Abstract
Plants have intercellular channels, plasmodesmata (PD), that span the cell wall to enable cell-to-cell transport of micro- and macromolecules. We identified an Arabidopsis thaliana embryo lethal mutant increased size exclusion limit 1 (ise1) that results in increased PD-mediated transport of fluorescent tracers. The ise1 mutants have a higher frequency of branched and twinned PD than wild-type embryos. Silencing of ISE1 in mature Nicotiana benthamiana leaves also leads to increased PD transport, as monitored by intercellular movement of a GFP fusion to the tobacco mosaic virus movement protein. ISE1 encodes a putative plant-specific DEAD-box RNA helicase that localizes specifically to mitochondria. The N-terminal 100 aa of ISE1 specify mitochondrial targeting. Mitochondrial metabolism is compromised severely in ise1 mutant embryos, because their mitochondrial proton gradient is disrupted and reactive oxygen species production is increased. Although mitochondria are essential for numerous cell-autonomous functions, the present studies demonstrate that mitochondrial function also regulates the critical cell non-cell-autonomous function of PD.
Keywords: intercellular, traffic, Arabidopsis, embryogenesis, Redox
Plasmodesmata (PD) are plasma membrane-lined channels that span the cell walls between adjacent plant cells (reviewed in ref. 1). Plasmodesmata facilitate cell-to-cell communication essential for intercellular signaling of micro- and macromolecules during plant growth and development. Because plant cells are immobile and encased in cell walls, PD provide symplastic continuity between cells. Plasmodesmata are bounded by the plasma membrane and have a core of modified endoplasmic reticulum at their center. Transport through PD occurs primarily through the cytoplasmic space between the plasma membrane and the modified endoplasmic reticulum.
An important measure of PD permeability is the size exclusion limit (SEL), the upper limit of the size of macromolecules capable of freely diffusing from cell to cell in a tissue. The SEL is regulated temporally, spatially, and physiologically throughout development (2, 3). That plant viruses (much larger than the SEL of PD channels) pirate these passageways during infectious spread implies that PD can expand their aperture or SEL to allow transport of viral genomes and proteins. Numerous reports have demonstrated intercellular movement of endogenously expressed macromolecules through these channels, including transcription factors (4), mRNA (5), and siRNA (6).
There are two pathways for movement via PD: facilitated movement and diffusion. Proteins such as viral movement proteins (7), class I KNOTTED1-like homeobox (KNOX) domain transcription factors (8), and non-cell-autonomous proteins (9) are transported actively and can increase the PD SEL. Other proteins, such as the exogenous tracer GFP, traffic via simple diffusion through the cytoplasmic sleeve (10). Rapid inhibition of PD transport occurs via deposition of the linear β-glucan callose around the neck regions of PD in response to pathogen invasion, wounding, and other acute stresses (11, 12). Callose at PD can be removed by β-1,3-glucanases localized to the PD (13). Anaerobic stress increases the PD SEL (14). However, the genetic mechanisms that regulate PD structure and function are not well known.
Because PD are essential cellular components, PD mutants are likely to have severe growth defects manifested as early as embryogenesis. Thus, we screened embryo defective lines of Arabidopsis for altered, specifically increased, intercellular transport via PD; two mutants with increased size exclusion limit (ise) phenotypes, ise1 and ise2, were identified (15). Such embryo defective lines can be propagated stably as heterozygotes, and 25% homozygous mutant embryos are detected segregating in their siliques.
Here, we identify the INCREASED SIZE EXCLUSION LIMIT 1 (ISE1) gene. ISE1 encodes a DEAD-box RNA helicase that localizes to mitochondria and is essential for mitochondrial function. The data suggest that wild-type mitochondrial function is critical to regulate cell-to-cell transport via PD.
Results
Identification of Mutants Affecting PD Transport.
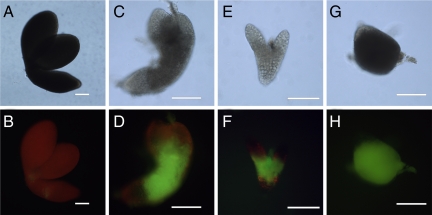
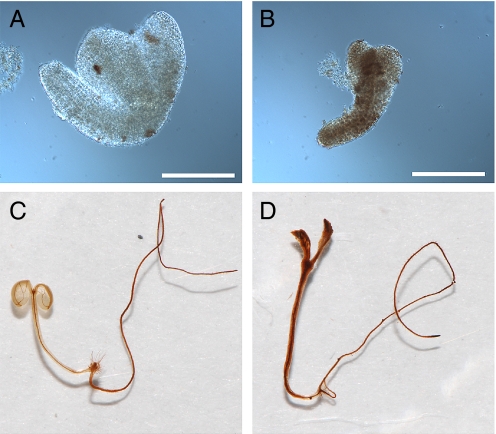
We developed an assay for alterations in PD-mediated transport of fluorescent tracers during embryogenesis (15). Basically, when embryos are extruded from their seed coats, small breaks arise in a few of the external cell walls of embryos. These breaks provide sites of entry for fluorescent tracers, and depending on the SEL of the cells at the break site, tracers can move either into the internal cells of the embryo or not. During early stages of embryogenesis, tracers move readily into the internal cell layers. However, wild-type embryos stop trafficking 10-kDa fluorescein (F)-conjugated dextran at the midtorpedo stage (Fig. 1 A and B). In contrast, sibling ise1–1 mutant embryos sustain trafficking of 10-kDa F-dextran at this developmental stage (Fig. 1 C and D). The recessive ise1–1 mutant allele was derived from ethyl methanesulfonate mutagenesis that induces point mutations. Single-nucleotide polymorphism markers were used to construct a detailed physical map to identify the ISE1 gene (SI Materials and Methods). Sequencing of DNA from heterozygous ise1 plants compared with wild type revealed a guanine to adenine substitution at nucleotide 683 of locus At1g12770 resulting in the substitution of glutamic acid for glycine at codon 228. ISE1 consists of 1656 nt encoding a 551-aa protein with a calculated molecular mass of 60.7 kDa. There are no introns in the coding sequence; however, there is an intron in the 5′ UTR (Fig. 2A).
