Abstract
Plasma cells daily secrete their own mass in antibodies, which fold and assemble in the endoplasmic reticulum (ER). To reach these levels, cells require pERp1, a novel lymphocyte-specific small ER-resident protein, which attains expression levels as high as BiP when B cells differentiate into plasma cells. Although pERp1 has no homology with known ER proteins, it does contain a CXXC motif typical for oxidoreductases. In steady state, the CXXC cysteines are locked by two parallel disulfide bonds with a downstream C(X)6C motif, and pERp1 displays only modest oxidoreductase activity. pERp1 emerged as a dedicated folding factor for IgM, associating with both heavy and light chains and promoting assembly and secretion of mature IgM.
Key to an effective antibody-mediated immune system is that under no condition antibody “leaks” from nonactivated B lymphocytes, whereas on activation, massive secretion starts, and then only of completely assembled antibody. Although they do produce subunits, resting B cells do not secrete antibody. Only when cells are activated by antigen or mitogen do they differentiate into plasma cells, which secrete their own mass in antibody molecules per day (1). The conversion to an antibody-secreting plasma cell requires a “total makeover” of the lymphocyte: All cellular machineries are reorganized for the single purpose of bulk antibody production (2–4). Most striking is the change in volume of endoplasmic reticulum (ER), because this organelle accommodates the biosynthesis and assembly of antibody.
The ER is the first compartment of the secretory pathway; it supports disulfide bond formation, folding, and oligomerization of newly synthesized proteins. Efficiency in the folding process is accomplished through assistance by an abundance of both “generic” and tissue- or substrate-specific chaperones and folding enzymes (5, 6). The ER harbors a single prominent and highly conserved HSP70 family member BiP, but also contains a variety (>20) of PDI family oxidoreductases with CXXC active site motifs (7). They all seem to be involved in the oxidation, reduction, and/or isomerization of disulfide bonds, but how they divide or share these tasks and their substrates is largely unknown.
IgM is a challenging client for the plasma cell ER. The IgM subunits undergo oxidative folding and form interchain disulfide bonds during their stepwise assembly into mature secretory protein. In the end, IgM consists of at least 21 subunits [10 μ heavy (H) chains, 10 λ or κ light (L) chains, and a single J chain] and counts ≈75 intrachain and ≈25 interchain disulfide bonds (1). Besides the increase of “generic” ER folding factors that are present already in the resting B cell, specialized folding assistants may enrich the ER of plasma cells and even be required for efficient IgM maturation and secretion.
Here, we report on a previously undescribed dedicated folding assistant of IgM: the lymphocyte-specific ER-resident protein pERp1. In the course of B cell differentiation, pERp1 was up-regulated more than any other protein: from nearly undetectable to abundance in the same range as GRP94 and BiP in the plasma cell. It associated with IgM H and L chains, promoted their assembly, and thereby, the secretion of mature IgM polymers.
Results
The Novel 18-kDa Protein Is Strongly Up-Regulated During B Cell Differentiation.
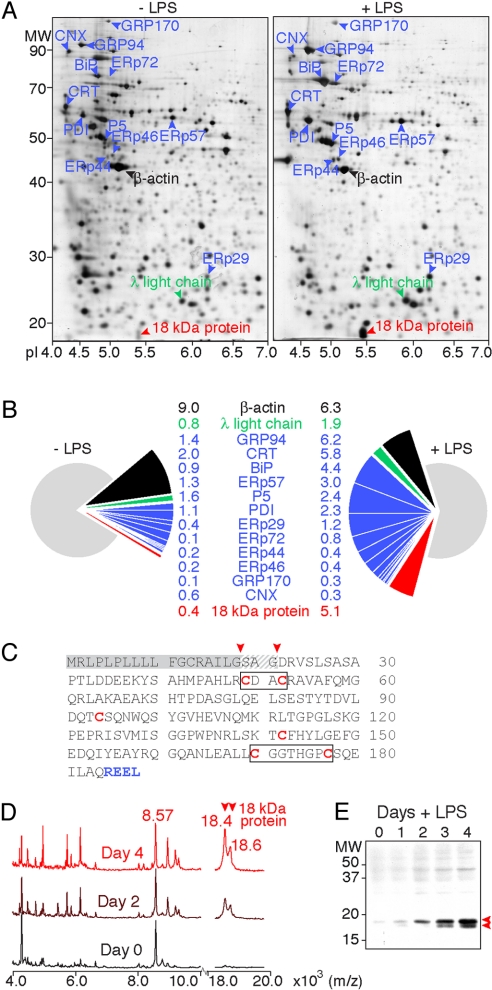
Using a dynamic proteomic approach on LPS-activated murine I.29μ+ (IgM, λ) lymphomas as model B lymphocytes (8), we found that, next to IgM subunits, the ER-resident proteins dramatically increased (3). The relationship between function and expression pattern identified a candidate ER-resident protein of 18 kDa (Fig. 1A) with expression levels that rose to ≈5% of total signal in the silver-stained gel after 5 days. Staining with SYPRO-Ruby gave similar results. This protein ranked among the most highly expressed proteins in plasma cells, with actin and the ER proteins GRP94, calreticulin, and BiP (Fig. 1B).
Fig. 1.
