Abstract
Although the basic components and mechanisms of mitochondrial transcription in mammals have been described, the components involved in mRNA processing, translation and stability remain largely unknown. In plants, pentatricopeptide domain RNA-binding proteins regulate the stability, expression and translation of mitochondrial transcripts; therefore, we investigated the role of an uncharacterized mammalian pentatricopeptide domain protein, (PTCD1), in mitochondrial RNA metabolism. We show that PTCD1 is a mitochondrial matrix protein which associates with leucine tRNAs and precursor RNAs that contain leucine tRNAs. Knockdown of PTCD1 in 143B osteosarcoma cells did not change mitochondrial mRNA levels; however, it increased the abundance precursor RNAs and of leucine tRNAs and PTCD1 overexpression led to a reduction of these RNAs. Lowering PTCD1 in cells increased levels of several mitochondria-encoded proteins and Complex IV activity, suggesting that PTCD1 acts as a negative regulator of leucine tRNA levels and hence mitochondrial translation.
INTRODUCTION
Human mitochondrial DNA (mtDNA) is a circular double-stranded genome that encodes 22 tRNAs, 2 rRNAs and 13 protein subunits of the electron transport chain complexes (1). Therefore, human mtDNA is dependent on nuclear-encoded proteins for replication, repair, transcription and translation. This dependency lies at the heart of several human diseases that are caused by defects of mtDNA maintenance and expression (2). Human mitochondrial diseases can encompass mutations of both mtDNA and nuclear DNA (3). However, the identification of mtDNA or OXPHOS abnormalities in Parkinson's disease, Alzheimer's disease, cancer and diabetes indicate that our understanding of mitochondrial function in disease is still in its infancy (4). The most significant gaps in our knowledge of mitochondrial function and disease are in the regulation of mitochondrial gene expression (5,6). This is particularly important since remarkable tissue-, cell- and disease-specific variations have been observed in the expression of different mitochondrial RNAs, but cannot be explained at present (7–9). Mitochondrial RNAs are transcribed as part of long polycistronic, precursor transcripts that generally encompass the entire mtDNA and are processed by removing interspersed tRNAs to release the individual mRNAs and rRNAs (10). Therefore, large variations in ratios of the 13 levels of individual mRNAs and their proteins must be controlled at a post-transcriptional level. Mitochondrial transcripts are subsequently polyadenylated at their 3′ ends to yield the mature mRNAs, which are translated on mitochondrial ribosomes, while tRNAs and rRNAs are subject to specific nucleotide modifications and CCA is added to the 3′ end of tRNAs (11,12). Little is known about how the levels of specific RNAs are regulated in mammalian mitochondria.
The best-characterized RNA-binding protein known to regulate mammalian mitochondrial gene expression is the leucine-rich pentatricopeptide repeat cassette (LRPPRC) protein (13–15). This was recently identified as a gene causing the rare French–Canadian variant of Leigh syndrome, a debilitating neurodegenerative condition resulting from mitochondrial cytochrome c oxidase deficiency (15). Mutation of the LRPPRC gene results in decreased levels of the cytochrome oxidase (COX) I and III mRNAs (14); however, the RNA sequence that this protein binds has not been identified. LRPPRC is one of the seven mammalian mitochondrial PPR domain proteins, along with the mitochondrial RNA polymerase (POLRMT), mitochondrial ribosomal protein of the small subunit 27 (MRPS27), pentatricopeptide repeat domain proteins (PTCD) 1, 2 and 3 and mitochondrial RNase P protein 3 (MRPP3), which is a recently identified subunit of the mammalian mitochondrial RNase P complex (16,17). PPR domain proteins were identified first in plants, where they constitute a large family of mitochondrial and chloroplast proteins involved in transcript processing, editing and translation (18). Recently, the PPR domain protein PTCD2 was shown to regulate cytochrome b RNA processing in mice (19), and we found that PTCD3 associates with the mitochondrial small ribosomal subunit and is important for protein synthesis (20). The functions of the remaining PPR domain proteins in mammalian mitochondria have not been elucidated to date.
Here we have investigated the role of the previously uncharacterized mammalian PTCD1 and show that it associates with and regulates the level of leucine tRNAs in mitochondria. We show that by decreasing PTCD1 we increase leucine tRNA levels in cells and consequently observe an increased expression of mitochondrial-encoded Complex I and IV subunits and increased Complex IV activity.
MATERIALS AND METHODS
Plasmid expression vectors
All expression vectors were based on pcDNA3 (Invitrogen). Full-length human PTCD1 (NCBI accession number NP_056360) was expressed with its native termination codon or fused to a tandem affinity purification tag (TAP, ABO76910) (21) or EYFP (BD Biosciences) at the C-terminus. All plasmids were tested for expression by transfection and immunoblotting.
Cell culture
143B osteosarcoma cells were cultured at 37°C under humidified 95% air/5% CO2 in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) containing glucose (4.5 g l–1), 1 mM pyruvate, 2 mM glutamine, penicillin (100 U ml–1), streptomycin sulphate (100 µg ml–1) and 10% fetal bovine serum (FBS). Rho0 143B cybrids were a gift from Dr Elizabeth Ledgerwood, University of Otago, New Zealand and MELAS A3243G 143B cybrids were a gift from Prof. Rudolf Wiesner, University of Köln, Germany, with permission from Dr Eric Shoubridge. These were grown in the same medium supplemented with uridine (50 mg l–1).