Fig. 1.
Dye loading phenotype of ise1 mutant embryos. (A–H) ise1 mutant embryos allow intercellular transport of 10-kDa fluorescein (F)-conjugated dextran. (A, B) Wild-type torpedo embryos. (C–H) Sibling mutant embryos: (C, D) ise1–1, (E, F) ise1–2, (G, H) ise1–3. (A, C, E, G) Bright-field images. (B, D, F, H) Corresponding images assaying for F-dextran movement. (Scale bars, 100 μm.)
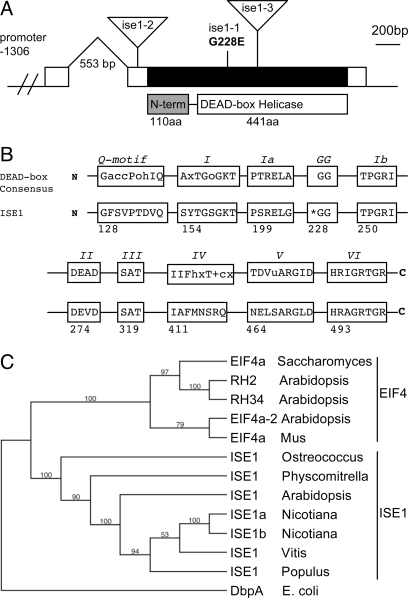
Fig. 2.
ISE1 encodes a conserved, plant-specific DEAD-box RNA helicase. (A) Gene structure of ISE1. ISE1 mRNA (white box) contains a single exon (black) with a 553-bp intron in the 5′ UTR. The relative positions of the ise1–1 single amino acid mutation and the T-DNA insertions in ise1–2 and ise1–3 are indicated. The ISE1 protein consists of a 441-aa DEAD-box RNA helicase domain and a unique 110-aa N-terminal domain. (B) ISE1 contains all of the domains conserved throughout the DExD family of RNA helicases. Asterisk indicates site of the ise1–1 mutation. (C) Homologs of ISE1 are found throughout the green plants and the unicellular algae Ostreococcus tauri. The most similar protein to ISE1 in yeast and animals is eukaryotic initiation factor 4a, although there is no shared homology outside the RNA helicase domains. There are two isoforms of ISE1 in Nicotiana benthamiana.
ISE1 Encodes a DEAD-Box RNA Helicase.
ISE1 (At1g12770) encodes a highly conserved 441-aa DEAD-box RNA helicase domain and a unique 110-aa N-terminal domain. ISE1 was described previously as RNA helicase 47 (16). ISE1 has all highly conserved motifs necessary for the RNA helicase function of DEAD-box proteins (17) (Fig. 2B). The missense mutation in the ise1–1 allele substitutes glutamic acid for the first glycine of the conserved “GG loop” of the DEAD-box domain. A similarly charged amino acid substitution of aspartic acid for the homologous glycine residue in the GG loop of eukaryotic initiation factor 4a in Saccharomyces cerevisiae results in a lethal loss of function (18). In the cocrystal structure of the Drosophila melanogaster DEAD-box protein Vasa with a single-stranded nucleic acid, the GG loop contacts the sugar–phosphate backbone of RNA (19). When GG is mutated to EG, the charged glutamic acid likely interferes with RNA binding. Thus, the ise1–1 mutant phenotype is expected to be the result of lost or reduced RNA helicase function due to disruption of the GG loop.
Fig. 2C summarizes the phylogeny of the ISE1 protein (details in Fig. S1). Homologs of ISE1 occur in all green plants for which quality sequence databases are available, such as the higher plants Arabidopsis, rice, and grape, the moss Physcomitrella, and the unicellular algae Ostreococcus tauri. ISE1 is present as a single or recently duplicated gene in each of these species, thus supporting a conserved function for ISE1 throughout the plant lineage.
ISE1 Is an Essential Gene.
Two additional mutant alleles, ise1–2 and ise1–3, were identified originally as Agrobacterium-mediated T-DNA insertions in locus EMB1586 (At1g12770) (20). Genetic tests confirmed allelism between ise1–1 and emb1586–1 (renamed ise1–2) and emb1586–2 (renamed ise1–3) (Fig. 2A). The T-DNA insert in ise1–2 is 75 bp upstream of the start of translation and likely interferes with transcription of ISE1. The T-DNA insert in ise1–3 is located in the ISE1 exon at the position corresponding to amino acid 283; this large insertion results in loss of the C-terminal portion of ISE1.
