 4A). Using this approach the minimal active element was found to reside in fragment 2a which spans a 180-nt region from 90 to 270 in the IR 5′-UTR (451–271 nt upstream of the translation start site) (Figure 4A). It is likely therefore that elements that are required for IRES function are present in this region of RNA and indeed there are two CCU motifs within polypyrimdine rich stretches of RNA are located at 93–95 and 172–174 (Figures 1 and 4Bi).
Figure 4.
The minimal active IR-IRES element contains two CCU motifs. (Ai) DNA fragments from insulin receptor were amplified by PCR using oligonucleotides at the position shown and inserted into the dicistronic construct pRF at the EcoRI and NcoI sites. (Aii) Dicistronic constructs containing the full-length 5′-UTR sequence or fragments were transfected into HeLa cells. All constructs were co-transfected with pcDNA3.1/HisB/lacZ. The ratio of Renilla luciferase and firefly luciferase activities were determined and normalized to β-galactosidase activities. (Bi) Potential PTB binding regions within the minimal active element (91–270) are indicated by underlining. (ii) EMSAs were performed using radiolabelled fragments of IR-IRES RNA incubated with 0.2 µg of PTB1 or nPTB. Products were separated on a 5% TBE polyacrylamide gel.
To characterize further the elements that bind PTB, EMSAs were performed using radiolabelled RNAs that spanned the 5′-UTR. Fragments 1c, 2a and 2d, containing both the 93–95 and 172–174 CCU motifs, interacted with PTB1 and nPTB as indicated by the retarded RNA species observed in each case, whilst fragments 3a, 4b and 5c did not interact with PTB or nPTB (Figure 4Bii). Fragments 4b and 5c lack both the 93–95 and 172–174 CCU motifs while fragment 3a, comprised of a 100-nt region from 170–270, does contain the second CCU motif (172–174), albeit at its extreme 5′-end.
To obtain further insight into the IR-IRES a structural model of the 5′-UTR was generated. RNAse V1 (which cleaves double-stranded RNA) and chemical modifying agents (DMS, kethoxal) were used to probe the RNA as described previously (29), and the data obtained (Supplementary Data 1) were used to constrain the Mfold algorithm. The resulting model (Figure 5) has extensive secondary structure, within which the proposed PTB binding sites at 93–95 and 172–174 are both present in stems that contain bulges. The model predicts that the structural context of the PTB binding site at 172–174 would likely be preserved in the minimal active fragment 2a, although the context of the site at 93–95 would be severely compromised.
Figure 5.
Predicted secondary structure of IR 5′-UTR. The IR-IRES RNA was probed with chemicals and enzymes (see Supplementary 1) and the data were used to constrain secondary structure predictions generated using the web implementation of the Mfold algorithm incorporating version 3.0 of the Turner rules. The positions of the two PTB binding sites (CCU motifs) are marked.
The IR-IRES activity is increased by cell density
Activity of cellular IRESs varies between cell lines presumably reflecting differences in the expression of IRES-trans-acting factors (3,29). Since the IR-IRES was shown to be activated in vitro by PTB isoforms and particularly by nPTB the IR-IRES function in a number of cell lines was determined particularly those of neuronal origin, which have been shown previously to express high levels of nPTB including SY5Y, N2a, NIE115, NB2a (29). These cell lines were transfected with pRIRF and luciferase activity determined (Figure 6A). The IR-IRES was found to be active in a range of cell lines but especially so in certain neuronal cell lines, e.g. in SY5Y cells the IR-IRES is twice as active as in HeLa cells (Figure 6A) and is similar in this regard to both the N-myc and Apaf-1 IRESs that also require nPTB for function (13,33). The exception to this was the cell line B50, originally derived from rat hippocampal neurons, in which IR-IRES activity was low. The reason for low IR-IRES function in these cells is not clear but could be due to species differences or differential expression of additional ITAFs that are required for IR-IRES activity.
Figure 6.