Mitochondrial isolation and subfractionation
Mitochondria were prepared from 107 cells grown overnight in 15 cm2 dishes and isolated as described before (22). Rat liver mitochondria were prepared by homogenization in 250 mM sucrose, 5 mM Tris–HCl pH 7.4 and 1 mM EGTA (STE) followed by differential centrifugation (23) and the protein concentration determined by the biuret assay using BSA as a standard (24). Rat liver mitochondria were washed and suspended in STE containing 0.1% (w/v) fat-free BSA (STE/BSA) instead of STE for the final spin. The final pellet was suspended in a minimal amount of STE/BSA and the protein concentration was measured by the biuret assay. The mitochondria were diluted with STE/BSA to a final concentration of 100 mg ml–1 and stirred on ice. One millilitre of digitonin (12.5 mg ml–1) was added dropwise over 2 min and the solution was kept on ice for a further 13 min. Three volumes of STE/BSA were added and the mixture was homogenized four times with a tight homogenizer. The mitoplasts were collected by centrifugation for 10 min at 10 000g and the concentration was measured using the biuret assay. The supernatant was kept and centrifuged at 144 000g for 20 min at 5°C to separate the intermembrane fraction (supernatant) from the outer membrane (pellet). The mitoplasts were suspended at 30 mg ml–1 in STE containing 0.16 mg lubrol mg–1 mitoplast protein and incubated on ice for 20 min. This solution was diluted with three volumes of STE and centrifuged at 144 000g for 50 min at 5°C to separate the matrix (supernatant) from the inner membrane (pellet). Both membrane fractions were suspended in 1% lauryl maltoside in STE. The protein concentration of each fraction was determined using the BCA assay (25) and immunoblotting was carried out to confirm the fractionation.
Transfections
143B cells were plated at 60% confluence in six-well plates or 10-cm dishes and transfected with annealed siRNAs or mammalian expression plasmids in OptiMEM media (Invitrogen). A total of 125 nM (for six-well plates) or 145 nM (for 10-cm dishes) of PTCD1 or control, off-target siRNAs (Dharmacon) were transfected using Lipofectamine 2000 (Invitrogen). A total of 158 ng/cm2 of PTCD1 or control EYFP plasmid DNA was transfected using Fugene HD (Roche). Cell incubations were carried out for up to 72 h following transfection. PTCD1 expression was detected by immunoblotting using a polyclonal PTCD1 antibody.
Fluorescence cell microscopy
For immunocytochemistry, 143B cells were plated onto 13-mm diameter glass coverslips and allowed to attach overnight. Non-transfected cells were fixed with 4% paraformaldehyde in Tris buffered saline (TBS) for 30 min, washed with TBS and incubated with 10% FCS/0.1% Triton X-100/TBS (TBST) for 10 min. The PTCD1 antibody (1 : 500) diluted in TBST was added and incubated for 3 h at room temperature. After washing with TBS (3 × 5 min) the cells were incubated with anti-rabbit IgG Oregon green secondary antibody (diluted 1 : 100 in TBS) (Molecular Probes) for 15 min in the dark. Cells were then washed in TBS. Alternatively, cells were transfected with pPTCD1-EYFP for 48 h, and at the end of the incubation treated with 100 nM Mitotracker Orange for 15 min, then fixed with 4% paraformaldehyde in TBS for 30 min and washed with TBS. Cells were mounted in DABCO/PVA medium. Images were acquired using an Olympus DP70 fluorescent inverted microscope using a Nikon 40× objective.
Affinity purification and RNA isolation
143B cells were lysed in 50 mM Tris/HCl (pH 7.5), 125 mM NaCl, 5% glycerol, 1% Igepal CA-630, 1.5 mM MgCl2, 1 mM DTT, 25 mM NaF, 1 mM Na3VO4, 1 mM EDTA, 1× complete protease inhibitors (Roche), 200 U ml–1 RNaseOUT (Invitrogen) at 4°C. The lysate was cleared by centrifugation and incubated with rabbit-IgG agarose (Sigma) at 4°C for 2 h. The agarose was washed with lysis buffer and then with 10 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 1% Igepal CA-630, 200 U.ml–1 RNaseOUT and protein was eluted by addition of 10 U of AcTEV protease (Invitrogen, RT for 1 h). RNA was isolated using the miRNeasy Mini kit (Qiagen) incorporating an on-column RNase-free DNase digestion to remove all DNA. For slot blotting, RNA was isolated from 5 × 107 143B cells expressing PTCD1-TAP or EYFP-TAP by affinity purification as described above. The entire sample of purified RNA was applied to a Hybond-N+ nitrocellulose membrane using a Bio-Dot SF microfiltration apparatus (BioRad) and probed as described for the northern blots.
Quantitative RT–PCR
The transcript abundance of mitochondrial genes and pre-processed junctions was measured on RNA isolated from 143B cells using the miRNeasy RNA extraction kit (Qiagen). Levels of PTCD1 mRNA were measured from RNA isolated from normal human tissues (Ambion). cDNA was prepared using ThermoScript reverse transcriptase (Invitrogen) and random hexamers and used as a template in the subsequent PCR that was performed using a Corbett Rotorgene 3000 using Platinum UDG SYBR Green mastermix (Invitrogen) and normalized to 18S ribosomal RNA. Primer sequences are listed in Supplementary Table 1.