Homozygous ise1–1 and ise1–2 mutant embryos maintain a large SEL and allow 10-kDa F-dextran transport (Fig. 1 D and F) when wild-type sibling embryos do not (Fig. 1B). The ise1–3 mutants typically arrest before the mid-torpedo stage; ise1–3 embryos traffic 10-kDa F-dextran as expected (Fig. 1 G and H), because wild-type embryos also traffic this tracer at all stages before the midtorpedo stage (15). The embryo shown in Fig. 1 G and H is a globular-shaped embryo that illustrates the lack of distinct morphogenesis in ise1–3 embryos. The more severe phenotype of ise1–3 is logical, because the ise1–3 lesion eliminates the expression of the C terminus of ISE1 (Fig. 2A). Because embryos lacking full-length ISE1 cannot survive past early stages of development, ISE1 is an essential gene product.
ISE1 Localizes to Mitochondria.
To provide insight into ISE1 function during embryogenesis and its role in PD regulation, we determined its cellular localization pattern. We produced transgenic Arabidopsis plants expressing ISE1 fused to GFP at the N (GFP-ISE1) or C terminus (ISE1-GFP) of ISE1. The native promoter region (1,800 bases upstream of the start of ISE1 translation) driving expression of ISE1-GFP produced viable transgenic plants and fully rescued the embryo defective phenotype of the strong ise1–3 allele; such ISE1-GFP/ise1–3 transgenic plants cannot be distinguished from wild-type plants. A PCR analysis of the ISE1-GFP T1 population in phenotypically normal plants identified plants homozygous for the ise1–3 mutant allele (see Materials and Methods for details). Further studies of T2 progeny of rescued ISE1-GFP/ise1–3 transgenic plants indicated the presence of a single copy of the ISE1-GFP transgene. In contrast, no rescue of the ise1–3 phenotype occurred in GFP-ISE1 transgenic plants.
Rescued embryos were fully wild type in morphology and developmental timing. The RT-PCR analyses of endogenous ISE1 transcripts reveal expression in all tissues and at all developmental times (Fig. S2). We also monitored the expression of ISE1-GFP in rescued ise1–3 transgenic Arabidopsis plants by fluorescence microscopy. The strongest expression of ISE1-GFP occurs in root and shoot meristematic regions (Fig. S3). In roots, this expression extends upward into the elongation zone. ISE1-GFP is also expressed in flowers, with the highest expression in the gynoecium. Note that ISE1 is incorrectly annotated as the immediately downstream gene (At1g12775) at Genevestigator.
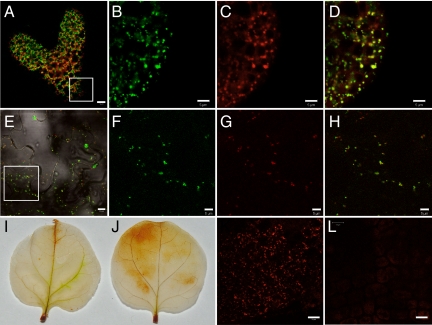
The transcomplementing ISE1-GFP fusion protein localized to distinct cytoplasmic foci ≈0.5–1 μm in diameter in late-heart Arabidopsis embryos (Fig. 3A). Fig. 3 A–D shows colocalization between ISE1-GFP foci and MitoTracker Red-labeled mitochondria in embryos, and Fig. S4 A–D shows similar colocalization patterns in cells of the root in Arabidopsis seedlings. Note that in the colocalization panels yellow fluorescence marks the most intensely colocalized mitochondria; however, the individual red or green fluorescence panels clearly reveal additional patterns of identical fluorescence.
Fig. 3.
ISE1 localizes to mitochondria and affects mitochondrial function. (A) Rescued Arabidopsis embryos expressing ISE1-GFP detected in the presence of (red) chlorophyll autofluorescence. An equivalent region shown as a square in A is enlarged in B, C, and D. (B) Green fluorescence of ISE1-GFP. (C) MitoTracker Red stain. (D) Colocalization of ISE1-GFP and MitoTracker Red. The N-terminal 100 aa of ISE1 are sufficient for targeting to the mitochondria. The square region in E is shown enlarged in F–H. 100-ISE1-GFP (E, F) was coexpressed transiently in N. benthamiana with mitochodria-targeted CFP (G), revealing mitochondrial colocalization (H). ISE1-silenced N. benthamiana leaves have increased production of the reactive oxygen species H2O2. 3,3′-Diaminobenzidine staining of control (I) and ISE1-silenced leaves (J). The membrane-potential-dependent mitochondrial stain MitoTracker Red stains wild-type embryo mitochondria in (K) but fails to stain mitochondria in the ise1–1 mutants (L). (Scale bars, B–D, F–H, 5 μm; A, E, 10 μm; K, L, 20 μm.)
We hypothesized that the unique N-terminal 110 aa of ISE1, just upstream of the DEAD-box RNA helicase domain, contain a mitochondrial transit peptide. Indeed, an N-terminal mitochondrial transit peptide is predicted weakly by protein sorting software. Agrobacterium carrying a construct to express the N-terminal 100 aa of ISE1-fused GFP (100-ISE1-GFP) was used to infect Nicotiana benthamiana leaves. We coexpressed 100-ISE1-GFP with a known mitochondrial transit peptide marker mitochodria-targeted CFP (mito-CFP) (21). Fig. 3 E–H documents that mito-CFP and the first 100 aa of ISE1 fused to GFP colocalize when transiently coexpressed. Finally, we transfected a stable transgenic N. benthamiana line expressing ISE1-GFP with mito-CFP; once again, ISE1-GFP and mito-CFP colocalize (Fig. S4 E–H). These studies together demonstrate that ISE1-GFP localizes to mitochondria in embryos, seedlings, and mature leaves and that targeting is mediated by a signal peptide in the first 100 aa.
