Abstract
Cell-surface pili are important virulence factors that enable bacterial pathogens to adhere to specific host tissues and modulate host immune response. Relatively little is known about the structure of Gram-positive bacterial pili, which are built by the sortase-catalyzed covalent crosslinking of individual pilin proteins. Here we report the 1.6-Å resolution crystal structure of the shaft pilin component SpaA from Corynebacterium diphtheriae, revealing both common and unique features. The SpaA pilin comprises 3 tandem Ig-like domains, with characteristic folds related to those typically found in non-pilus adhesins. Whereas both the middle and the C-terminal domains contain an intramolecular Lys–Asn isopeptide bond, previously detected in the shaft pilins of Streptococcus pyogenes and Bacillus cereus, the middle Ig-like domain also harbors a calcium ion, and the C-terminal domain contains a disulfide bond. By mass spectrometry, we show that the SpaA monomers are cross-linked in the assembled pili by a Lys–Thr isopeptide bond, as predicted by previous genetic studies. Together, our results reveal that despite profound dissimilarities in primary sequences, the shaft pilins of Gram-positive pathogens have strikingly similar tertiary structures, suggesting a modular backbone construction, including stabilizing intermolecular and intramolecular isopeptide bonds.
Keywords: crystal structure, polymerization, bacterial pilus, mass spectrometry, pilin motif
Pili are long, thin protein assemblies that extend from the cell surface of many bacteria and play pivotal roles in colonization and pathogenesis. Like Gram-negative bacteria, many Gram-positive pathogens express pili on their surface (1–3). These structures have aroused great interest because of their direct roles in infection and pathogenesis and their importance as vaccine candidates (2, 3). They also use covalent isopeptide (amide) bonds, both intermolecular and intramolecular, to give strength and stability, and thus present a new paradigm among protein polymers (4–6).
Unlike Gram-negative pili, whose subunits associate via noncovalent interactions, these Gram-positive pili are formed by covalent polymerization of pilin subunits, orchestrated by transpeptidase enzymes called sortases (6, 7). The general principles of assembly were first established through studies on the SpaA pili expressed by Corynebacterium diphtheriae. These pili, encoded by the gene cluster spaA-spaB-srtA-spaC, comprise a polymeric shaft formed by SpaA, SpaC located at the tip, and SpaB found at the base and occasionally along the shaft (6, 8–10). In our current model, successive major pilin SpaA subunits are joined by the action of the pilus-specific sortase SrtA. Cleavage between Thr-494 and Gly-495 of the LPXTG motif near the SpaA C terminus is followed by presumed amide bond formation between the new C terminus and Lys-190 from a conserved YPKN pilin motif in the next subunit (6). Finally, the entire assembly is covalently attached to the cell wall peptidoglycan by a housekeeping sortase (9, 11). Presumably, the tip pilin SpaC is linked to the SpaA shaft via the same reaction as recently identified for the tip pilin BcpB of Bacillus cereus (12). It remains unclear how the minor pilin SpaB is incorporated into the pilus structure, although recent evidence indicates that SpaB forms the basal subunit for tethering the pilus to the cell wall and that it, too, is attached to the polymer via a specific Lys residue (9).
A major step forward in understanding Gram-positive pilus structure and assembly came with the structural analysis of the major pilin Spy0128 from Streptococcus pyogenes (5). Spy0128 does not have a recognizable pilin motif, but the crystal structure revealed columns of molecules resembling a putative polymer assembly and identified a candidate lysine; this was then confirmed by mass spectral analysis of native S. pyogenes pili. The structure also revealed unexpected internal crosslinks in the form of self-generated isopeptide bonds, 1 in each domain of the 2-domain structure, joining Lys and Asn side chains. These are strategically located to give strength and stability to the pilus assembly.