Northern blotting
RNA (20 µg) was resolved on 1.2% agarose formaldehyde gels, then transferred to 0.45 µm Hybond-N+ nitrocellulose membrane (GE Lifesciences) and hybridized with biotinylated oligonucleotide probes specific to mitochondrial tRNAs (Supplementary Table 1). The hybridizations were carried out overnight at 50°C in 5× SSC, 20 mM Na2HPO4, 7% SDS and 100 µg ml–1 heparin, followed by washing. The signal was detected using a streptavidin-linked horseradish peroxidase (diluted 1 : 2000 in 3× SSC, 5% SDS, 25 mM Na2HPO4, pH 7.5) by enhanced chemiluminescence (GE Lifesciences).
Immunoblotting
Specific proteins were detected using a mouse ND1 monoclonal antibody (diluted 1 : 2000, Santa Cruz Biotechnology), NDUFA9, COXI, COXII and COXIV (MitoSciences, diluted 1 : 2000), Bcl-2 antibody (Zymed, diluted 1 : 500) in 1% skim milk powder in phosphate-buffered saline (PBS). Control immunoblots against porin (MitoSciences, diluted 1 : 2000) were used to confirm normalized protein loading. Control immunoblots of the cytosol fractions against the MnSOD antibody showed complete separation of the mitochondria from the cytosol.
Respiration
Permeabilized cell respiration on glutamate/malate was measured according to Kuznetsov et al. (26). Briefly, 1 × 106 cells were washed in PBS, resuspended in 0.25 ml mitomedium B (0.5 mM EGTA, 3 mM MgCl2, 20 mM taurine, 10 mM KH2PO4, 20 mM HEPES, 1 g l–1 fatty acid-free BSA, 60 mM lactobionate, 110 mM mannitol, 0.3 mM DTT, pH 7.1 with KOH) and added to 0.5 ml mitomedium B in a 1 ml Rank Brothers Oxygen Electrode thermostatically maintained at 37°C. The system was left to equilibrate for 5 min, before adding digitonin (50 µg ml–1) and waiting for 5 min for the oxygen consumption to decline. Respiration of the permeabilized cells was initiated with 10 mM glutamate and 5 mM malate.
Complex enzyme assays
Enzyme assays were carried out in a 1 ml cuvette at 30°C using a Perkin Elmer lambda 35 dual beam spectrophotometer. For mitochondrial oxidative phosphorylation complex assays, mitochondria in STE plus protease inhibitors were disrupted by two to four cycles of freezing in dry ice/ethanol and thawing at 30°C. Combined Complex II to Complex III activity was assayed as the myxothiazol-sensitive rate of reduction of ferricytochrome c by succinate (27). Complex III was measured as the myxothiazol-sensitive rate of reduction of ferricytochrome c by CoQ2 and Complex IV was measured as the cyanide-sensitive oxidation of ferrocytochrome c (27).
RESULTS
PTCD1 is a mitochondrial matrix protein with eight PPR domains
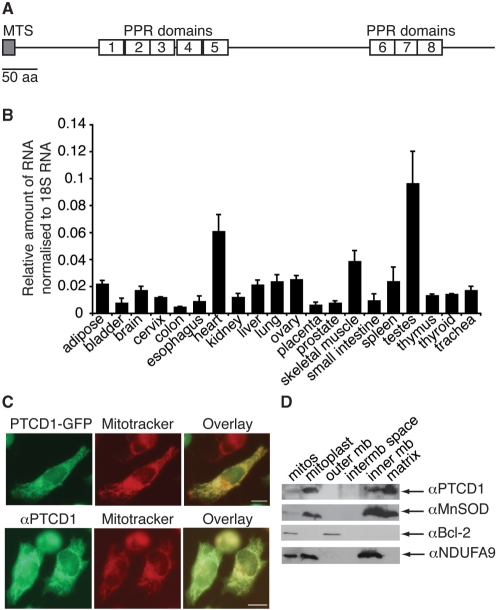
The uncharacterized PPR-domain protein PTCD1 has been noted previously to share homology with the Neurospora crassa protein CIA84 that is an assembly factor for mitochondrial Complex I. However, the reciprocal best match of PTCD1 in N. crassa is the uncharacterized protein NCU007871 (Supplementary Figure 1) and the most distinctive features of PTCD1 are its eight PPR domains and an N-terminal mitochondrial targeting sequence (Figure 1 A, Supplementary Figure 2).
Figure 1.
PTCD1 is a mitochondrial matrix protein with eight PPR domains. (A) Gene structure of PTCD1. (B) mRNA distribution in human tissues analysed by Q-PCR. (C) 143B cells transfected with pPTCD1-EYFP or untreated143B cells were incubated with 100 nM Mitotracker Orange and fixed. PTCD1-EYFP (green) was co-localized to mitochondria stained with Mitotracker Orange (red) directly by fluorescence microscopy and untreated cells were incubated with an antibody against PTCD1 (green). In the overlaid images yellow indicates co-localization of PTCD1 with mitochondria. Scale bar is 5 µm. (D) Isolated mitochondria were fractionated into four compartments; proteins from each fraction were resolved by SDS–PAGE and marker proteins for each mitochondrial compartment were detected by immunoblotting. We detected Bcl-2 as a marker for the outer mitochondrial membrane, NDUAF9 as the marker for the inner mitochondrial membrane and MnSOD as a marker for the mitochondrial matrix. PTCD1 is associated predominantly with the mitochondrial matrix.