Plasmodesmata Are Altered in ise1–1 Embryos.
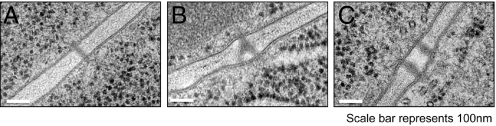
To determine if PD function or regulation is altered in ise1 mutants due to alterations in PD morphology, we compared the ultrastructure of PD in wild-type, ise1, and ise2 embryos. We found three types of PD structures in developing midtorpedo-stage embryos: simple PD, branched (H-, Y-, and X-shaped) PD, and twinned PD (Fig. 4 A–C). Table 1 reveals that ise1 mutant embryos have a higher frequency of branched (7.2%) PD compared with their wild-type siblings (1.0%). For comparison and to ensure the accuracy of our characterization of PD ultrastructure, we measured modified PD in the ise2 mutant; previously, we found 15% modified PD in ise2 (22). Here, we find 14.3% modified PD in ise2, in close agreement with our earlier results. Here, we also measured twinned PD, defined as PD that are within 100 nm of each other. Twinned PD likely represent newly arising PD in close juxtaposition (23). Wild type and ise2 mutants have a similar frequency of twinned PD, 4.9% and 6.0% respectively; ise1 embryos have an increase in twinned PD (7.9%), suggesting that there may be an increase in PD synthesis in ise1 embryos. Because ultrastructural analyses only detect major morphological changes, minor alterations that nevertheless significantly affect PD function in ise1 (or ise2) likely are missed.
Fig. 4.
ise1–1 mutant embryos have a higher frequency of branched and twinned plasmodesmata (PD) than wild-type embryos. (A) Simple PD. (B) Branched PD. (C) Twinned PD. (Scale bars, 100 nm.)
Table 1.
Branched and twinned plasmodesmata
| Genotype | Total | Simple | Branched | Twinned |
|---|---|---|---|---|
| Wild type | 284 | 267 | 3 (1.0) | 14 (4.9) |
| ise2 | 217 | 173 | 31 (14.3)* | 13 (6.0) |
| ise1 | 290 | 246 | 21 (7.2)* | 23 (7.9) |
*P < 0.05.
Silencing of ISE1 in Mature Leaves Recapitulates the Increased Intercellular Transport Observed in ise1 Embryos.
Because ise1 mutants arrest during embryonic development, we cannot examine the requirement for ISE1 in mature mutant tissues. Instead, we induced loss of ISE1 function in mature leaves by an RNA silencing strategy based on viral-induced gene silencing (VIGS) and the widely used tobacco rattle virus (TRV) system (24). Because the TRV system uses N. benthamiana as its host, we first identified two homologs of ISE1 in N. benthamiana. Fig. S5 compares ISE1 from Arabidopsis and N. benthamiana.
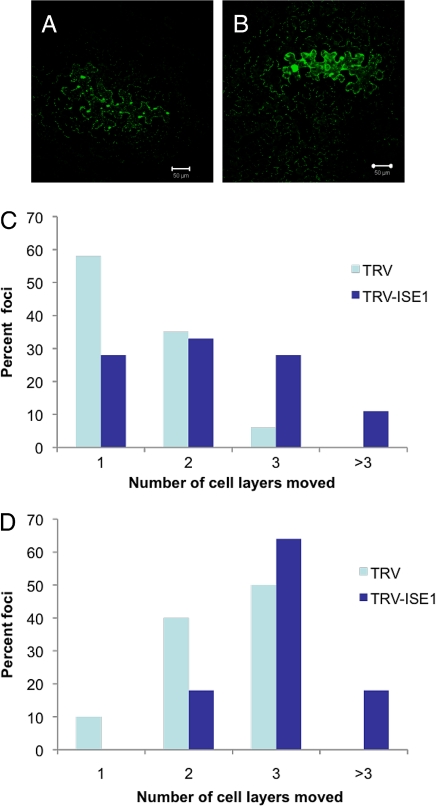
Two weeks after induction of ISE1 silencing, the newly arising upper leaves are slightly chlorotic, which is not unexpected for the silencing of an essential gene. Transcript levels from both N. benthamiana ISE1 homologs are significantly reduced by VIGS (Fig. S6). Then, PD transport was assayed in these newly arising ISE1-silenced leaves by Agrobacterium mediated expression of the tobacco mosaic virus (TMV) movement protein, P30, fused to 2XGFP. TMV P30–2XGFP was expressed in isolated leaf epidermal cells by infiltrating dilute cultures of Agrobacterium. TMV P30 is a well-established marker for PD-mediated cell-to-cell spread in tobacco tissues. P30 fused to GFP forms distinct puncta in cell walls and allows higher resolution and better quantification of cell-to-cell movement patterns than free GFP that forms a diffuse pattern. P30–2XGFP moves less extensively than P30–1XGFP and better reveals alterations in movement patterns.