The major pilins of different Gram-positive bacteria show wide variations in size and sequence, making it difficult to predict whether the structural principles seen for S. pyogenes apply also to other Gram-positive pili. Here we present the high-resolution crystal structure of SpaA, the archetypal major pilin from C. diphtheriae. This reveals a modular structure comprising 3 tandem Ig-like domains, 2 of which contain internal Lys–Asn isopeptide bonds like those in Spy0128. We also confirm, by mass spectrometry, the identity of the lysine used in polymerization and note a pilus-like assembly of SpaA molecules in the crystal. These results point to a likely common architecture for the backbones of many Gram-positive pili and consolidate a new paradigm for the structure, stability, and assembly of these remarkable covalent polymers.
Results
Structure Determination.
A construct comprising residues 53–486 of C. diphtheriae SpaA was expressed in Escherichia coli, purified, and crystallized. This lacks residues 1–52 encompassing the signal peptide and ends 4 residues before the sortase-recognition LPXTG motif. The crystal structure, with 1 SpaA molecule per asymmetric unit, was solved by single wavelength anomalous dispersion methods and refined at 1.6-Å resolution (R = 19.3%, Rfree = 22.0%) [supporting information (SI) Table S1]. Only the N-terminal Glu-53, an external loop 69–79, and the C-terminal residues 485–486 could not be modeled for lack of interpretable electron density.
Modular Structure of SpaA.
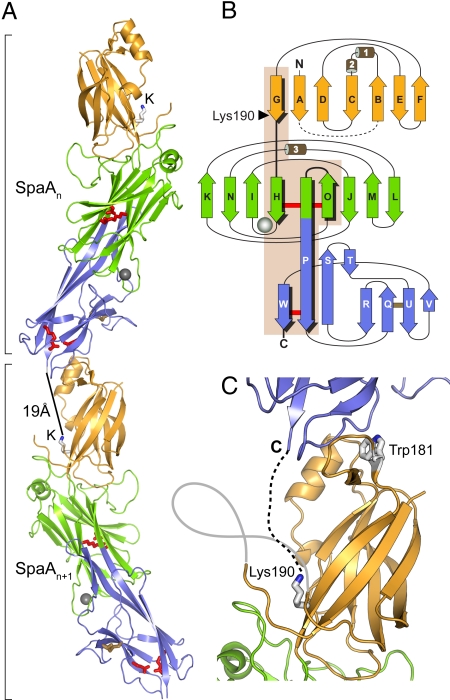
SpaA is folded into 3 tandem Ig-type domains, giving an elongated molecule of ≈105 Å in length. Whereas the middle domain (M-domain; residues 193–351) and the C-terminal domain (C-domain; residues 352–484) are arranged linearly and share an extended strand P (Fig. 1), the N-terminal domain (N-domain; residues 54–192) sits on the M-domain at an angle of ≈20° to the long axis of the molecule. The SpaA molecules pack in columns through the crystal, in a manner resembling a pilus assembly; the N-domain of each molecule abuts against the C-domain of the next (Fig. 1A).
Fig. 1.
Crystal structure of SpaA. (A) End-to-end stacking of successive molecules, in which the C-domain of one molecule, SpaAn, packs against the N-domain of the next, SpaAn + 1. For each molecule, the N-domain is in gold, M-domain green, and C-domain blue. The M-domain Ca2+ ion is shown as a gray sphere. Residues that form isopeptide bonds in the M- and C-domains are shown in red stick mode, and the disulfide bond in the C-domain is in yellow. The pilin motif lysine, Lys-190, in each molecule is labeled K. A black bar shows the distance between the C terminus of one molecule and Lys-190 on the next. (B) Schematic representation of the fold of each domain colored as in A. Helices are labeled 1 to 3, and β-strands A to W. The isopeptide bond crosslinks are shown with red bars and the disulfide bond with a yellow bar. An arrowhead points to Lys-190. Strands and loops connected by inter- or intramolecular isopeptide bonds are highlighted on a beige background. (C) Close view of the packing of adjacent molecules. Lys-190 projects upward, between the disordered AB loop (gray line) and the N-domain, toward the C terminus of the next molecule (C). A broken line shows where the 10 missing C-terminal residues would bridge the ≈19-Å gap to Lys-190 on the next molecule. The conserved Trp-181 of the pilin motif is in the interface between molecules.