We measured the mRNA levels of PTCD1 in human tissues and found that it is most abundant in testes, skeletal muscle and heart (Figure 1B). We fused the C-terminus of the protein to EYFP and showed that PTCD1 localizes specifically to mitochondria in 143B cells (Figure 1C). We used an antibody to PTCD1 to confirm its localization to mitochondria (Figure 1C). Fractionation of isolated mitochondria from rat liver into the four compartments, followed by immunoblotting of each fraction against a marker protein, indicated that PTCD1 was predominantly a mitochondrial matrix protein (Figure 1D).
PTCD1 associates with mitochondrial leucine tRNAs
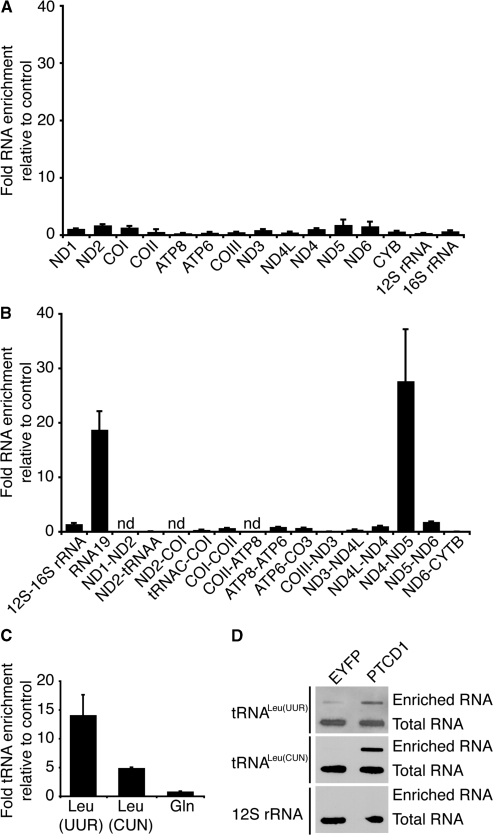
We fused the C-terminus of PTCD1 to a TAP tag and expressed this protein in 143B osteosarcoma cells. We used the TAP tag to isolate the PTCD1-TAP protein from cells and measured the enrichment of the 13 mitochondrial RNA transcripts by quantitative reverse transcriptase PCR (QRT-PCR) to determine the RNA targets of PTCD1. There was no significant enrichment observed of these transcripts compared to the control EYFP-TAP protein (Figure 2 A). Because many plant PPR domain proteins are involved in processing of organelle transcripts, we investigated if PTCD1 bound any of the mitochondrial precursor RNAs. We designed primers around the junctions between each gene and most of these areas included tRNAs which are thought to act as ‘punctuation marks’ for most of the mammalian mRNA transcripts. We found that two junction transcripts were enriched in RNA associated with PTCD1-TAP: RNA19 and ND4–5 (Figure 2B). The RNA19 transcript encompasses the 16S rRNA, tRNALeu(UUR) and the ND1 RNA. The ND4–5 precursor transcript encompasses the ND4 RNA, tRNAHis, tRNASer, tRNALeu(CUN) and the ND5 RNA. We quantitated the enrichment of the tRNAs in each junction and found that the leucine tRNAs (tRNALeu(UUR) and tRNALeu(CUN)) were targets for PTCD1 (Figure 2C). To confirm that the association of PTCD1 with mitochondrial leucine tRNAs was specific we isolated the enriched RNA from cells expressing PTCD1-TAP or EYFP-TAP and detected the presence of tRNALeu(UUR), tRNALeu(CUN) and 12S rRNA by hybridization with biotinylated oligonucleotide probes against these transcripts. We found that tRNALeu(UUR) and tRNALeu(CUN) were specifically enriched in RNA associated with PTCD1-TAP but not in the control RNA isolated from cells expressing EYFP-TAP (Figure 2D).
Figure 2.
PTCD1 is a mitochondrial RNA-binding protein. PTCD1-TAP protein expressed in 143B cells was purified using rabbit IgG agarose and eluted by tobacco etch virus (TEV) protease cleavage. RNA associated with the eluted PTCD1-TAP was reverse transcribed and analysed by Q-PCR. Values are expressed as a ratio of PTCD1-TAP compared to purification from cells that express a control, EYFP-TAP protein. (A) Enriched mitochondrial mRNA transcripts compared to control. (B) Enrichment of mitochondrial RNA junction transcripts compared to control (n.d., not detected). (C) Enrichment of leucine tRNAs compared to control. Data are means ± SD of three separate experiments. (D) Enrichment of leucine tRNAs but not 12S rRNA, confirmed by slot blotting using biotinylated oligonucleotide probes.