We monitored the relative cell-to-cell spread of P30–2XGFP within the epidermis of ISE1-silenced and nonsilenced control leaves (see Materials and Methods). P30–2XGFP spread is increased dramatically in ISE1-silenced leaves compared with control leaves (Fig. 5 A and B). Counting the number of surrounding epidermal cells away from the initial Agrobacterium-transformed cell that displays fluorescent punctae in its cell walls provides a quantitative measure of P30–2XGFP movement. Fig. 5 C and D compares P30-GFP spread at 48 and 72 h postinfection in control and ISE1-silenced leaves; the results are derived from counting >50 foci for each type of leaf. These data reveal that P30–2XGFP moves more extensively in ISE1-silenced leaves compared with control leaves, recapitulating the phenotype of ise1–1 mutants. These data imply that PD function or regulation have been altered by silencing of ISE1 in mature tissue. Thus, loss of ISE1 function in Arabidopsis embryos or N. benthamiana leaves leads to an identical phenotype of increased intercellular movement via PD.
Fig. 5.
Intercellular movement of tobacco mosaic virus (TMV) P30-2XGFP is increased in ISE1-silenced leaves. Transient expression and intercellular movement TMV P30-2XGFP in control (A) and ISE1-silenced (B) leaves. Quantitative measurements of the number of cells to which P30-2XGFP had moved were assessed at 48 (C) and 72 (D) h postinfiltration.
Mitochondrial Function Is Disrupted in ise1 Mutants and ISE1-Silenced Plants.
The hallmarks of defective mitochondrial function are the increased production of reactive oxygen species (ROS) and reduced respiration. In fact, ise1–1 mutant embryos and seedlings (Fig. 6) and ISE1-silenced tissues (Fig. 3 I and J) exhibit increased production of the ROS H2O2, as would be expected in tissue with defective mitochondrial electron transport (25). Although ise1–1 mutants contain intact mitochondria as observed by transmission electron microscopy, respiration likely is disrupted in ise1–1 mutant embryos, because ise1–1 mutant mitochondria do not stain with MitoTracker Red (Fig. 3 K and L), a dye that is dependent upon mitochondrial transmembrane potential for specific accumulation in mitochondria (26). This latter result indicates that the electrochemical proton gradient is disrupted in the mitochondria of ise1–1 mutants.
Fig. 6.
3,3′-Diaminobenzidine staining detects increased production of H2O2 in ise1–1 mutants. Wild-type embryos (A) and seedlings (C) stain less than ise1–1 embryos (B) or seedlings (D). Seedlings were grown on Murashige and Skoog media containing 1% sucrose for 5 (C) and 10 (D) days. (Scale bars, A, B, 200 μm.)
Discussion
We show that the ISE1 gene encodes a mitochondria-localized DEAD-box RNA helicase. Absence of functional ISE1 leads to increased intercellular transport of large dextrans during Arabidopsis embryogenesis. In support of the role of ISE1 in PD-mediated intercellular transport, PD are altered in ise1 mutant embryos; specifically, ise1 mutants have more branched and twinned PD than wild-type embryos, suggesting that PD biogenesis may be up-regulated in ise1–1 mutants. Further, the ise1 phenotype can be recapitulated in mature leaf tissues by silencing the ISE1 gene; ISE1-silenced tissues exhibit increased intercellular movement of TMV P30–2XGFP. Thus, the pathway or process disrupted by the loss of ISE1 function affects transport via PD in mature and embryonic tissues. We show that ISE1 is localized to mitochondria and that the N terminus of ISE1 contains a mitochondria-targeting sequence. The disruption of mitochondrial function in ise1 mutants is supported by the failure of their mitochondria to stain with the dye MitoTracker Red, a stain that is dependent upon the proton gradient for accumulation in mitochondria. The ise1 mutants and ISE1-silenced tissues produce increased levels of ROS, likely caused by compromised mitochondrial function. We propose that the ise1 mutant phenotype is caused by defects in mitochondrial gene expression, resulting in dysfunctional mitochondria, and that when mitochondrial function is compromised, PD transport increases.
Why might mitochondrial function be defective without the putative ISE1 RNA helicase? DEAD-box RNA helicases are implicated, by biochemical, genetic and structural studies, in virtually every cellular process involving RNA unwinding or rearrangement, including transcription, ribosomal biogenesis, pre-mRNA splicing, RNA export, translation, RNA degradation, and the polar localization of developmental mRNAs (17). Thus, ISE1 may play a role in several RNA processing events in plant mitochondria. The need for RNA helicases in plant mitochondrial gene expression is acute, because the production of functional mitochondrial mRNAs is especially complex. Mitochondrial transcripts are made without systematic initiation or termination sites so that large, mostly noncoding transcripts are produced (27, 28) and noncoding regions must be degraded. Many genes have group II introns, some of which are spliced in trans. Recently, two additional (non-ISE1) DEAD-box RNA helicases have been purified from Arabidopsis mitochondria in large RNA-dependent complexes (29). Given the substantial need for RNA editing in plant mitochondria and that ISE1 is a plant-specific mitochondria-localized RNA helicase, it is tempting to suggest that ISE1 facilitates mitochondrial RNA processing or translation is tempting. Loss of these critical functions would lead to defective mitochondria and an embryo lethal phenotype.