The activity of the IR-IRES is dependent upon cell type and cell density. (A) The cell lines shown were transfected with IR-IRES and luciferase activity was determined. IRES activity is expressed using the ratio of the downstream cistron expression to upstream cistron with any differences in transfection efficiencies corrected for using the β-galactosidase transfection control. All experiments were performed in triplicate on three independent occasions. (B) To determine whether differences in cell density affect IR-IRES function HeLa (i), B50 (ii), SY5Y (iii) and NIE115 (iv) were transfected with the IR IRES at the cell densities shown. IRES activity is expressed using the ratio of the downstream cistron expression to upstream cistron relative to the β-galactosidase transfection control. There was no change in Renilla luciferase activity in sub-confluent cells. All experiments were performed in triplicate on three independent occasions.
There was a considerable variation in IR-IRES mediated translation between experiments and the IRES appeared to be more active in confluent cultures. To investigate this phenomenon further HeLa, B50, SY5Y and NIE115 cells were plated at a range of densities and were transfected with the IR-IRES (Figure 6B). Activity of the IR-IRES increased with cell density in all cases, the greatest effect being observed in SY5Y cells with an eight fold difference in the amount of firefly luciferase produced in confluent cells when compared to sub-confluent cells (Figure 6Biii). There was no change in Renilla luciferase activity in sub-confluent cells.
We also investigated whether IRES activity was altered during differentiation of murine 3T3-L1 fibroblasts into adipocytes, as it is known that IR expression increases dramatically during differentiation (36). Confluent fibroblasts transfected with the dicistronic reporter were stimulated to differentiate by adding a cocktail of insulin, isobutylmethylxanthine and dexamethasone. Although there were some changes in control (Renilla) luciferase expression during the course of differentiation, IRES activity as indicated by the ratio of firefly to Renilla activity did not change (data not shown).
The IR-IRES is stimulated by insulin
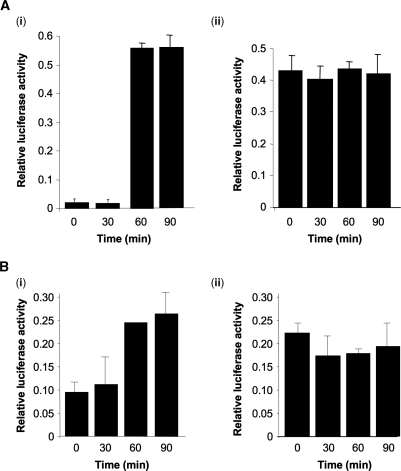
We have shown previously that increased cell contact of B-cells stimulates signalling through the mTOR pathway and the translational up-regulation of c-myc (37). The role of insulin in signalling and cell proliferation is well established and its effects on translation are mediated through the mTOR pathway (38). Therefore experiments were performed to determine whether it was possible to mimic the cell density effect by the addition of insulin to cultured cells (Figure 7). SY5Y and HeLa cells were seeded at either low or high density, transfected with the IR-IRES-containing construct and then treated with insulin and luciferase activities determined at the times shown. There was a rapid induction in the amount of luciferase produced in all cases in the sub-confluent cells to a level that was the equivalent that observed in confluent cells (Figure 7Ai, Bi and Ci) yet there was no stimulation of firefly luciferase production from IR-IRES in confluent cultures (Figure 7Aii, Bii, Cii). This effect was specific to the IR-IRES and no stimulation by insulin was observed for the IRESs found in c-myc, Apaf-1 or BAG-1 (data not shown). The relative firefly luciferase activity stimulated by insulin did not reach the levels observed in the cells grown at 120 000 per well suggesting that additional pathways may be involved in this response.
Figure 7.
The IR-IRES is stimulated by insulin in subconfluent cells. SY5Y (A) or HeLa (B) cells were seeded at a density of either 3250 cells per well (Ai and Bi) or 6000 cells per well (Aii and Bii). Cells transfected with the IR-IRES-containing construct were treated with 100 nM insulin and luciferase activities determined at the times shown. Luciferase activity is expressed as the ratio of Firefly/Renilla luciferase relative to the transfection control β-galactosidase. There was no change in Renilla luciferase activity in insulin treated cells. All experiments were performed on at least four independent occasions.
In confluent, differentiated 3T3-L1 adipocytes, IRES activity indicated by the ratio firefly/Renilla luciferase was not significantly affected by incubation with insulin (100 nM), phorbol myristoyl acetate (1 µM) or fetal bovine serum (10%) for 1h or 4 h (data not shown).