A novel 18-kDa protein is heavily up-regulated during B cell differentiation. (A) Two-dimensional gel analysis of the I.29μ+ B cell proteome before or after activation with LPS for 5 days. Proteins were first separated by pI using isoelectric focusing in the horizontal direction (x axis, pI) and next separated by molecular mass using standard 10% SDS PAGE in the vertical direction (y axis, MW). ER-resident chaperones and folding enzymes [blue, calnexin (CNX) and calreticulin (CRT)], λ L chain (green), the most abundant protein β-actin (black), and the novel 18-kDa protein (red) are indicated with arrowheads. (B) Relative abundance of proteins annotated in A as determined by densitometry of silver stained gels and quantitation by PDQuest. The percentage within the total proteome before or after activation is given and depicted in pie diagrams; color-coding as in A. (C) Sequence of the novel 18-kDa protein. Indicated are signal peptide (gray), cysteines (red), and the C-terminal peptide that conveys ER retention (blue). Arrowheads mark the two alternative signal peptide cleavage sites. The CXXC and C(X)6C motifs are boxed. (D) Profiling of I.29μ+ B lymphomas. Crude lysates were analyzed by using MALDI-ToF, either before or after stimulation with LPS for 2 or 4 days. Spectra were normalized to the highest peak in the spectrum (8.6 kDa). (E) Immunoblot analysis of lysates from I.29μ+ B lymphomas, before or after activation with LPS for 1, 2, 3, or 4 days with an antiserum raised against the novel 18-kDa protein.
Sequencing of tryptic fragments by MS revealed the identity of the 18.4-kDa (188-residue) protein (Swiss-Prot: Q9D8I1; Fig. 1C). It was annotated before by homology with the human gene MGC29506, which frequently is down-regulated transcriptionally in intestinal-type gastric cancer (9), and as the PACAP protein, implicated in apoptosis (10). However, the latter protein was deviant from residue 59 because of a frame shift (9). No sequence homology existed with any known family of ER proteins.
We then compared the proteome at various days of differentiation by direct profiling of lysates from I.29μ+ lymphomas by using MALDI-ToF-MS. This technique is complementary to 2D gel electrophoresis, as it allows detection of low molecular mass proteins only (11). The spectra displayed several ion signals that increased with time (Fig. 1D). In line with the novel protein's highly abundant expression, its signal rose from undetectable levels to the most prominent amid all proteins smaller than 20 kDa in the cell at 4 days after activation. Western blotting confirmed that the novel protein was heavily up-regulated in the course of B cell differentiation (Fig. 1E).
The Novel 18-kDa Protein Is ER Resident.
The hydrophobic N terminus of the novel protein (Fig. 1C) was predicted (9) to be a cleavable signal peptide. Two partly overlapping fragments corresponding to residues D22-K38 and S19-R48 did not arise from trypsin digestion alone, because neither D22 nor S19 flanks a K or R residue. The hydrophobic N terminus of the protein, thus, was cleaved off at either of two sites (Fig. 1C), which explains why the “main” 18.4-kDa peak (Fig. 1D) was accompanied by an 18.6-kDa “shoulder” peak, and why the protein spot in the 2D gel was spread out in the vertical (mass) direction (Fig. 1A) and appeared as a doublet in Fig. 1E. Further confirmation came from the exact mass of the main and shoulder peaks as determined by ESI-Q-ToF MS (Fig. S1 and Table S1). To biochemically validate that the signal peptide mediates ER entry, we in vitro translated and radiolabeled the novel protein in the absence or presence of semipermeabilized HT1080 cells as a source of ER membrane (12). The novel protein indeed was translocated across the ER membrane, whereupon its signal peptide was removed by the signal peptidase (Fig. S2A).
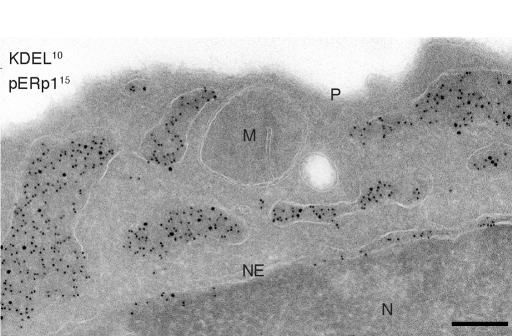
Whereas its N terminus mediates entry, the C-terminal REEL sequence (Fig. 1C) may well serve as ER retention signal, like the canonical KDEL (13). Indirect immunofluorescence indeed showed colocalization with the ER marker PDI (Fig. S2B). We confirmed its ER residency in LPS treated I.29μ+ cells by ImmunoGold labeling and electron microscopy. The novel protein exclusively populated ER cisternae, identified through its content of KDEL-containing resident ER proteins (Fig. 2). We named the novel protein plasma cell induced ER-resident protein 1 (pERp1).
Fig. 2.
The novel 18-kDa protein is resident to the ER. Subcellular localization of the 18-kDa protein by immunoelectron microscopy. I.29μ+ B cells were activated for 4 days with LPS before fixation and ImmunoGold labeling with protein A-gold conjugated α-pERp1 (15 nm) and α-KDEL antisera (10 nm), which recognizes several ER-resident proteins (including BiP, PDI, and CRT). Cryosections were analyzed by electron microscopy; the nucleus (N), nuclear envelope (NE), mitochondrion (M), and plasma membrane (P) are indicated. The empty structure is an endocytic vesicle, the ER is dilated and labeled with the two sizes gold. (Scale bar, 200 nm.)
Expression of pERp1 Is Lymphocyte Specific.