Why might mitochondrial function regulate PD aperture or function? Potentially, tissue homeostasis requires that cells communicate stress and energy status to neighboring cells by either increasing or decreasing transport of signaling molecules or metabolites via PD. In fact, anaerobiosis, a stress specifically affecting mitochondrial oxidative phosphorylation, leads to an increase in PD SEL from <1 kDa to between 5 and 10 kDa in wheat roots (14). The ROS are candidate signal molecules, because their production increases in ise1 mutants and ISE1-silenced tissues and during anaerobiosis (30). One might then deduce that increased ROS leads to increased PD aperture.
However, a recent study demonstrated that increased ROS results in callose deposition at PD, leading to decreased PD aperture (31). These latter studies are complementary to our own, because the authors used a genetic screen to identify mutants with reduced PD aperture, designated GFP arrested trafficking (gat) mutants. gat1 is defective in a plastid thioredoxin, and gat1 mutants have increased production of ROS. Earlier studies on a maize mutant, sucrose export defective 1 (sxd1), with altered PD morphology and decreased intercellular transport of sucrose identified a chloroplast-localized protein called SXD1 (32) involved in vitamin E synthesis (33) that regulates oxidative stress. Thus, two reports show that alteration of chloroplast redox leads to decreased transport via PD.
Despite the common phenotype of increased production of ROS in gat1 and ise1, mutations in these genes have opposite effects on PD aperture. These opposing effects may be explained by the overall cellular response to different levels of ROS, where high ROS may lead to PD closure and low ROS may lead to PD opening. [Note also that chloroplasts produce at least 30 times more ROS than mitochondria (34).] Or, there are likely fundamental differences in plant cells' responses to ROS production in different organelles. Production of ROS may be perceived locally within each organelle where short-lived ROS species such as superoxide and singlet oxygen alter redox-sensitive proteins. There is a vast literature on ROS signaling, where ROS networks affect growth, cell cycle, programmed cell death, hormone signaling, biotic and abiotic stresses, and development (reviewed in ref. 35). An elegant strategy to induce H2O2 in planta identified >700 differentially expressed transcripts, including transcription factors and protein kinases that may mediate ROS signaling (36).
In summary, cellular homeostasis mediated by chloroplasts or mitochondria dramatically affects non-cell-autonomous pathways for transport of molecules between plant cells. Future studies will address the precise role of the ISE1 RNA helicase in editing and production of mitochondrial mRNAs to determine which mitochondrial gene products are affected by loss of ISE1. These studies in turn will lead to long-term studies to identify nuclear gene products that are altered in ise1 and thereby affect PD function. This study highlights how global cell physiology plays a critical role in regulating how cells interact with their neighbors by PD-mediated signal transport.
Materials and Methods
Mutant Rescue.
The ise1–3 heterozygotes were transformed with Agrobacterium carrying plasmids containing pISE1::ISE1-GFP and pISE1::GFP-ISE1. Transformants were screened on basal medium containing (20 μg/mL) hygromycin B (A.G. Scientific) (37). The T1 plants were screened for the ise1–3 T-DNA insertion using a three-primer PCR that produces a 1,500-bp product from the wild-type ISE1 allele and an 1,800-bp product from the ise1–3 mutant allele. This PCR strategy allowed detection of rescued ise1–3 homozygotes in the T1 population. Segregation of the ise1–3 mutant phenotype was tracked in the T2 generation by examining the segregation of developing mutant embryos in the siliques of T1 plants.
Transgenic N. benthamiana.
Transgenic N. benthamiana plants constitutively expressing 35S::ISE1-GFP were generated by the Ralph M. Parsons Foundation Plant Transformation Facility at University of California, Davis.
Agroinfiltration.
Nicotiana benthamiana was transformed with Agrobacterium tumefaciens GV3101 carrying plant expression constructs. After growth in liquid LB at 28 °C, bacteria were pelleted and resuspended in 100 mM MES, 100 mM MgCl2, and 200 μM acetosyringone (AS) at A600 densities of 0.001 for cell-to-cell movement assays and 1.0 for colocalization studies and induction of VIGS. Cultures were induced in AS for 2–6 h before infiltration. Nicotiana benthamiana plants were grown <24 h in light at room temperature. Cultures were injected into leaves with needleless 1-mL syringes.
Imaging.
Bright-field and epifluorescent microscopy of embryos was performed on a Zeiss Axio Imager M1 microscope. Seedlings and mature plants expressing fluorescent fusion proteins were imaged with a Zeiss SteREO Lumar epifluorescence dissecting microscope. Each microscope was fitted with a CCD camera (QImaging), and images were captured using Ivision software (BioVision Technologies) Confocal microscopy was performed on a Zeiss 510 Meta UV-vis microscope equipped with argon ion (458/488 nm) and helium/neon (543 nm) lasers. The GFP was excited with the 488-nm laser band, and emitted light was collected between 505 and 550 nm. The CFP was excited at 458 nm, and emitted light was collected between 470 and 500 nm. For colocalization images, CFP fluorescence was false colored in red to allow better visualization of its fluorescence in the presence of GFP. MitoTracker Red was excited at 543 nm, and emitted light was collected between 560 and 615 nm. Chlorophyll autofluorescence was excited at 543 nm and collected using a 560-nm long-pass filter. Images of stained leaves and seedlings were acquired with a D60 digital camera (Nikon).
Dye Loading and Staining.