PTCD1 lowers the abundance of mitochondrial leucine tRNAs
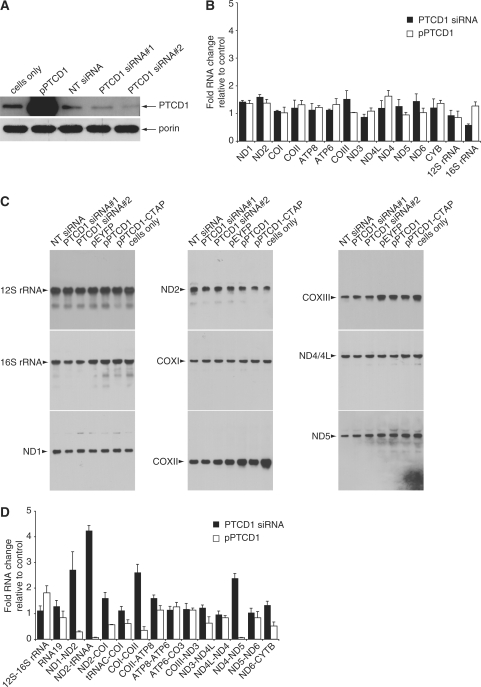
We investigated the effects of PTCD1 knockdown and overexpression in 143B cells. We used two distinct siRNA duplexes that significantly lowered the protein levels of PTCD1 but did not affect the abundance of porin, which was used as a loading control (Figure 3 A). Next, we found that the mature mRNA and rRNA transcripts were not affected by knocking down or overexpressing PTCD1 in 143B cells (Figure 3B) suggesting that PTCD1 does not affect processing of the polycistronic mitochondrial mRNA in a way that changes the abundance of the mature mRNAs. We investigated the levels of mitochondrial mature transcripts by northern blotting and found that they were not changed when PTCD1 was knocked down or overexpressed (Figure 3C). Then we investigated the effect of PTCD1 knockdown and overexpression on the mitochondrial RNA junctions and found that increasing PTCD1 abundance reduced the precursor ND4–5 transcripts, whereas decreasing PTCD1 30 abundance resulted in an increase of the ND4–5 transcript (Figure 3D). RNA19 levels were not significantly affected with the changes in PTCD1 abundance. However, we observed that the levels of transcript junctions ND1-2, ND2-tRNAAla and COI-COII increase when PTCD1 is knocked down in cells (Figure 3D). Also, we observed that the level of these transcript junctions is decreased significantly when the levels of PTCD1 is increased (Figure 3D).
Figure 3.
PTCD1 changes the level of precursor RNAs in 143B cells. (A) Protein expression of PTCD1 following knockdown by specific and non targeting siRNA in cells was detected by immunoblotting. Porin was detected to check for equal loading of total protein. (B) Changes in PTCD1 levels do not affect mature mRNA and rRNA abundance. RNA isolated from cells transfected with PTCD1 siRNA, non-targeting (NT) siRNA, a plasmid that overexpresses PTCD1 (pPTCD1) or a plasmid that overexpresses EYFP (pEYFP) was reverse transcribed and analysed by Q-PCR. The data is expressed as a ratio of transcripts from experimental samples compared to control samples. Amplification of mature mitochondrial mRNA transcripts is shown. (C) The abundance of mature mitochondrial transcripts was analysed by northern blotting. The data are typical of results repeated on two separate RNA preparations. (D) Amplification of precursor RNA transcripts by Q RT-PCR. Data are means ± SD of three separate experiments.
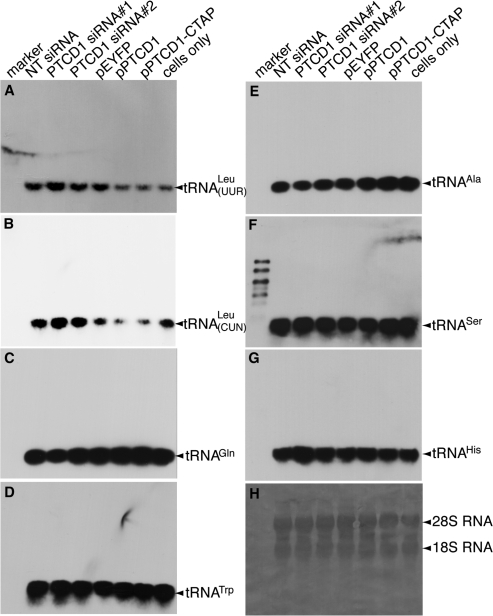
We investigated changes in mitochondrial tRNAs in cells where PTCD1 was knocked down or overexpressed by northern blotting. We observed a decrease of tRNALeu(UUR) and tRNALeu(CUN) when PTCD1 was overexpressed in cells either wild type or as a TAP fusion, respectively (Figure 4 A and B). Knockdown of PTCD1 in cells led to increased accumulation of both leucine tRNAs using two distinct siRNA duplexes (Figure 4A and B). Changes in cellular PTCD1 levels did not affect the abundance of other tRNAs such as tRNAGln, tRNATrp, tRNAAla (Figure 4C–E), nor the levels of tRNASer(AGY) and tRNAHis associated with the ND4–5 precursor transcript (Figure 4F–G).
Figure 4.
PTCD1 lowers the abundance of mitochondrial leucine tRNA in 143B cells. RNA isolated from cells transfected with PTCD1 siRNA, NT siRNA, pPTCD1, pEYFP and from untransfected cells was analysed for tRNA abundance by northern blotting. (A) tRNALeu(UUR). (B) tRNALeu(CUN). (C) tRNAGln. (D) tRNATrp. (E) tRNAAla. (F) tRNASer(AGY). (G) tRNAHis. (H) Total RNA was detected using methylene blue. The data are typical of results repeated on four separate RNA preparations. Each northern blot was probed for all the tRNAs by stripping the blot and re-probing it.