The N- and C-domains both have the inverse IgG fold first described for the CnaB domains of the collagen binding protein Cna from Staphylococcus aureus (13). This comprises a β-sandwich of 7 strands. When superimposed, the 2 CnaB-type domains of SpaA show an rmsd in Cα positions of 2.2 Å over 88 equivalent residues and share 14% sequence identity. A distinctive feature of the N-domain is the presence of 2 helices between strands B and C that partially cover one side of the core, whereas the C-domain uniquely contains an elongated β-ribbon, formed by strands S and T, running toward the M-domain. In contrast, the M-domain of SpaA has the CnaA fold, first seen in the N2 domain of S. aureus CnaA (14). This comprises 9 β-strands that form a partially open β-barrel.
The closest structural homologues of the N- and C-domains of SpaA are the 2 CnaB-type domains of the Streptococcus agalactiae minor pilin GBS52 (15). In particular, the SpaA C-domain has significant sequence identity (25%) and structural similarity (rmsd 1.7 Å over 91 equivalent Cα atoms) with the N2 domain of GBS52. The 2 domains of the S. pyogenes major pilin Spy0128 also share the same CnaB-type fold, albeit with some elaborations (5). Structural superpositions of the Spy0128 domains on to the SpaA N- and C-domains give rmsds in Cα positions ranging from 2.5 to 3.1 Å and sequence identities ranging from 3% to 17%. The M-domain shows strong structural homology with the N2 domain of CnaA, despite minimal sequence identity (≈8%); the rmsd is 3.4 Å over 123 equivalent Cα atoms. Other similar CnaA-type domains include the N3 domain of S. aureus clumping factor A and the N2 domain of the Enterococcus faecalis collagen-binding protein Ace (16, 17).
Internal Isopeptide Bonds and Other Stabilizing Features.
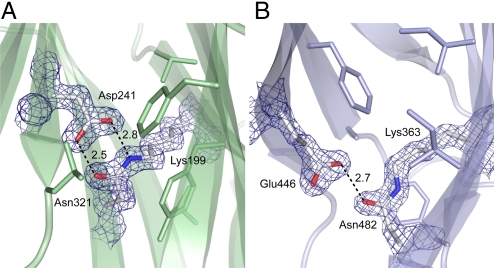
The M- and C-domains of SpaA both contain stabilizing internal isopeptide bonds, formed by intramolecular reaction between the Lys ε-amino group and the carboxyamide group of Asn. These were clearly apparent in the initial experimentally phased electron density map, in which continuous density linked the side chains of Lys-199 and Asn-321 in the M-domain and Lys-363 and Asn-482 in the C-domain (Fig. 2). The existence of these Lys–Asn isopeptide bonds was confirmed by electrospray ionization–time-of-flight mass spectrometry. The protein Mr was measured as 46,795.4 Da, ≈34 Da less than that calculated from the amino acid sequence, 46,829.6 Da, consistent with loss of 2 units of NH3 through formation of 2 isopeptide bonds. Confirmation was obtained by digestion of the recombinant protein and analysis by liquid chromatography–tandem mass spectrometry (LC-MS/MS). Parent ions with mass-to-charge ratio (m/z) 996.83+ and 1067.53+ contained the M-domain isopeptide bond between Lys-199 and Asn-321, and a parent ion with m/z 751.73+ contained the C-domain linkage between Lys-363 and Asn-482 (Tables S2 and S3).
Fig. 2.