Fluorescent dye loading of mutant embryos was performed as in ref. 15. 3,3′-Diaminobenzidine (DAB) staining was performed by vacuum infiltrating DAB staining solution (1 mg/mL DAB in 50 mM Tris, pH 3.9) into plant tissue. Staining was allowed to proceed overnight before removal of chlorophyll with 95% ethanol. Mitochondria were stained by incubating seedlings or embryos in 100 nM MitoTracker Red CMXRos (Invitrogen) in 1× Murashige and Skoog media in the dark for 30 min before imaging.
Transmission Electron Microscopy.
Wild-type, ise1, and ise2 embryos were prepared for transmission electron microscopy as described in ref. 22. Samples were viewed with a Philips/FEI Tecnai 12 microscope. For measurement of PD structure, at least three different sections were examined.
VIGS and Monitoring Intercellular Transport.
The NbISE1 sequence was amplified from N. benthamiana total cDNA using primers 5′-TTTCTCGAGGTGATTCAGTCGTACACAGGT-3′ and 5′-TACGGATCCGAAAAAGGCACTGTTGCAGA-3′. This fragment was cloned into pYL156, i.e., pTRV2 (24), to generate pTBS16. For a nonsilencing control, a fragment of the GUS gene was cloned into pYL156 with primers 5′TTCGAATTCTCCCCAGATGAACATGGCAT-3′ and 5′TGAGGATCCCCATCAAAGAGATC-3′ to generate pYC1. pTBS16 and pYC1 then were introduced separately into Agrobacterium GV3101. Viral-induced gene silencing then was performed on 2- to 3-week-old N. benthamiana plants according to a standard protocol (38). Tobacco rattle virus containing GUS sequences instead of the ISE1 silencing trigger acts as the nonsilencing control. Fourteen days after infiltration of VIGS constructs, the upper silenced leaves of N. benthamiana plants were agroinfiltrated with cultures for expression of TMV MP30–2XGFP. Samples from these leaves then were observed at 48 or 72 h postinfiltration on a Zeiss510 Meta confocal laser scanning microscope. The MP30-GFP foci were located, and the number of cells in each focus was counted. Approximately 50 lesions and >4 experiments were scored for each treatment.
Supplementary Material
Acknowledgments.
We thank Steve Ruzin and Denise Schichnes of the College of Natural Resources Biological Imaging Facility for excellent advice and support and Bob Buchanan and Frank Van Breusegem for critical discussions of redox regulation. This work was supported by a National Science Foundation predoctoral fellowship (to S.S.), a UCB Miller postdoctoral fellowship (to T.B.-S.), Syngenta and David Patton (to D.M.), National Institutes of Health Grant HG-000205 to Ron Davis (to M.M.), and National Institutes of Health Grant GM45244 (to P.Z.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909229106/DCSupplemental.
References
- 1.Cilia ML, Jackson D. Plasmodesmata form and function. Curr Opin Cell Biol. 2004;16:500–506. doi: 10.1016/j.ceb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Kim I, Cho E, Crawford K, Hempel FD, Zambryski PC. Cell-to-cell movement of GFP during embryogenesis and early seedling development in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:2227–2231. doi: 10.1073/pnas.0409193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crawford KM, Zambryski PC. Non-targeted and targeted protein movement through plasmodesmata in leaves in different developmental and physiological states. Plant Physiol. 2001;125:1802–1812. doi: 10.1104/pp.125.4.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurata T, Okada K, Wada T. Intercellular movement of transcription factors. Curr Opin Plant Biol. 2005;8:600–605. doi: 10.1016/j.pbi.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Lucas WJ, et al. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science. 1995;270:1980–1983. doi: 10.1126/science.270.5244.1980. [DOI] [PubMed] [Google Scholar]
- 6.Kalantidis K, Schumacher HT, Alexiadis T, Helm JM. RNA silencing movement in plants. Biol Cell. 2008;100:13–26. doi: 10.1042/BC20070079. [DOI] [PubMed] [Google Scholar]
- 7.Ashby J, et al. Tobacco mosaic virus movement protein functions as a structural microtubule-associated protein. J Virol. 2006;80:8329–8344. doi: 10.1128/JVI.00540-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JY, Rim Y, Wang J, Jackson D. A novel cell-to-cell trafficking assay indicates that the KNOX homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev. 2005;19:788–793. doi: 10.1101/gad.332805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JY, et al. Selective trafficking of non-cell-autonomous proteins mediated by NtNCAPP1. Science. 2003;299:392–396. doi: 10.1126/science.1077813. [DOI] [PubMed] [Google Scholar]
- 10.Crawford KM, Zambryski PC. Subcellular localization determines the availability of non-targeted proteins to plasmodesmatal transport. Curr Biol. 2000;10:1032–1040. doi: 10.1016/s0960-9822(00)00657-6. [DOI] [PubMed] [Google Scholar]
- 11.Rinne PLH, van der Schoot C. Plasmodesmata at the crossroads between development, dormancy, and defense. Botany. 2003;81:1182–1197. [Google Scholar]