DISCUSSION
We have shown that the 5′-UTR of mRNA encoding human IR contains an IRES, which allows protein synthesis to be initiated by cap-independent mechanisms (Figure 2). The 5′-UTR of IGF-1R mRNA also contain an IRES (18,39), as does the Drosophila insulin/IGF-like receptor (dINR) mRNA (22). Despite a high degree of sequence similarity between the IR and IGF-1R coding regions, there is no discernable sequence similarity between the respective 5′-UTRs, although for both receptors the 5′-UTRs are well conserved between species (∼70% identity, human vs rat/mouse). The human IR 5′-UTR is ∼540 nt in length, and deletion analysis indicated that a region 90–270 from the transcription start site (or 451–271 nt upstream of the translation start site) contained the minimal active IRES element (Figure 4A). The IGF-1R mRNA has an even longer 5′-UTR (1038 nt in human, 943 nt in rat) and has the potential to form stable secondary structure which is inhibitory to scanning by the 43S ribosomal complex (18,40,41). The IGF-1R IRES has been localized to a region of 85 nt immediately upstream of the translation start site and downstream of the elements that are proposed to be inhibitory to cap-dependent ribosomal scanning (39). The 3′-end of the 5′-UTR is highly pyrimidine-rich and predicted to form a stem loop structure (18).
Like many cellular IRESs the IR-IRES requires PTB for activity in vivo and in vitro (Figure 3). Within cellular IRESs PTB interacts with polypyrimidine rich sequences that contain the motif CCU (4). The hIR 5′-UTR contains several such motifs, of which those at 6, 30, 93, 172, 387 and 456 relative to the transcription start site (−535, −511, −448, −369, −154 and −85, respectively relative to the translation start site) are conserved in the rat (Figure 1). Moreover, two of these CCU motifs lie within the minimal IRES-containing region (90–270) defined by deletion mapping, which was also shown by EMSA to bind PTB (Figure 4). A model of the structure of the IR 5′-UTR, derived by constraining the Mfold algorithm with experimental data identifying double- and single-stranded regions, predicts that the CCU motifs at 93 and 172 are both situated in bulges within stem structures (Figure 5) and that the structural context of the motif at 172 would be preserved in the minimal active IRES fragment. Thus, it is highly likely that binding of PTB at one or both of the CCU motifs located at 93–95 and 172–174 (448–446 and 369–367 nt upstream of translation start site) is critical for IRES function.
The IGF-1R IRES also binds PTB (18) and several other proteins, including the ELAV RNA stability factor HuR, which is inhibitory to both cap-dependent scanning and especially IRES-mediated translation, and hnRNP C which competes with HuR for binding and enhances IRES-mediated translation (39,41,42). It remains to be seen whether these or other proteins also bind to the IR IRES, although the differences in location and sequence of the IR and IGF-1R IRESs are strongly suggestive of different mechanisms of regulation.
The IR-IRES is similar to IRESs in the Apaf-1 and N-myc 5′-UTRs in that it is activated in vitro by nPTB (Figure 4). Moreover, it is highly active in cell lines of neuronal origin (Figure 6) which we have shown previously to express high levels of nPTB (29). Both IR and IGF-1R are expressed in the nervous system and have been proposed to function both during neuronal development and in the adult brain (43–45). Indeed, the IR is widely expressed throughout the adult brain (43), with highest levels in the hypothalamus, olfactory bulb and cerebellum, and a specific receptor-mediated transport system exists for insulin at the blood brain barrier (46). Numerous functions have been proposed for neuronal insulin receptors, including roles in neurogenesis (47), learning and memory (48), maintenance of energy balance and reproductive function (49,50), and both protection from and promotion of age-related neurodegenerative diseases (49,51,52). It would not be surprising therefore if sophisticated mechanisms exist to regulate the expression of brain IR in response to diverse stimuli.
It is known that in both fully differentiated tissues, such as the brain, and in cells grown to high confluency there is a reduction in cap-dependent ribosomal scanning of mRNAs due to changes in the levels and/or phosphorylation states of the eIFs (53). IRESs can potentially be used to maintain expression of key proteins in terminally differentiated tissues. For instance, it has been shown that the FGF-2-IRES (but not the c-myc-IRES) functions in adult mouse brain (54). It is possible therefore that the IR-IRES is active in certain (but not all) cells arrested in G0 and in some terminally differentiated tissues, such as adult brain, to maintain expression of the IR and/or to allow regulation of that expression in response to external stimuli.