Internal isopeptide bonds in SpaA. Residues involved in bond formation are in stick mode, colored by atom type, with surrounding hydrophobic residues also shown. Hydrogen bonds are shown with broken lines, distances in Å. The electron density is from an (Fobs - Fcal) · Phical map, contoured at 3σ. (A) The M-domain isopeptide bond formed between Lys-199 and Asn-321, with catalytic Asp-241. (B) The C-domain isopeptide bond between Lys-363 and Asn-482, with catalytic Glu-446.
Similar Lys-Asn isopeptide bonds were first observed in the structure of Spy0128, where an associated Glu residue was shown to be essential for the intramolecular reaction to occur (5). In SpaA, the same role is played by Asp-241 for the M-domain isopeptide bond and Glu-446 for the C-domain bond (Fig. 2).
Two isomeric forms of isopeptide bond are found in SpaA. The M-domain bond (Lys-199–Asn-321) has a cis configuration, as for both isopeptide bonds of Spy0128, allowing its NH and O moieties to form a bidentate hydrogen-bonded interaction with the Asp-241 carboxyl group (Fig. 2). In contrast, the C-domain bond (Lys-363–Asn-482) has a trans configuration and only a single hydrogen bond with the carboxyl group of Glu-446. The hydrogen bonding patterns imply that both carboxyl groups are protonated. Both isopeptide bonds are located in the interior of their respective domains, surrounded by hydrophobic residues. Both also stack against aromatic residues, Tyr-219 in the M-domain and Phe-365 in the C-domain (Fig. 2). Other surrounding hydrophobic residues include Phe-306, Val-352, Val-221, and Leu-243 in the M-domain and Phe-378, Ile-361, Ile-480, and Ala-376 in the C-domain. This hydrophobic environment favors a nonprotonated Lys amino group and protonated Glu/Asp carboxyl group, thus facilitating the intramolecular reaction (5, 18).
The SpaA structure has 2 other notable stabilizing features. In the M-domain a metal binding site is formed by the loop joining strands H and I, with the metal ion coordinated by 8 oxygen atoms, from Asp-204, Asp-205, Gln-208, Gly-210, Glu-215, and 2 water molecules (Fig. 1). The coordination environment and average metal-ligand bond length (2.48 Å) are indicative of a Ca2+ ion, presumably cell derived. This is a unique feature, not seen before in any other CnaB- or CnaA-like domain, and its persistence despite the use of 1 mM EDTA in buffers implies a high affinity. In the C-domain, a disulfide bond joins Cys-383 on strand Q to Cys-443 on strand U (Fig. 1). The electron density shows that this bond is incompletely formed, with approximately 40% of molecules having both Cys reduced. This may result from the DTT needed for tag cleavage, and we anticipate that the disulfide would be fully formed in vivo, in the oxidizing extracellular environment.
Interestingly, whereas SpaA has no internal isopeptide bond in its N-domain, such a bond is present in the N-domain of the 3-domain major pilin BcpA from B. cereus, joining Lys-37 and Asn-163 (4). These residues are conserved in other pilins but replaced by Ala-61 and His-191, respectively, in SpaA (Fig. S1). In the SpaA structure, the Ala and His side chains are close enough such that if replaced by Lys and Asn, as in other pilins, an isopeptide bond could be formed. A conserved Glu that could catalyze Lys–Asn bond formation is present in the other pilins but in SpaA is replaced by Gln-153, positioned close to Ala-61 and His-191 (Fig. S2).
Sequence Elements Implicated in Pilus Assembly.