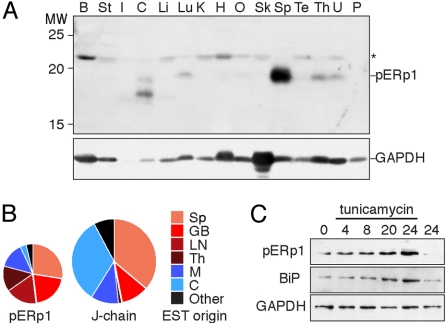
When comparing levels of pERp1 by tissue immunoblotting, we found that pERp1 expression was restricted to spleen and (to a far lesser extent) thymus, lung, and uterus (Fig. 3A). Spleen and thymus typically harbor significant populations of lymphocytes, and pERp1-specific ESTs surfaced most in cDNA libraries derived from germinal B cells or from tissues rich in lymphocytes: spleen, lymph nodes, or thymus (Fig. 3B). In human cancerous tissues, most pERp1 ESTs were from leukemia or lymphoma derived cDNA libraries (data not shown). Whereas J chain is an exclusive subunit of antibodies, expressed only in B lymphocytes, it was less exclusively enriched in lymphoid tissues than pERp1. We found it also in mammary and intestinal tissue ESTs, because J chain is a subunit of both IgM and IgA, the antibody type secreted by the majority of plasma cells that reside in these tissues. Thus, pERp1 expression appears most prominently up-regulated in Ig secreting plasma cells. A Northern blotting on human tissues by Shimizu et al. (14) showed a prominent pERp1 signal in thymus, suggesting that pERp1 in mammals is specific not only to B, but also to T lymphocytes.
Fig. 3.
pERp1 is a lymphocyte-specific protein with highest abundance in the spleen. (A) Immunoblot analysis using α-pERp1 antiserum of a commercially available (Zyagen) blot with normalized amounts of proteins from various murine tissues (75 μg per tissue). Tissues are: B, brain; St, stomach; I, intestine; C, colon; Li, liver; Lu, lung; K, kidney; H, heart; O, ovary; Sk, skeletal muscle; Sp, spleen; Te, testis; Th, thymus; U, uterus; and P, placenta. The asterisk indicates a background band. The same blot was analyzed with an α-GAPDH antiserum for comparison. (B) All murine ESTs corresponding with pERp1 or J-chain were retrieved from the National Center for Biotechnology Information EST database on September 15, 2008. We categorized ESTs by tissue origin represented in pie diagrams. ESTs from cancerous or mixed origin were omitted from the analysis. Differences in size of pie diagrams are in proportion to the number of ESTs found for pERp1 (n = 29) and J chain (n = 64). Slices representing ESTs from lymphoid tissues are color coded in shades of red, from other secretory tissues in shades of blue, and from other sources in black. Tissues are abbreviated as in A except GB, germinal B cell; LN, lymph node; and M, mammary gland. (C) Immunoblot analysis using α-pERp1, α-BiP, or α-GAPDH antisera of lysates from I.29μ+ B lymphomas, before or after indicated time (hours) in the presence of the ER stressor tunicamycin (3 μg/mL) or after 24 h without the drug. The UPR target BiP serves as positive reference and GAPDH as loading control.
As pERp1 rose to abundant levels in the course of B cell differentiation, similar to known ER proteins (Fig. 1B), we tested whether pERp1 expression was regulated by the unfolded protein response (UPR). This signaling pathway is turned on when cells suffer from ER stress, but also when precursor cells (like B cells) differentiate into professional secretory cells (15–17). We treated cells with tunicamycin, and found that pERp1 expression was up-regulated in I.29μ+ cells (Fig. 3C), but remained undetectable in control (HT1080) cells (data not shown). Based on these and similar results of Shimizu et al. (14), we considered that pERp1 is not a “conventional” UPR target gene, but instead a conditional one: it becomes a UPR target in the context of the B cell.
pERp1 Has a Putative Redox-Active CXXC Motif That Interacts with a C(X)6C Motif.
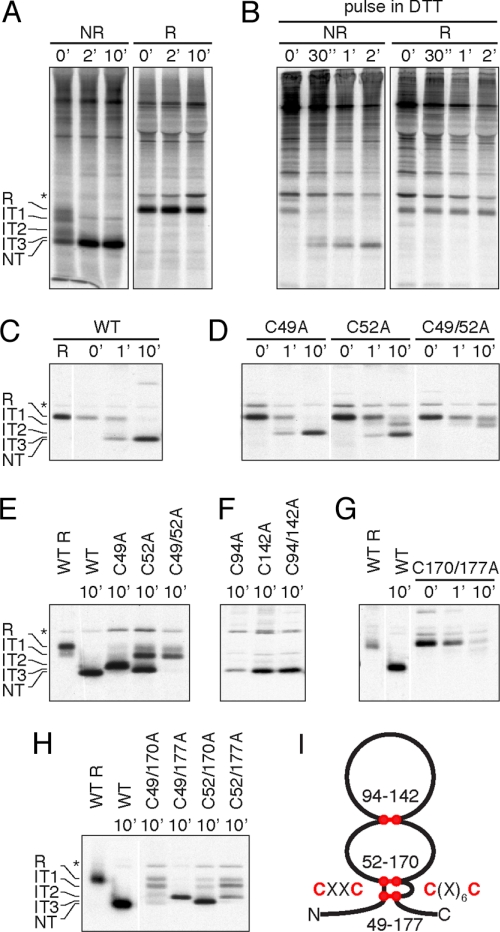
After signal peptide removal, pERp1 counts six cysteines (Fig. 1C) that are strictly conserved among pERp1 homologs (9), suggesting that they are important for structure and/or function. Of particular interest is the CXXC motif formed by the two N-terminal cysteines (Fig. 1C), which may be a redox-active site like the CXXC motifs in the otherwise unrelated PDI family members. From the exact mass determination data, we deduced that in I.29μ+ cells pERp1 was fully oxidized, because oxidation of six sulfhydryl groups into three disulfide bonds entails loss of 6 Da (Table S1). In pulse–chase analysis, pERp1 displayed faster mobility in nonreducing gels than in reducing gels, and thus, acquired a native disulfide bonded, more compact state, already during the pulse (Fig. 4A). Even when disulfide bond formation was postponed by adding the reducing agent DTT in the pulse, reduced pERp1 folded into an oxidized native form within minutes of chase (Fig. 4B).