We found that the activity of the IR-IRES was dependent not only on cell type but also on cell density. In common with the PKCδ IRES (55) the IR-IRES was most active at high cell density (Figure 5B), when levels of total protein synthesis will be reduced. However, cellular IRESs found in the 5′-UTRs of Apaf-1, c-myc or Bag-1 are not sensitive to changes in cell density (unpublished data). Insulin itself stimulated IRES activity in sub-confluent but not in confluent cells (Figure 7). In contrast, in Drosophila it was proposed that cap-dependent translation is used for dINR synthesis when nutrients and insulin-like peptides are available, and that the dINR IRES is used to drive increased receptor synthesis under conditions of low availability of nutrients and insulin-like peptides, when transcription of the dINR gene is enhanced while cap-dependent translation is inhibited by dephosphorylation of d4E-BP (22). Interestingly, nutrient and growth factor deprivation of cultured mouse muscle cells was similarly accompanied by a 6- to 8-fold increase in IR protein and 2-fold up-regulation of IR mRNA, suggestive of an analogous FOXO-mediated coupled transcription/translation response (26).
There is little published information describing regulation of expression of mammalian IR at the level of mRNA translation. The human IR mRNA is one of only a very small number of transcripts that remain associated with polysomes when cap-dependent translation is inhibited by poliovirus infection (6), consistent with IRES-dependent translation initiation under these conditions. However, studies of factors determining IR expression in different mammalian tissues and under different patho-physiological conditions have focused on mRNA transcription and protein degradation as regulatory mechanisms. It is possible that the IR-IRES simply functions to maintain expression of IR under conditions where cap-dependent initiation is compromised, including perhaps in some differentiated cells. However, in the case of the IGF-1R there is evidence that regulation of expression does occur at a translational level. The IGF-1R was among a number of transcripts shown by cDNA array screening of polysome-associated mRNAs to be translationally regulated during antigenic stimulation of human T-lymphocytes, although underlying mechanisms were not investigated (56). Recently, it was shown that binding of inhibitory HuR and stimulatory hnRNP C to the IGF-1R IRES was influenced by changes in cellular environment or proliferative status, and it was suggested that dysregulation of the IGF-1R-IRES through changes in expression of RNA-binding translational-regulatory proteins could contribute to over-expression of IGF-1R in some tumours (39). It will be interesting now to investigate whether the IR-IRES is similarly capable of dynamic regulation, and whether this influences the expression of IR protein under different patho-physiological conditions, especially in neuronal tissues.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
UK Biotechnology and Biological Sciences Research Council (to L.C.C., M.D., A.B., A.E.W., K.S.); Diabetes UK (R.D. Lawrence Fellowship to S.H.R.); European Union COST Action BM0602. Funding for open access charge: Biotechnology and Biological Sciences Research Council grant.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Pain VM. Initiation of protein synthesis in eukaryotic cells. Eur. J. Biochem. 1996;236:747–771. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- 2.Komar AA, Hatzoglou M. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J. Biol. Chem. 2005;280:23425–23428. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- 3.Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 4.Mitchell SA, Spriggs KA, Bushell M, Evans JR, Stoneley M, Le Quesne JP, Spriggs RV, Willis AE. Identification of a motif that mediates polypyrimidine tract-binding protein-dependent internal ribosome entry. Genes Dev. 2005;19:1556–1571. doi: 10.1101/gad.339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sawicka K, Bushell M, Spriggs KA, Willis AE. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem. Soc. Trans. 2008;36:641–647. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- 6.Johannes G, Carter MS, Eisen MB, Brown PO, Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl Acad. Sci. USA. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoneley M, Chappell SA, Jopling CL, Dickens M, MacFarlane M, Willis AE. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol. Cell Biol. 2000;20:1162–1169. doi: 10.1128/mcb.20.4.1162-1169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin X, Sarnow P. Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J. Biol. Chem. 2004;279:13721–13728. doi: 10.1074/jbc.M312854200. [DOI] [PubMed] [Google Scholar]
- 9.Subkhankulova T, Mitchell SA, Willis AE. Internal ribosome entry segment-mediated initiation of c-Myc protein synthesis following genotoxic stress. Biochem. J. 2001;359:183–192. doi: 10.1042/0264-6021:3590183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coldwell MJ, deSchoolmeester ML, Fraser GA, Pickering BM, Packham G, Willis AE. The p36 isoform of BAG-1 is translated by internal ribosome entry following heat shock. Oncogene. 2001;20:4095–4100. doi: 10.1038/sj.onc.1204547. [DOI] [PubMed] [Google Scholar]
- 11.Spriggs KA, Stoneley M, Bushell M, Willis AE. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol. Cell. 2008;100:27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- 12.Jopling CL, Spriggs KA, Mitchell SA, Stoneley M, Willis AE. L-Myc protein synthesis is initiated by internal ribosome entry. RNA. 2004;10:287–298. doi: 10.1261/rna.5138804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jopling CL, Willis AE. N-myc translation is initiated via an internal ribosome entry segment that displays enhanced activity in neuronal cells. Oncogene. 2001;20:2664–2670. doi: 10.1038/sj.onc.1204404. [DOI] [PubMed] [Google Scholar]
- 14.Nanbru C, Lafon I, Audigier S, Gensac MC, Vagner S, Huez G, Prats AC. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J. Biol. Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- 15.Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 16.Cobbold LC, Spriggs KA, Haines SJ, Dobbyn HC, Hayes C, de Moor CH, Lilley KS, Bushell M, Willis AE. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol. Cell Biol. 2008;28:40–49. doi: 10.1128/MCB.01298-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Velden AW, Thomas AA. The role of the 5′ untranslated region of an mRNA in translation regulation during development. Int. J. Biochem. Cell Biol. 1999;31:87–106. doi: 10.1016/s1357-2725(98)00134-4. [DOI] [PubMed] [Google Scholar]
- 18.Giraud S, Greco A, Brink M, Diaz JJ, Delafontaine P. Translation initiation of the insulin-like growth factor I receptor mRNA is mediated by an internal ribosome entry site. J. Biol. Chem. 2001;276:5668–5675. doi: 10.1074/jbc.M005928200. [DOI] [PubMed] [Google Scholar]
- 19.Martin MM, Garcia JA, McFarland JD, Duffy AA, Gregson JP, Elton TS. Translation of the human angiotensin II type 1 receptor mRNA is mediated by a highly efficient internal ribosome entry site. Mol. Cell Endocrinol. 2003;212:51–61. doi: 10.1016/j.mce.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 20.Rabadan-Diehl C, Volpi S, Nikodemova M, Aguilera G. Translational regulation of the vasopressin v1b receptor involves an internal ribosome entry site. Mol. Endocrinol. 2003;17:1959–1971. doi: 10.1210/me.2001-0336. [DOI] [PubMed] [Google Scholar]
- 21.Dobson T, Minic A, Nielsen K, Amiott E, Krushel L. Internal initiation of translation of the TrkB mRNA is mediated by multiple regions within the 5′ leader. Nucleic Acids Res. 2005;33:2929–2941. doi: 10.1093/nar/gki605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marr MT, II, D'Alessio JA, Puig O, Tjian R. IRES-mediated functional coupling of transcription and translation amplifies insulin receptor feedback. Genes Dev. 2007;21:175–183. doi: 10.1101/gad.1506407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakae J, Kido Y, Accili D. Distinct and overlapping functions of insulin and IGF-I receptors. Endocr. Rev. 2001;22:818–835. doi: 10.1210/edrv.22.6.0452. [DOI] [PubMed] [Google Scholar]