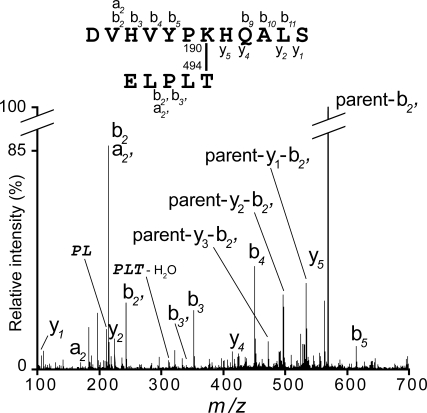
Sequence comparisons and mutagenesis have identified 2 conserved sequence motifs that contain residues essential for pilus assembly. The pilin motif (consensus WxxxVxVYPK) contains the lysine that is joined to the C terminus of the next molecule during polymerization; Lys-190 in SpaA. We sought to confirm the role of Lys-190 in SpaA polymer formation, using mass spectrometry. Purified SpaA pilus polymers were separated by SDS-PAGE and digested in-gel with trypsin and AspN endopeptidase. The digestion products were analyzed by LC-MS/MS and the intersubunit amide bond identified from a peptide peak with m/z 585.73+. The Mr of this peptide corresponded exactly with that expected, allowing for the loss of 18 Da due to water elimination during amide bond formation. The fragment ion spectra uniquely identified the peptides surrounding the pilin motif Lys-190 and the sortase-cleaved C-terminal Thr-494, respectively (Fig. 3 and Table S4).
Fig. 3.
Intersubunit isopeptide bonds in SpaA. Fragmentation spectra of the parent ion at m/z 585.73+ containing the intersubunit bond between SpaA Lys-190 and Thr-494 are shown. Ion types are indicated, and internal ions are shown in italics. The structure of crosslinked peptide is shown above the spectra. Daughter ions produced during MS/MS of these peptides are summarized in Table S4.
In the SpaA structure, the pilin motif is located on βG, the last strand of the N-domain. Lys-190 is close to the point where βG crosses to the M-domain, becoming βH (Fig. 1). Nine residues before Lys-190 is the conserved Trp-181, the first residue of the pilin motif, and 9 residues after is Lys-199, which forms the M-domain isopeptide bond. The side chain of Lys-190 projects into a cleft between the main body of the N-domain and a mobile loop, residues 63–83 (Fig. 1C). Head-to-tail packing of molecules in the crystal places the ε-amino group of Lys-190 ≈19 Å from the C terminus of the next molecule, ample distance to accommodate the 10 missing residues between the last modeled residue, Lys-484, and the true sortase cleavage site, Thr-494. It also places Trp-181 in the interface between the 2 molecules (Fig. 1C); the conservation of this residue supports the idea that the crystal packing models the true biologic assembly. The fact that the M-domain isopeptide bond closely follows Lys-190, on the same extended β-strand, suggests why polymer formation is abrogated by deletion of the equivalent isopeptide bond in B. cereus BcpA (4); local structural destabilization could prevent proper presentation of the essential lysine to the sortase.
The second sequence motif implicated in assembly is the E-box motif (consensus YxLxETxAPxGY). This contains a conserved glutamate, Glu-446 in SpaA, which is essential for the incorporation of the minor pilins SpaB and SpaC (7). Intriguingly, Glu-446 proves to be the catalytic Glu that mediates formation of the Lys-363–Asn-462 intramolecular bond. Because Glu-446 is internal, we infer that it is the stability imparted by the intramolecular crosslink that is essential for minor pilin incorporation.
Discussion
The discovery of thin, hair-like pili on the surface of C. diphtheriae in 2003, and their characterization as covalent polymers, was a milestone in understanding colonization and infection by Gram-positive bacteria (6). Similar pilus assemblies are found for such important human pathogens as Group A and B streptococci, Streptococcus pneumoniae, and B. cereus (19–21), but the pilin subunits involved show large variations in size and sequence that mask any possible structural homology.
The present structure for the C. diphtheriae major pilin SpaA resolves this question, revealing a modular assembly that utilizes Ig-like domains similar to those used in the S. pyogenes major pilin Spy0128, despite very low sequence identity. These domains correspond to 2 types of Ig-like domain, the CnaB and CnaA folds that are widely used in the cell-surface adhesins known as MSCRAMMS (microbial surface components recognizing adhesive matrix molecules) (22). Prototype CnaB and CnaA domains are present in the multidomain S. aureus Cna protein, which has a structural B-region with repeating doublets of CnaB domains, preceded by a collagen-binding A-region with 2 CnaA domains (22, 23).