Fig. 4.
Disulfide-bonded structure of pERp1. (A) Oxidative folding of pERp1. I.29μ+ B lymphomas were activated for 4 days with LPS and pulse-labeled for 2 min and chased for indicated times followed by nonreducing and reducing SDS/PAGE. Reduced pERp1 (R) rapidly oxidizes via a series of folding intermediates (IT1–IT3) with incomplete set of disulfide bonds to a native (NT) fully disulfide-bonded form. The asterisk indicates a background band. (B) Postponed oxidative folding of pERp1. Analysis of pERp1 folding was performed as in A with DTT present during the pulse to postpone disulfide bond formation until the chase. (C) Oxidative folding of pERp1 in HeLa cells. Cells were infected with vaccinia virus VVT7 and transfected with pBS-pERp1 WT, followed by postponed oxidative folding analysis as in B. The nonreducing gel is shown with a reduced sample (WT R) for reference. (D) Oxidative folding analysis as in C of single cysteine mutants C49A, C52A, and the corresponding double mutant C49/52A. (E) Folding “endpoint” analysis of pERp1 for WT and mutants as in D. Oxidative folding was analyzed as in A. The 10-min chase samples were run next to each other on gel to compare the endpoint of oxidative folding. Note that the final, most oxidized form of various mutants corresponds with different folding intermediates of the WT. (F) Folding endpoint analysis as in E of C94A, C142A, and C94/142A mutants. (G) Oxidative folding as in C of the C170/177A double mutant. (H) Folding endpoint analysis as in E of “cross-motif” mutants C49/170A, C49/177A, C52/170A, and C52/177A. (I) Schematic representation of the disulfide-bonded structure of native pERp1 with disulfide bonds shown in red. Note the parallel disulfide bonds between the CXXC and C(X)6C motifs.
To map the disulfide bonded structure of pERp1, we constructed a series of cysteine to alanine mutants. Because the high levels of endogenous pERp1 in I.29μ+ cells would confound analysis of mutants, we expressed them in HeLa cells and validated the expression system by radioactive pulse–chase analysis (Fig. 4C). Although the single CXXC mutations, C49A and C52A, did not prevent pERp1 from reaching native-like mobility in nonreducing gels, mobility of the double C49/52A mutant was markedly lower (Fig. 4 D and E). This finding implies that a long-range disulfide bond was disrupted, which rules out the existence of a native disulfide bond between the CXXC cysteines. Mutation of C94 or C142 alone or in combination did not prevent pERp1 from forming this long-range disulfide bond. Instead, all three mutants displayed a similar, minor loss in mobility (Fig. 4F), which suggests that these two cysteines together form native disulfide bond 94–142.
Therefore, the CXXC cysteines must team up with the two remaining cysteines close to the C terminus, which form a C(X)6C motif (Fig. 1C). Indeed, the double mutant of the C(X)6C cysteines, C170/177A, failed to form long-range disulfide bonds (Fig. 4G), similar to the CXXC double mutant C49/52A (Fig. 4D). To establish which cysteines in the CXXC and C(X)6C motifs form pairs, we analyzed all four combinations of cross-motif double mutants. Long-range disulfide bonds formed only in the C49/177A and the C52/170A mutants (Fig. 4H), implying native disulfide bonds 52–170 and 49–177.
The arrangement of disulfide bonds is in keeping with data obtained on the steady state expression and activity of various mutant pairs (14) and with MS. Analysis of tryptic fragments of nonreduced versus reduced recombinant pERp1 confirmed that C94 forms a disulfide bond with C142, and that the CXXC and C(X)6C motifs were connected via two disulfide bonds (data not shown). This technique did not allow confirmation of the arrangement of the cross-motif disulfide bonds because of a lack of trypsin cleavage sites within these motifs. Thus, we mapped the disulfide-bonded structure of pERp1 as schematically represented in Fig. 4I.
pERp1 Displays Modest Oxidoreductase Activity on Model Substrates.
The fact that cysteines of pERp1 form long-range disulfide bonds does not exclude that they can serve a role in thiol oxidoreductase function. For example, cysteines of the two active site motifs in Ero1α cooperate in the thiol oxidation of PDI, but are also important for structure and stability of the protein (18). Because pERp1 can form mixed disulfide bonds with IgM monomers (14), it may similarly shuttle between a disulfide bonded “locked” and a thiol-active “open” conformation. To test whether pERp1 has oxidoreductase activity, we examined its influence on oxidative folding in vivo by pulse–chase analysis of model ER clients, influenza hemagglutinin, HIV-1 envelope glycoprotein, and the LDL receptor, but found no clear effects.