PTCD1 knockdown leads to an increase in mitochondrial and nuclear-encoded subunits of Complexes I and IV
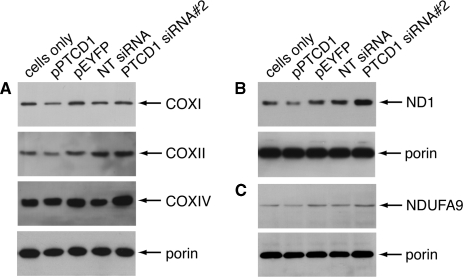
Changes in mitochondrial tRNA levels have been shown to affect the translation of mitochondrial-encoded genes (28). Therefore, we investigated, by immunoblotting, differences in mitochondrial-encoded proteins in 143B cells where PTCD1 was knocked down or overexpressed. We found increased protein levels of subunits I and II of Complex IV when PTCD1 was knocked down and decreased levels of these proteins when PTCD1 was overexpressed in the cells (Figure 5 A). We observed the same trend with the ND1 subunit of Complex I (Figure 5B). In addition, there are possibly subtle changes in the levels of nuclear-encoded subunits subunit IV of Complex IV and NDUFA9 of Complex I (Figure 5A and C). We used porin as a loading control to show that the mitochondrial protein abundance was equal under all treatment conditions (Figure 5A–C).
Figure 5.
PTCD1 knockdown leads to an increase in mitochondrial and nuclear-encoded subunits of Complexes I and IV. Mitochondria were isolated from cells transfected with PTCD1 siRNA, NT siRNA, pPTCD1, pEYFP and from untransfected cells. Mitochondrial proteins were resolved by SDS–PAGE and specific proteins were detected by immunoblotting. (A) COXI, COXII, COXIV and porin. (B) ND1 and porin (C) NDUFA9 and porin. The data are typical of results repeated on four separate mitochondrial preparations (one of which is shown in the supplementary Figure 6).
Functional effects of PTCD1 knockdown and overexpression
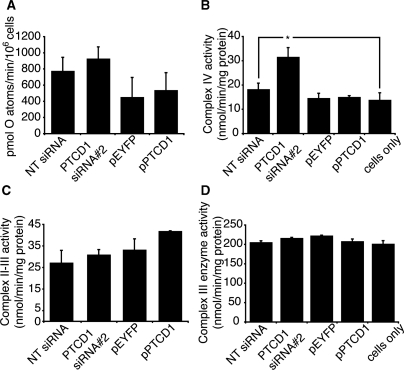
Because we observed changes in several subunits of Complexes I and IV, we measured if their activities were affected when PTCD1 was knocked down or overexpressed in 143B cells. The activity of Complex I measured by respiration of digitonin-permeabilized cells on glutamate/malate was not affected significantly when PTCD1 was knocked down in 143B cells (Figure 6 A). However, enzyme assays showed an increased Complex IV activity when PTCD1 was knocked down in cells (Figure 6B). The enzyme activities of Complexs II to III and Complex III were unaffected by the changes in PTCD1 abundance (Figure 6C and D).
Figure 6.
Functional effects of PTCD1 knockdown and overexpression. (A) State 4 respiration on glutamate/malate was measured in digitonin-permeabilized 143B cells transfected with PTCD1 siRNA, NT siRNA, pPTCD1 and pEYFP using an oxygen electrode. The enzyme activities of cytochrome oxidase (B), Complexes II–III (C) and Complex III (D) were measured spectrophotometrically. Data are means ± SEM of four separate experiments; *P < 0.05 compared with control treatments by a two-tailed paired Student's t-test.
DISCUSSION
Here we show that PTCD1 is a mitochondrial matrix protein that is more abundant in mitochondria-rich tissues. Comparative genomic analyses have predicted PTCD1 to be an assembly factor for Complex I because of some shared homology with the N. crassa protein CIA84. Since PTCD1 is associated with both the mitochondrial matrix and the inner membrane, it could be considered that PTCD1 may have two functions within mitochondria, as an RNA-binding protein and a Complex I assembly factor. In this study we found that PTCD1 has an important role as an RNA regulator.
The eight predicted PPR domains most likely give PTCD1 RNA-binding affinity for the leucine tRNAs (tRNALeu(CUN) and tRNALeu(UUR)) and the precursor RNAs that contain them. This suggests that the binding of PTCD1 to the tRNAs does not require aminoacylation nor CCA addition at the 3′ end of the tRNAs. Overexpression of PTCD1 reduced the levels of leucine tRNAs in cells, while siRNA knockdown of PTCD1 increased the levels of these tRNAs. In addition, we show that increasing the abundance of PTCD1 in cells decreased the levels of precursor ND4–5 RNA as a result of reduced tRNALeu(CUN) levels. The tRNALeu(UUR) levels have a more subtle decrease which is not apparent at the RNA19 level. Changing PTCD1 levels affected some other precursor RNAs; however, the levels of the tRNAs contained within these transcripts that we investigated were not changed. It is not clear if these represent genuine PTCD1 targets or whether these changes are a secondary effect of PTCD1 at the regions containing the leucine tRNAs which affects the rate or hierarchy of processing of primary mtDNA transcripts. Because altered levels of PTCD1 modulate the levels of leucine tRNAs without affecting the levels of the mRNAs that flank them, it seems unlikely that PTCD1 acts solely at the level of RNA processing. Whether PTCD1 induces cleavage of the leucine tRNAs before or after RNase P and RNase Z cleavage remains to be determined. The observation that mRNA levels are not changed implies that either PTCD1 has an effect after tRNA processing or that mRNA processing can proceed even if the intervening tRNA is cleaved. This suggests a negative regulatory function of PTCD1 at the tRNA level that does not severely affect the processing and translation of mitochondrial transcripts. This contrasts with a recent study where mice lacking the PTCD2 gene showed an increase in a precursor RNA containing cytochrome b and a concomitant decrease in mature cytochrome b mRNA (19).