- 24.Mamula PW, McDonald AR, Brunetti A, Okabayashi Y, Wong KY, Maddux BA, Logsdon C, Goldfine ID. Regulating insulin-receptor-gene expression by differentiation and hormones. Diabetes Care. 1990;13:288–301. doi: 10.2337/diacare.13.3.288. [DOI] [PubMed] [Google Scholar]
- 25.McKeon C. Transcriptional regulation of the insulin receptor gene promoter. Adv. Exp. Med. Biol. 1993;343:79–89. doi: 10.1007/978-1-4615-2988-0_9. [DOI] [PubMed] [Google Scholar]
- 26.Puig O, Tjian R. Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev. 2005;19:2435–2446. doi: 10.1101/gad.1340505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knutson VP. Cellular trafficking and processing of the insulin receptor. FASEB J. 1991;5:2130–2138. doi: 10.1096/fasebj.5.8.2022311. [DOI] [PubMed] [Google Scholar]
- 28.Tewari DS, Cook DM, Taub R. Characterization of the promoter region and 3′ end of the human insulin receptor gene. J. Biol. Chem. 1989;264:16238–16245. [PubMed] [Google Scholar]
- 29.Mitchell SA, Spriggs KA, Coldwell MJ, Jackson RJ, Willis AE. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell. 2003;11:757–771. doi: 10.1016/s1097-2765(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 30.Urso B, Cope DL, Kalloo-Hosein HE, Hayward AC, Whitehead JP, O'Rahilly S, Siddle K. Differences in signaling properties of the cytoplasmic domains of the insulin receptor and insulin-like growth factor receptor in 3T3-L1 adipocytes. J. Biol. Chem. 1999;274:30864–30873. doi: 10.1074/jbc.274.43.30864. [DOI] [PubMed] [Google Scholar]
- 31.Jordan M, Schallhorn A, Wurm FM. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 1996;24:596–601. doi: 10.1093/nar/24.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner EJ, Garcia-Blanco MA. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell. 2002;10:943–949. doi: 10.1016/s1097-2765(02)00645-7. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell SA, Brown EC, Coldwell MJ, Jackson RJ, Willis AE. Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol. Cell Biol. 2001;21:3364–3374. doi: 10.1128/MCB.21.10.3364-3374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobbyn HC, Hill K, Hamilton TL, Spriggs KA, Pickering BM, Coldwell MJ, de Moor CH, Bushell M, Willis AE. Regulation of BAG-1 IRES-mediated translation following chemotoxic stress. Oncogene. 2008;27:1167–1174. doi: 10.1038/sj.onc.1210723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoneley M, Subkhankulova T, Le Quesne JP, Coldwell MJ, Jopling CL, Belsham GJ, Willis AE. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 2000;28:687–694. doi: 10.1093/nar/28.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Modan-Moses D, Janicot M, McLenithan JC, Lane MD, Casella SJ. Expression and function of insulin/insulin-like growth factor I hybrid receptors during differentiation of 3T3-L1 preadipocytes. Biochem. J. 1998;333:825–831. doi: 10.1042/bj3330825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.West MJ, Stoneley M, Willis AE. Translational induction of the c-myc oncogene via activation of the FRAP/TOR signalling pathway. Oncogene. 1998;17:769–780. doi: 10.1038/sj.onc.1201990. [DOI] [PubMed] [Google Scholar]
- 38.Avruch J, Hara K, Lin Y, Liu M, Long X, Ortiz-Vega S, Yonezawa K. Insulin and amino-acid regulation of mTOR signaling and kinase activity through the Rheb GTPase. Oncogene. 2006;25:6361–6372. doi: 10.1038/sj.onc.1209882. [DOI] [PubMed] [Google Scholar]
- 39.Meng Z, Jackson NL, Choi H, King PH, Emanuel PD, Blume SW. Alterations in RNA-binding activities of IRES-regulatory proteins as a mechanism for physiological variability and pathological dysregulation of IGF-IR translational control in human breast tumor cells. J. Cell Physiol. 2008;217:172–183. doi: 10.1002/jcp.21486. [DOI] [PubMed] [Google Scholar]