Spy0128, the major pilin of S. pyogenes, is one of the smallest shaft pilins (32.5 kDa) and comprises 2 tandem CnaB domains (5). SpaA, in contrast, is significantly larger (47 kDa) and has a single CnaA-type domain, the M-domain, inserted between 2 CnaB-type domains. This mosaic architecture suggests an evolutionary process in which copies or pieces of older genes are assembled to form new genes. It seems likely that all of the major pilins of sortase-assembled Gram-positive pili conform to the same structural principles. In many cases, for example the major pilins SpaD and SpaH that form the 2 other types of pilus produced by C. diphtheriae, sufficient sequence identity exists to infer similar structures; these share ≈24% identity with SpaA, including the intramolecular isopeptide bond-forming residues, the pilin motif, and the Cys residues. In others, such as Spy0128, there is much less sequence similarity.
Sequence comparisons with the major pilins from other Gram-positive bacteria, such as S. pneumoniae, S. agalactiae, and B. cereus, reveal N- and C-terminal regions that are similar to the N- and C-domains of SpaA, including the N-domain pilin motif and the C-domain isopeptide bond-forming residues. The middle regions are more variable; some may form CnaA-type domains like the SpaA M-domain, with substantial insertions/deletions, whereas others may have CnaB-type middle domains or adopt entirely different folds.
There is growing evidence that this modular architecture extends also to the minor pilin subunits. GBS52, a minor pilin from S. agalactiae that seems to correspond functionally to SpaB, comprises 2 CnaB-type domains (15), and its C-terminal N2 domain closely resembles the SpaA C-domain, including the internal isopeptide bond. Sequence comparisons show that the S. pyogenes minor pilin Cpa also has a C-terminal domain homologous with the C-domain of the S. pyogenes major pilin Spy0128, again including an internal isopeptide bond (5). This leads to the concept that incorporation of the minor pilins is facilitated by their structural resemblance to the pilins that comprise the polymeric shaft.
The 2 internal isopeptide bonds in SpaA confirm that such crosslinks are a common feature of Gram-positive pili. First discovered in the crystal structure of Spy0128 and confirmed by mass spectral analysis of native GAS pili (5), they have also been found in the B. cereus major pilin BcpA, which has 3 such bonds (4). The combined sequence and structural data from these 3 characterized major pilins now enable internal isopeptide bonds to be inferred from the sequences of other major pilins. Re-evaluation of the crystal structures of GBS52 and the A- and B-domains of S. aureus Cna further showed that similar isopeptide bonds are also present in minor pilins and adhesins (5).
The SpaA structure further defines the determinants of intramolecular isopeptide bond formation. Mutagenesis has shown that a catalytic carboxyl group is essential for bond formation (4, 5, 24); this can be Asp or Glu, as shown by the use of Asp-241 in the M-domain and Glu-446 in the C-domain. The isopeptide moiety can have either cis or trans configuration, with corresponding bidentate or monodentate interaction with the essential carboxyl side chain. The main requirement is proximity of the Lys–Asn pair and a hydrophobic environment in which both the Lys and Asp/Glu are uncharged. The locations of these isopeptide bonds seem to be characteristic of the folds of the domains in which they occur, and hence probably reflect their evolutionary history. In the CnaB-like C-domain of SpaA, the isopeptide bond joins the first and last β-strands, as in both CnaB-type domains of Spy0128. In contrast, in the CnaA-type M-domain, the isopeptide bond bridges the first and second-last strands, linking 2 opposing β-sheets.