The enrichment of leucine tRNAs with PTCD1 from cells was detected using primers that bind only within the tRNAs, which are present at higher amounts than the precursor RNAs that contain them. Although a high-resolution structure of a PPR domain has not yet been described, structural modelling has led to the proposal that PPR domains most likely bind single-stranded RNA (29). Since tRNAs have mostly double-stranded structures, there could be two possible modes of association between PTCD1 and leucine tRNAs; PTCD1 could either bind sequences found in the loop regions of properly folded tRNA or it could bind in regions that are double stranded in the properly folded tRNA by associating before the tRNA has folded or by displacing the non-target strand to enable its association. The degradation of leucine tRNAs could result from nuclease activity of PTCD1; however, we could not identify any regions of primary sequence with similarity to known nuclease domains. Alternatively, PTCD1 may recruit nucleases that cleave the leucine tRNAs or rearrange the tRNA structure to produce a substrate for a nuclease activity not directly associated with PTCD1. We have found that PTCD1 is insoluble when expressed in Escherichia coli and in wheat germ extract, which has made functional in vitro studies difficult. The ability to generate soluble recombinant PTCD1 will be critical for functional analysis of the RNA-binding affinities and minimal recognition sites. Towards this goal we are investigating the binding affinity of individual PTCD1 domains for the leucine tRNAs.
The targeted destruction of tRNAs has been observed in the cytoplasm of a variety of diverse organisms ranging from bacteria and plants to mammalian cells (30,31). To the best of our knowledge this phenomenon has been observed here in an organelle for the first time. The depletion of tRNA provides a rapid way to lower protein translation in response to cellular stresses such as oxidative stress, nutrient deprivation and amino acid starvation (31). Increased leucine tRNAs in response to PTCD1 knockdown increased mitochondrial-encoded Complexes I and IV subunits, suggesting that leucine tRNAs may be rate-limiting for mitochondrial protein synthesis. The subtle increases in nuclear-encoded subunits of Complexes I and IV are most likely a retrograde response to the increased mitochondria-encoded subunits. We have investigated the effects of PTCD1 overexpression and knockdown on de novo mitochondrial protein synthesis in 143B cells and found that there were subtle but not significant differences in the abundance of the 13 mitochondria-encoded proteins (Supplementary Figure 5). The subtle protein changes over 1 h of de novo synthesis may accumulate over time to yield significant differences in the steady state levels of the proteins after knockdown or overexpression of PTCD1, as detected by immunoblotting. This is consistent with the observed functional increase in the activity of Complex IV following PTCD1 knockdown. We have observed that cybrids devoid of mitochondrial DNA or cybrids with a mutation in the tRNALeu(UUR) that causes MELAS, have increased levels of PTCD1 (Supplementary Figure 7). Although the physiological inducers of PTCD1 remain to be identified, this suggests that PTCD1 may play a role in the regulation of mitochondrial gene expression in response to stress.
In mammals mitochondrial transcripts lack introns and untranslated regions and rely on tRNA processing to generate mature RNAs, whereas in yeast mitochondrial transcripts have 5′ untranslated regions that regulate translation and plant mitochondrial transcripts are edited and have introns. The diverse regulatory functions of PPR proteins may have evolved in different organisms as a result of the idiosyncratic features of organelle gene expression between different kingdoms. Mammalian PPR domain proteins studied to date regulate mitochondrial gene expression by different mechanisms: POLRMT transcribes mtDNA (5), PTCD2 processes specific precursor RNAs (19), LRPPRC stabilizes specific mRNAs (14), PTCD3 associates with the small ribosomal subunit and is necessary for efficient translation (20) and MRPP3 is a subunit of the RNase P complex needed for processing of tRNAs (16). We have expanded the repertoire of PPR protein function by showing that PTCD1 acts as a negative regulator of specific mitochondrial tRNAs. Further characterization of mammalian PPR domain proteins will provide insights into the regulation of mitochondrial gene expression at a post-transcriptional level.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Health and Medical Research Council. Funding for open access charge: Royal Perth Hospital Medical Research Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank A/Prof. Michael Ryan from La Trobe University for the PTCD1 antibody and Prof. Ian Small from UWA for critical discussion and advice.