- 40.Cooke DW, Casella SJ. The 5′-untranslated region of the IGF-I receptor gene modulates reporter gene expression by both pre- and post-transcriptional mechanisms. Mol. Cell Endocrinol. 1994;101:77–84. doi: 10.1016/0303-7207(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 41.Meng Z, Snyder RC, Shrestha K, Miller DM, Emanuel PD, Blume SW. Evidence for differential ribonucleoprotein complex assembly in vitro on the 5′-untranslated region of the human IGF-IR transcript. Mol. Cell Endocrinol. 2003;200:127–140. doi: 10.1016/s0303-7207(02)00381-7. [DOI] [PubMed] [Google Scholar]
- 42.Meng Z, King PH, Nabors LB, Jackson NL, Chen CY, Emanuel PD, Blume SW. The ELAV RNA-stability factor HuR binds the 5′-untranslated region of the human IGF-IR transcript and differentially represses cap-dependent and IRES-mediated translation. Nucleic Acids Res. 2005;33:2962–2979. doi: 10.1093/nar/gki603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adamo M, Raizada MK, LeRoith D. Insulin and insulin-like growth factor receptors in the nervous system. Mol. Neurobiol. 1989;3:71–100. doi: 10.1007/BF02935589. [DOI] [PubMed] [Google Scholar]
- 44.D'Ercole AJ, Ye P. Expanding the mind: insulin-like growth factor I and brain development. Endocrinology. 2008;149:5958–5962. doi: 10.1210/en.2008-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kitamura T, Kahn CR, Accili D. Insulin receptor knockout mice. Annu. Rev. Physiol. 2003;65:313–332. doi: 10.1146/annurev.physiol.65.092101.142540. [DOI] [PubMed] [Google Scholar]
- 46.Woods SC, Seeley RJ, Baskin DG, Schwartz MW. Insulin and the blood-brain barrier. Curr. Pharm. Des. 2003;9:795–800. doi: 10.2174/1381612033455323. [DOI] [PubMed] [Google Scholar]
- 47.Bateman JM, McNeill H. Insulin/IGF signalling in neurogenesis. Cell Mol. Life Sci. 2006;63:1701–1705. doi: 10.1007/s00018-006-6036-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Heide LP, Ramakers GM, Smidt MP. Insulin signaling in the central nervous system: learning to survive. Prog. Neurobiol. 2006;79:205–221. doi: 10.1016/j.pneurobio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 49.Plum L, Schubert M, Bruning JC. The role of insulin receptor signaling in the brain. Trends Endocrinol. Metab. 2005;16:59–65. doi: 10.1016/j.tem.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 50.Porte D, Jr, Baskin DG, Schwartz MW. Insulin signaling in the central nervous system: a critical role in metabolic homeostasis and disease from C. elegans to humans. Diabetes. 2005;54:1264–1276. doi: 10.2337/diabetes.54.5.1264. [DOI] [PubMed] [Google Scholar]
- 51.de la Monte SM, Wands JR. Review of insulin and insulin-like growth factor expression, signaling, and malfunction in the central nervous system: relevance to Alzheimer's disease. J. Alzheimers Dis. 2005;7:45–61. doi: 10.3233/jad-2005-7106. [DOI] [PubMed] [Google Scholar]
- 52.Taguchi A, White MF. Insulin-like signaling, nutrient homeostasis, and life span. Annu. Rev. Physiol. 2008;70:191–212. doi: 10.1146/annurev.physiol.70.113006.100533. [DOI] [PubMed] [Google Scholar]
- 53.Gerlitz G, Jagus R, Elroy-Stein O. Phosphorylation of initiation factor-2 alpha is required for activation of internal translation initiation during cell differentiation. Eur. J. Biochem. 2002;269:2810–2819. doi: 10.1046/j.1432-1033.2002.02974.x. [DOI] [PubMed] [Google Scholar]
- 54.Creancier L, Morello D, Mercier P, Prats AC. Fibroblast growth factor 2 internal ribosome entry site (IRES) activity ex vivo and in transgenic mice reveals a stringent tissue-specific regulation. J. Cell Biol. 2000;150:275–281. doi: 10.1083/jcb.150.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morrish BC, Rumsby MG. The 5′ untranslated region of protein kinase Cdelta directs translation by an internal ribosome entry segment that is most active in densely growing cells and during apoptosis. Mol. Cell Biol. 2002;22:6089–6099. doi: 10.1128/MCB.22.17.6089-6099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mikulits W, Pradet-Balade B, Habermann B, Beug H, Garcia-Sanz JA, Mullner EW. Isolation of translationally controlled mRNAs by differential screening. FASEB J. 2000;14:1641–1652. doi: 10.1096/fj.14.11.1641. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.