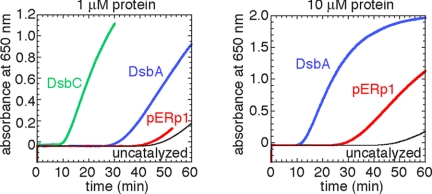
We then tested disulfide isomerase and reductase activity of pERp1 in vitro. In the reductase assay using insulin as substrate, pERp1 displayed low disulfide reductase activity compared with the generic dithioloxidase DsbA and the generic disulfide isomerase DsbC from Escherichia coli (Fig. 5). The results were similar in the scrambled RNase A isomerase assay (data not shown). These findings indicate that pERp1 does have oxidoreductase activity, although very modest. Shimizu et al. (14) showed that mutation of the CXXC cysteines did not affect in vivo activity, suggesting that either the CXXC motif is not enzymatically active or that other cysteines contribute to activity. Unfortunately, it has not been possible yet to distinguish a structural role from a role in oxidoreductase activity for all disulfide bonds (14).
Fig. 5.
Thiol reductase activity of pERp1 in vitro. Activity was assayed by using 140 mM insulin as substrate and 1 μM (Left) or 10 μM (Right) pERp1 and, for reference, E. coli DsbA and DsbC as catalysts. The onset of aggregation (increased optical density at 650 nm) is a measure for reductase efficiency.
pERp1 Interacts with IgM Subunits, Stimulates Their Assembly, and Promotes Secretion of Mature IgM.
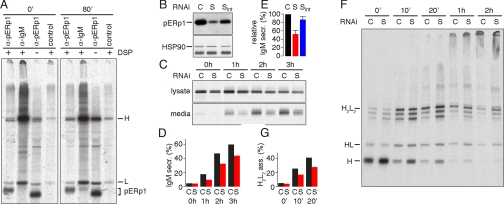
The exclusive, but highly abundant expression of pERp1 in activated B cells prompted us to test whether pERp1 serves as a folding assistant in the maturation of IgM, the bulk secretory product in plasma cells. We pulse-labeled differentiated I.29μ+ cells for 5 min, chased for 80 min or not, lysed cells in detergent in the presence or absence of the cross-linker dithiobis(succinimidylpropionate) (DSP), and immunoprecipitated proteins with antisera against pERp1 or IgM. Both H and L chain coimmunoprecipitated with pERp1 after the pulse, but also still after the chase, implying long-term interactions between pERp1 and IgM subunits (Fig. 6A). The interaction with pERp1 was strong enough to withstand immunoprecipitation conditions, because cross-linking did not enhance this effect. The complementary experiment showed that only 80-min-old and not newly synthesized radiolabeled pERp1 associated with IgM, consistent with maturation of pERp1 being required for the interaction. At early time points, associating pERp1 is unlabeled and, thus, undetectable. Despite the modest oxidoreductase activity of pERp1 (Fig. 5), interaction with the majority of IgM subunits was independent of any putative active site cysteines, because coimmunoprecipitation was identical in the presence of DTT (Fig. S3). This finding suggests that pERp1 interacts with IgM subunits in a chaperone-like manner.
Fig. 6.
pERp1 interacts with H and L chains and promotes assembly and secretion of IgM. (A) pERp1 associates with IgM subunits in I.29μ+ cells. Cells were activated with 20 mg/mL LPS for 3 days, pulse-labeled for 5 min, and lysed before or after a chase of 80 min in the presence or absence of cross-linker DSP. Lysates were immunoprecipitated with α-pERp1, α-IgM, or control antibodies. pERp1, IgM H and L chain are indicated. Note that the covalent association of pERp1 with the cross-linker causes a slight mobility shift. (B) Down-regulation of pERp1 in I.29μ+ lymphomas by using RNAi. Immunoblot analysis with α-pERp1 or, for reference, with α-HSP90 antibodies of lysates from 3-day activated I.29μ+ cells in which pERp1 expression was stably silenced at two different levels: (S) and (Sint). A control cell line (C) was generated in parallel with luciferase as target gene instead of pERp1. Selection and puromycin concentrations were identical for all cell lines. (C) Decreased IgM secretion from I.29μ+ lymphomas with stably silenced pERp1. Control cells and the cells with strongest silencing (S) were pulse labeled for 5 min and chased for indicated periods (hours). Culture media were harvested and cells were lysed. Equivalent samples from culture media and lysates were immunoprecipitated with α-IgM. Samples were analyzed by reducing 10% SDS/PAGE. (D) Quantitation of IgM secretion. Signals from C were quantified, and the percentage of secretion per sample is represented in a histogram showing quantization for control cells in black and for pERp1 silenced cells in red. (E) Decrease of IgM secretion by pERp1 silencing. Percentage of secretion of the silenced clone (S) was compared with that of the control cell at identical time points, setting the control at 100% (black bar). Samples from 30 min, 1, and 2 h chase from experiments shown in D and Fig. S3, as well as from replicate experiments were used for the analysis. Means and SEM are shown; n = 6 (S) in red; n = 2 (Sint) in blue. (F) Decrease in IgM assembly in I.29μ+ cells with stably silenced pERp1. Pulse–chase analysis as in C for indicated chase periods (minutes). Immunoprecipitates from lysates were analyzed by nonreducing 4–20% gradient SDS/PAGE. Assembly intermediates HL and H2L2 are indicated. (G) Quantitation of H2L2 assembly. Signals from F, showing the early phases of IgM assembly (0-, 10-, and 20-min chase), were quantified, specifically the monomeric H2L2 and submonomeric intermediates H, HL, and the two other bands below H2L2 that likely represent H2 and H2L. Percentage of H2L2 among this collection of subunits and intermediates is shown in a histogram; color-coding as in D.