REFERENCES
- 1.Smeitink J, van den Heuvel L, DiMauro S. The genetics and pathology of oxidative phosphorylation. Nat. Rev. Genet. 2001;2:342–352. doi: 10.1038/35072063. [DOI] [PubMed] [Google Scholar]
- 2.Shoubridge EA. Nuclear genetic defects of oxidative phosphorylation. Hum. Mol. Genet. 2001;10:2277–2284. doi: 10.1093/hmg/10.20.2277. [DOI] [PubMed] [Google Scholar]
- 3.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 4.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 6.Gagliardi D, Stepien PP, Temperley RJ, Lightowlers RN, Chrzanowska-Lightowlers ZM. Messenger RNA stability in mitochondria: different means to an end. Trends Genet. 2004;20:260–267. doi: 10.1016/j.tig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 7.Anisimov SV. A large-scale screening of the normalized mammalian mitochondrial gene expression profiles. Genet. Res. 2005;86:127–138. doi: 10.1017/S0016672305007718. [DOI] [PubMed] [Google Scholar]
- 8.Connor MK, Takahashi M, Hood DA. Tissue-specific stability of nuclear- and mitochondrially encoded mRNAs. Arch. Biochem. Biophys. 1996;333:103–108. doi: 10.1006/abbi.1996.0369. [DOI] [PubMed] [Google Scholar]
- 9.Ostronoff LK, Izquierdo JM, Enriquez JA, Montoya J, Cuezva JM. Transient activation of mitochondrial translation regulates the expression of the mitochondrial genome during mammalian mitochondrial differentiation. Biochem. J. 1996;316(Pt 1):183–191. doi: 10.1042/bj3160183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 11.Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J. Biol. Chem. 2005;280:19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- 12.Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 13.Mili S, Pinol-Roma S. LRP130, a pentatricopeptide motif protein with a noncanonical RNA-binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Mol. Cell Biol. 2003;23:4972–4982. doi: 10.1128/MCB.23.14.4972-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu F, Morin C, Mitchell G, Ackerley C, Robinson BH. The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA. Biochem. J. 2004;382:331–336. doi: 10.1042/BJ20040469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, Delmonte T, Villeneuve A, Sladek R, Xu F, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl Acad. Sci. USA. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: Identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 17.Lightowlers RN, Chrzanowska-Lightowlers ZM. PPR (pentatricopeptide repeat) proteins in mammals: important aids to mitochondrial gene expression. Biochem. J. 2008;416:e5–e6. doi: 10.1042/BJ20081942. [DOI] [PubMed] [Google Scholar]
- 18.Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Xu F, Ackerley C, Maj MC, Addis JB, Levandovskiy V, Lee J, Mackay N, Cameron JM, Robinson BH. Disruption of a mitochondrial RNA-binding protein gene results in decreased cytochrome b expression and a marked reduction in ubiquinol-cytochrome c reductase activity in mouse heart mitochondria. Biochem. J. 2008;416:15–26. doi: 10.1042/BJ20080847. [DOI] [PubMed] [Google Scholar]
- 20.Davies SMK, Rackham O, Shearwood A-MJ, Hamilton KL, Narsai R, Whelan J, Filipovska A. Pentatricopeptide repeat domain protein 3 associates with the mitochondrial small ribosomal subunit and regulates translation. FEBS Lett. 2009;583:1853–1858. doi: 10.1016/j.febslet.2009.04.048. [DOI] [PubMed] [Google Scholar]
- 21.Burckstummer T, Bennett KL, Preradovic A, Schutze G, Hantschel O, Superti-Furga G, Bauch A. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods. 2006;3:1013–1019. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- 22.Rackham O, Nichols SJ, Leedman PJ, Berners-Price SJ, Filipovska A. A gold(I) phosphine complex selectively induces apoptosis in breast cancer cells: Implications for anticancer therapeutics targeted to mitochondria. Biochem. Pharmacol. 2007;74:992–1002. doi: 10.1016/j.bcp.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 23.Chappell JB, Hansford RG. In: Subcellular Components: Preparation and Fractionation. Birnie GD, editor. London: Butterworths; 1972. pp. 77–91. [Google Scholar]
- 24.Gornall AG, Bardawill CJ, David MM. Determination of serum protein by means of the biuret reaction. J. Biol. Chem. 1949;177:751–766. [PubMed] [Google Scholar]
- 25.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 26.Kuznetsov AV, Veksler V, Gellerich FN, Saks V, Margreiter R, Kunz WS. Analysis of mitochondrial function in situ in permeabilized muscle fibers, tissues and cells. Nat. Protoc. 2008;3:965–976. doi: 10.1038/nprot.2008.61. [DOI] [PubMed] [Google Scholar]
- 27.James AM, Wei Y-H, Pang C-Y, Murphy MP. Altered mitochondrial function in fibroblasts containing MELAS or MERRF mitochondrial DNA mutations. Biochem. J. 1996;318:401–407. doi: 10.1042/bj3180401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayashi J, Ohta S, Kagawa Y, Takai D, Miyabayashi S, Tada K, Fukushima H, Inui K, Okada S, Goto Y, et al. Functional and morphological abnormalities of mitochondria in human cells containing mitochondrial DNA with pathogenic point mutations in tRNA genes. J. Biol. Chem. 1994;269:19060–19066. [PubMed] [Google Scholar]
- 29.Delannoy E, Stanley WA, Bond CS, Small ID. Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem. Soc. Trans. 2007;35:1643–1647. doi: 10.1042/BST0351643. [DOI] [PubMed] [Google Scholar]
- 30.Haiser HJ, Karginov FV, Hannon GJ, Elliot MA. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 2008;36:732–741. doi: 10.1093/nar/gkm1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.