- 12.Sivaguru M, et al. Aluminum-induced 1→3-β-d-glucan inhibits cell-to-cell trafficking of molecules through plasmodesmata. A new mechanism of aluminum toxicity in plants. Plant Physiol. 2000;124:991–1005. doi: 10.1104/pp.124.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy A, Erlanger M, Rosenthal M, Epel BL. A plasmodesmata-associated β-1,3-glucanase in Arabidopsis. Plant J. 2007;49:669–682. doi: 10.1111/j.1365-313X.2006.02986.x. [DOI] [PubMed] [Google Scholar]
- 14.Cleland RE, Fujiwara T, Lucas WJ. Plasmodesmal-mediated cell-to-cell transport in wheat roots is modulated by anaerobic stress. Protoplasma. 1994;178:81–85. doi: 10.1007/BF01404123. [DOI] [PubMed] [Google Scholar]
- 15.Kim I, Hempel FD, Sha K, Pfluger J, Zambryski PC. Identification of a developmental transition in plasmodesmatal function during embryogenesis in Arabidopsis thaliana. Development. 2002;129:1261–1272. doi: 10.1242/dev.129.5.1261. [DOI] [PubMed] [Google Scholar]
- 16.Mingam A, et al. DEAD-box RNA helicases in Arabidopsis thaliana: Establishing a link between quantitative expression, gene structure and evolution of a family of genes. Plant Biotechnol J. 2004;2:401–415. doi: 10.1111/j.1467-7652.2004.00084.x. [DOI] [PubMed] [Google Scholar]
- 17.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 18.Schmid SR, Linder P. Translation initiation factor 4A from Saccharomyces cerevisiae: Analysis of residues conserved in the D-E-A-D family of RNA helicases. Mol Cell Biol. 1991;11:3463–3471. doi: 10.1128/mcb.11.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 20.McElver J, et al. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics. 2001;159:1751–1763. doi: 10.1093/genetics/159.4.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niwa Y, Hirano T, Yoshimoto K, Shimizu M, Kobayashi H. Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J. 1999;18:455–463. doi: 10.1046/j.1365-313x.1999.00464.x. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi K, Otegui MS, Krishnakumar S, Mindrinos M, Zambryski P. INCREASED SIZE EXCLUSION LIMIT 2 encodes a putative DEVH box RNA helicase involved in plasmodesmata function during Arabidopsis embryogenesis. Plant Cell. 2007;19:1885–1897. doi: 10.1105/tpc.106.045666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faulkner C, Akman OE, Bell K, Jeffree C, Oparka K. Peeking into pit fields: A multiple twinning model of secondary plasmodesmata formation in tobacco. Plant Cell. 2008;20:1504–1518. doi: 10.1105/tpc.107.056903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato. Plant J. 2002;31:777–786. doi: 10.1046/j.1365-313x.2002.01394.x. [DOI] [PubMed] [Google Scholar]
- 25.Igamberdiev AU, Hill RD. Plant mitochondrial function during anaerobiosis. Ann Bot. 2009;103:259–268. doi: 10.1093/aob/mcn100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hawes CR, Satiat-Jeunemaitre B. Plant Cell Biology: A Practical Approach. New York: Oxford Univ Press; 2001. [Google Scholar]
- 27.Kubo T, Mikami T. Organization and variation of angiosperm mitochondrial genome. Physiol Plant. 2007;129:6–13. [Google Scholar]
- 28.Holec S, Lange H, Canaday J, Gagliardi D. Coping with cryptic and defective transcripts in plant mitochondria. Biochim Biophys Acta. 2008;1779:566–573. doi: 10.1016/j.bbagrm.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Matthes A, et al. Two DEAD-box proteins may be part of RNA-dependent high-molecular-mass protein complexes in Arabidopsis mitochondria. Plant Physiol. 2007;145:1637–1646. doi: 10.1104/pp.107.108076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blokhina OB, Chirkova TV, Fagerstedt KV. Anoxic stress leads to hydrogen peroxide formation in plant cells. J Exp Bot. 2001;52:1179–1190. [PubMed] [Google Scholar]
- 31.Benitez-Alfonso Y, et al. Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc Natl Acad Sci USA. 2009;106:3615–3620. doi: 10.1073/pnas.0808717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Provencher LM, Miao L, Sinha N, Lucas WJ. Sucrose Export Defective 1 encodes a novel protein implicated in chloroplast-to-nucleus signaling. Plant Cell. 2001;13:1127–1141. doi: 10.1105/tpc.13.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Porfirova S, Bergmuller E, Tropf S, Lemke R, Dormann P. Isolation of an Arabidopsis mutant lacking vitamin E and identification of a cyclase essential for all tocopherol biosynthesis. Proc Natl Acad Sci USA. 2002;99:12495–12500. doi: 10.1073/pnas.182330899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foyer CH, Noctor G. Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol Plant. 2003;119:355–364. [Google Scholar]
- 35.Gechev TS, Van Breusegem F, Stone JM, Denev I, Laloi C. Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays. 2006;28:1091–1101. doi: 10.1002/bies.20493. [DOI] [PubMed] [Google Scholar]
- 36.Vandenabeele S, et al. A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proc Natl Acad Sci USA. 2003;100:16113–16118. doi: 10.1073/pnas.2136610100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakazawa M, Matsui M. Selection of hygromycin-resistant Arabidopsis seedlings. Biotechniques. 2003;34:28–30. doi: 10.2144/03341bm02. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, et al. Virus induced gene silencing of a DEFICIENS ortholog in Nicotiana benthamiana. Plant Mol Biol. 2004;54:701–711. doi: 10.1023/B:PLAN.0000040899.53378.83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.