To test whether pERp1 assists folding and assembly of IgM, we silenced expression of pERp1 in I.29μ+ cells by shRNA and selected a series of clones from two different silencing constructs. In the silenced clone (S) with the strongest down-regulation of pERp1, <10% of endogenous levels remain (Fig. 6B). We monitored kinetics of antibody secretion by pulse–chase analysis and found that, compared with the luciferase control (C), pERp1 silencing lowered IgM secretion (Fig. 6 C and D). The difference was most pronounced ≈1 h of chase with the silenced cells secreting 55.6 ± 5.7% IgM compared with control cells (Fig. 6E). Another clone with intermediate silencing (Sint) showed almost normal secretion of IgM (Fig. 6E; Fig. S4), suggesting that pERp1 levels in the plasma cell are in excess of need, and that a complete lack of pERp1 expression will have an even stronger diminishing effect on IgM secretion.
The decrease in IgM secretion was accounted for by slower assembly kinetics of the subunits. With time, assembly progresses from H via HL to H2L2 and finally to “multimers” of H2L2“monomers” before secretion of mature IgM (2). Immediately after the pulse, the disulfide-linked assembly intermediates were present in control and pERp1 silenced cells in similar ratios, with H being the most prominent intermediate (Fig. 6F). The pace of the assembly process was markedly lower in pERp1 silenced cells than in control cells. Within 10–20 min, the most prominent intermediate in control cells was H2L2, but in pERp1 silenced cells H was more abundant still (Fig. 6F). At later times, the signal for monomeric H2L2 and “submonomeric” intermediates substantially decreased (Fig. 6E) because of secretion of mature IgM, in particular from control cells. Because secretion confounds assessment of the fraction H2L2 assembled from its subunits, we calculated values only for the earlier time points (Fig. 6G). We concluded that pERp1 promotes IgM assembly and secretion, because pERp1 down-regulation significantly impeded these processes.
Discussion
We identified pERp1, a lymphocyte-specific ER-resident protein that reaches levels on a par with the most abundant known ER-resident chaperones once B lymphocytes differentiate into plasma cells. Its up-regulation is >15-fold, which is at least 3-fold higher than that of any other detectable protein, indicative of its importance in plasma cells. pERp1 associates with Ig H and L chains and stimulates disulfide-linked assembly and secretion of mature IgM.
Consistent with the notion that pERp1 is key for IgM production in plasma cells, it appears to be lymphocyte specific. Although we found highest protein expression in the spleen of mice, Shimizu et al. (14) found the highest expression of pERp1 transcripts in thymus of humans. Irrespective of differences being due to species or protein versus mRNA, pERp1 expression is highest in lymphocyte rich tissues. Consistent with lymphocyte-specific expression and a role in acquired immunity, pERp1 has closely related homologs only in mammals or birds, but not in species with a less sophisticated immune system (data not shown; ref. 9). Accordingly, pERp1 expression in human cancers was almost exclusive to leukemia and lymphoma (data not shown), but down-regulated in stomach cancers (9). pERp1 could well serve as diagnostic marker for these immunological cancers, considering its abundant, but exclusive expression.
Tissue specificity for ER-resident folding factors is not unprecedented. The PDI family member PDIp is exclusively expressed in the pancreas (19) and PDILT, in testis (20). As a lymphocyte-specific protein, pERp1 likely is under control of a transcription factor specific for lymphocyte lineages. Interestingly, in B cells, pERp1 is a target of the UPR, similar to generic ER-resident proteins. This finding is compatible with a model in which pERp1 has UPR-specific promoter elements that are repressed in nonlymphoid cells, and that become responsive to ER stress only when the repression is lifted in lymphoid cells, although the fact that another UPR inducer, thapsigargin, did not up-regulate pERp1 (14) suggests that its regulation is likely to be quite complex. Other tissue-specific ER proteins may be similarly regulated as conditional UPR targets, i.e., via the interplay of tissue-specific signaling pathways with the UPR.
The list of novel chaperones and folding assistants in the ER is growing, in particular the PDI family (7). Although pERp1 does have a CXXC motif, which is the signature motif in the thioredoxin-like redox-active domains of PDI, pERp1 is no PDI/thioredoxin family member, since pERp1 is mainly α-helical (our unpublished observations; ref.14) unlike the thioredoxin fold. No relation was found with any other known protein either. Instead, pERp1 is the founding member of a previously undescribed protein family.
The complexity of IgM subunit folding and multimeric assembly is mirrored by the intricacy of its assembly cascade, which is lined with many chaperones and folding assistants in the ER: BiP (21), GRP94 (22), PDI, ERp72 (23), which together form a complex with P5 (also known as CaBP1), ERdj3, cyclophilin B, and UDP-glucosyltransferase (24). Shimizu et al. (14) identified pERp1 as part of this complex. Recently, also ERp44 and ERGIC53 were added to the list of assistants of IgM assembly (25). During folding and assembly of the IgM chains, all these ER or ERGIC proteins bind transiently, some shorter and some for longer periods, either simultaneously or in sequence. For example, early in the process, GRP94 acts after BiP (22), by promoting folding of H chains and assembly with L chains to HL “hemimers,” whereas much later, ERGIC53 acts after ERp44, when they assist assembly of H2L2 monomers into multimers (25).
IgM does mature in the absence of pERp1, but the reduction of IgM secretion on pERp1 down-regulation is impressive for any ER folding factor. For example, maturation of few glycoproteins is affected by deletion of either calnexin or calreticulin. A severe loss in ER function only becomes manifest once both lectins are absent (26). The absence of ERGIC53 leads to deficiencies in clotting factors V and VIII, but not to Ig deficiency (27). A link between pERp1 and disease has not been found, but its deficiency may lead to selective IgM deficiency, a condition linked to high susceptibility to persistent and life-threatening infections.
The question is how pERp1 exerts its effects. It may serve the IgM maturation process as a molecular chaperone, because it associates noncovalently to most assembly intermediates. As alternative scenario, or in addition, pERp1 may act as oxidoreductase. It has modest oxidoreductase activity, does form an interchain disulfide with IgM monomers and stimulates oxidation of Ig domains (14). It may have a role in the formation of the ≈100 disulfide bonds in IgM during folding and assembly, or in the B-cell-specific redox switch that releases the μ tailpiece cysteines from ER-resident oxidoreductases like PDI and ERp57 in the differentiating B cell (23, 28). In either case, the modest basal oxidoreductase activity of pERp1 may become enhanced by the unique conditions in the plasma cell. The preservation of activity in the SXXC mutant (14) shows that this protein is far from a standard oxidoreductase.
The interaction of pERp1 with many assembly intermediates from the moment of synthesis (our results; ref. 14) suggests pERp1 to be among the longest interacting folding assistants up to now, together with BiP. Although BiP is absolutely required for IgM folding (1), as it is for ER function in general (29), pERp1 may sustain the efficiency in IgM production that the plasma cell requires as a dedicated antibody factory.
Methods
Detailed methods for all approaches used are provided in the SI Methods.
B Cell and Protein Analyses.
I.29μ+ cells were activated with 20 μg/mL LPS and cells and harvested before or after different days of differentiation for various analyses (SI Methods). Cells lysates were analyzed by immunoblotting or 2D gel electrophoresis, and proteins were identified as described (3). Immunoelectron microscopy was done as described (30).
Pulse–Chase Analyses.
Pulse–chases were performed as described (31) in I.29μ+ cells or in HeLa cells expressing pERp1 or its mutants by using the vaccinia virus T7 infection-transfection system (32). Detergent lysates of radiolabeled cells or media were subjected to immunoprecipitation with protein A-Sepharose-coupled antisera. Immunoprecipitates were washed and heated in sample buffer in the presence (reducing) or absence (nonreducing) of DTT before SDS/PAGE analysis.
Silencing.
I.29μ+ cells were transduced with shRNA lentiviral vectors to silence pERp1, with an isogenic control targeting the reporter luciferase.
In Vitro Thiol Oxidoreductase Assay.
Recombinant pERp1 was compared with E. coli DsbA and DsbC as catalyst of insulin reduction with DTT by measuring the increase in turbidity at 650 nm, as described (33).
Supplementary Material
Acknowledgments.
We thank Yuichiro Shimizu, Linda Hendershot, Stefan Rüdiger, and members of the Braakman lab, in particular Mieko Otsu, for fruitful discussions. The I.29μ+ B cell line was a kind gift from Roberto Sitia (DiBiT-HSR, Milan, Italy). This work was supported by grants from EuroSCOPE (I.B. and A.O.), Zon-MW (I.B.), NWO-CW (I.B. and A.J.R.H.), The Netherlands Proteomics Centre (A.J.R.H.), the Swiss National Science Foundation (J.L.), the National Institute of Allergy and Infectious Diseases (J.L.), and the Sigrid Jusélius Foundation (A.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903036106/DCSupplemental.
References
- 1.Hendershot LM, Sitia R. In: Molecular Biology of B Cells. Honjo TAF, Neuberger MS, editors. Amsterdam: Elsevier; 2005. pp. 261–273. [Google Scholar]
- 2.Romijn EP, et al. Expression clustering reveals detailed co-expression patterns of functionally related proteins during B cell differentiation: A proteomic study using a combination of one-dimensional gel electrophoresis, LC-MS/MS, and stable isotope labeling by amino acids in cell culture (SILAC) Mol Cell Proteomics. 2005;4:1297–1310. doi: 10.1074/mcp.M500123-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.van Anken E, et al. Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity. 2003;18:243–253. doi: 10.1016/s1074-7613(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 4.Wiest DL, et al. Membrane biogenesis during B cell differentiation: Most endoplasmic reticulum proteins are expressed coordinately. J Cell Biol. 1990;110:1501–1511. doi: 10.1083/jcb.110.5.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 6.van Anken E, Braakman I. Versatility of the endoplasmic reticulum protein folding factory. Crit Rev Biochem Mol Biol. 2005;40:191–228. doi: 10.1080/10409230591008161. [DOI] [PubMed] [Google Scholar]
- 7.Appenzeller-Herzog C, Ellgaard L. The human PDI family: Versatility packed into a single fold. Biochim Biophys Acta. 2008;1783:535–548. doi: 10.1016/j.bbamcr.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Alberini C, Biassoni R, DeAmbrosis S, Vismara D, Sitia R. Differentiation in the murine B cell lymphoma I. 29: Individual mu + clones may be induced by lipopolysaccharide to both IgM secretion and isotype switching. Eur J Immunol. 1987;17:555–562. doi: 10.1002/eji.1830170419. [DOI] [PubMed] [Google Scholar]
- 9.Katoh M, Katoh M. MGC29506 gene, frequently down-regulated in intestinal-type gastric cancer, encodes secreted-type protein with conserved cysteine residues. Int J Oncol. 2003;23:235–241. [PubMed] [Google Scholar]
- 10.Bonfoco E, Li E, Kolbinger F, Cooper NR. Characterization of a novel proapoptotic caspase-2- and caspase-9-binding protein. J Biol Chem. 2001;276:29242–29250. doi: 10.1074/jbc.M100684200. [DOI] [PubMed] [Google Scholar]
- 11.Dalluge JJ. Mass spectrometry for direct determination of proteins in cells: Applications in biotechnology and microbiology. Fresenius J Anal Chem. 2000;366:701–711. doi: 10.1007/s002160051564. [DOI] [PubMed] [Google Scholar]
- 12.Wilson R, et al. The translocation, folding, assembly and redox-dependent degradation of secretory and membrane proteins in semi-permeabilized mammalian cells. Biochem J. 1995;307:679–687. doi: 10.1042/bj3070679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munro S, Pelham HR. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 14.Shimizu Y, et al. pERp1 is significantly upregulated during plasma cell differentiation and contributes to the oxidative folding of immunoglobulin. 2009 doi: 10.1073/pnas.0811591106. 10.1073/pnas.0811591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Anken E, Braakman I. Endoplasmic reticulum stress and the making of a professional secretory cell. Crit Rev Biochem Mol Biol. 2005;40:269–283. doi: 10.1080/10409230500315352. [DOI] [PubMed] [Google Scholar]
- 16.Brewer JW, Hendershot LM. Building an antibody factory: A job for the unfolded protein response. Nat Immunol. 2005;6:23–29. doi: 10.1038/ni1149. [DOI] [PubMed] [Google Scholar]
- 17.Iwakoshi NN, et al. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- 18.Bertoli G, et al. Two conserved cysteine triads in human Ero1alpha cooperate for efficient disulfide bond formation in the endoplasmic reticulum. J Biol Chem. 2004;279:30047–30052. doi: 10.1074/jbc.M403192200. [DOI] [PubMed] [Google Scholar]
- 19.Desilva MG, et al. Characterization and chromosomal localization of a new protein disulfide isomerase, PDIp, highly expressed in human pancreas. DNA Cell Biol. 1996;15:9–16. doi: 10.1089/dna.1996.15.9. [DOI] [PubMed] [Google Scholar]
- 20.van Lith M, Hartigan N, Hatch J, Benham AM. A divergent testis-specific PDI with a non-classical SXXC motif that engages in disulfide dependent interactions in the endoplasmic reticulum. J Biol Chem. 2004;280:1376–1383. doi: 10.1074/jbc.M408651200. [DOI] [PubMed] [Google Scholar]
- 21.Haas IG, Wabl M. Immunoglobulin heavy chain binding protein. Nature. 1983;306:387–389. doi: 10.1038/306387a0. [DOI] [PubMed] [Google Scholar]
- 22.Melnick J, Dul JL, Argon Y. Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature. 1994;370:373–375. doi: 10.1038/370373a0. [DOI] [PubMed] [Google Scholar]
- 23.Reddy P, Sparvoli A, Fagioli C, Fassina G, Sitia R. Formation of reversible disulfide bonds with the protein matrix of the endoplasmic reticulum correlates with the retention of unassembled Ig light chains. EMBO J. 1996;15:2077–2085. [PMC free article] [PubMed] [Google Scholar]
- 24.Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anelli T, et al. Sequential steps and checkpoints in the early exocytic compartment during secretory IgM biogenesis. EMBO J. 2007;26:4177–4188. doi: 10.1038/sj.emboj.7601844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molinari M, et al. Contrasting functions of calreticulin and calnexin in glycoprotein folding and ER quality control. Mol Cell. 2004;13:125–135. doi: 10.1016/s1097-2765(03)00494-5. [DOI] [PubMed] [Google Scholar]
- 27.Nichols WC, et al. Mutations in the ER-Golgi intermediate compartment protein ERGIC-53 cause combined deficiency of coagulation factors V and VIII. Cell. 1998;93:61–70. doi: 10.1016/s0092-8674(00)81146-0. [DOI] [PubMed] [Google Scholar]
- 28.Sitia R, et al. Developmental regulation of IgM secretion: The role of the carboxy-terminal cysteine. Cell. 1990;60:781–790. doi: 10.1016/0092-8674(90)90092-s. [DOI] [PubMed] [Google Scholar]
- 29.Hendershot LM. The ER chaperone BiP is a master regulator of ER function. Mt Sinai J Med. 2004;71:289–297. [PubMed] [Google Scholar]
- 30.Slot JW, Geuze HJ. Cryosectioning and immunolabeling. Nat Protoc. 2007;2:2480–2491. doi: 10.1038/nprot.2007.365. [DOI] [PubMed] [Google Scholar]
- 31.Braakman I, Hoover-Litty H, Wagner KR, Helenius A. Folding of influenza hemagglutinin in the endoplasmic reticulum. J Cell Biol. 1991;114:401–411. doi: 10.1083/jcb.114.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuerst TR, Niles EG, Studier FW, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frickel EM, et al. ERp57 is a multifunctional thiol-disulfide oxidoreductase. J Biol Chem. 2004;279:18277–18287. doi: 10.1074/jbc.M314089200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.