Given the widespread occurrence of internal isopeptide crosslinks in the cell-surface proteins of Gram-positive bacteria, what is their structural and functional importance? We have shown that the intramolecular isopeptide bonds in Spy0128 strongly enhance thermodynamic stability and resistance to proteolysis (24). SpaA additionally contains a disulfide bond, conserved in the other C. diphtheriae major pilins, which might be expected to further enhance stability. The major pilins of S. pyogenes, B. cereus, and S. pneumoniae lack Cys residues, however, and given that many Gram-positive bacteria lack the disulfide formation machinery of Gram-negative bacteria (25, 26), we speculate that isopeptide bonds have evolved as an alternative means of stabilization. As amide bonds they would also be less prone to chemical disruption than disulfide bonds, a property that may be important for such thin, exposed assemblies, which do not seem to form higher-order bundles.
We further hypothesize that their strategic location gives mechanical (force-bearing) stability. Both in these pili, typified by Spy0128 and SpaA, and in multidomain adhesins such as Cna, an almost linear chain of covalent connectivity can be traced along the long axis (5, 27). In SpaA this begins with Lys-190, the site of attachment to the preceding subunit, and extends through the M- and C-domains to the next intermolecular linkage, possibly explaining why the N-domain does not require an isopeptide bond. The attachment of adhesins to host cells subjects them to significant tensile stress, and their structures are thought to have evolved both to withstand stress and to use it to optimize binding (28).
There is also growing evidence that the structural stabilization conferred by the internal isopeptide bonds can be critical for molecular recognition. Defective sortase recognition or failure to present the Lys residue of the pilin motif appropriately would explain the loss of pilus assembly when intramolecular isopeptide bonds in SpaA or BcpA are deleted by mutation of the catalytic Asp/Glu. The isopeptide bonds in CnaA and GBS52 may similarly influence the binding of partner molecules, because their isopeptide bond-containing domains are important for specific binding of collagen (CnaA) and capable of binding to human pulmonary epithelial cells (GBS52) (15, 29).
Finally, an intriguing feature of the crystal structures of both SpaA and Spy0128 is the way the pilin molecules pack end-to-end in columns in the crystal, resembling assembled pili. The intermolecular contacts do not seem to be particularly extensive (buried surface 850 Å2 for Spy0128 and 814 Å2 for SpaA), yet in each case the molecules pack such that the critical lysine residue (Lys-161 in Spy0128, Lys-190 in SpaA) is brought close to the C terminus of the next molecule. Importantly, in SpaA the conserved Trp-181 of the pilin motif forms part of this interface, consistent with a role in oligomer formation. The roles of the conserved Tyr-188 and Pro-189 of the pilin motif are less clear, but patches of electron density around them may represent parts of the unmodeled AB loop. These residues may interact transiently with this loop, which could in turn mediate the pilin–pilin and/or pilin–sortase interactions.
Materials and Methods
Cloning and Protein Purification.
DNA encoding amino acids 53–486 of SpaA from C. diphtheriae was amplified by PCR from genomic DNA, cloned, over-expressed in E. coli as an N-terminally His-tagged protein, and purified by nickel-affinity chromatography (30). After His-tag removal and final size-exclusion chromatography, the protein was concentrated to 100 mg/mL in 10 mM Tris-HCl (pH 8.0) and 50 mM NaCl. Selenomethionine (SeMet)-substituted SpaA was produced using the methionine biosynthesis inhibition method (31) and similarly purified, but with 5 mM DTT and 1 mM EDTA in the final gel filtration buffer.
Crystallization and Structure Determination.
Crystals were grown in sitting drops comprising 100 nL protein (100 mg/mL) and 100 nL precipitant. The best native SpaA crystals were obtained with 20% PEG 3350, 0.1 M NaI, and 0.1 M NaF as precipitant and SeMet-SpaA crystals with 20% PEG 3350 and 0.2 M Na formate. Crystals were flash-cooled without further cryoprotection. X-ray diffraction data were collected on beamline PX1 at the Australian Synchrotron, to 1.6 Å and 1.8 Å resolutions, respectively, for native and SeMet-SpaA crystals. Data were processed and scaled with MOSFLM and SCALA (32). All 4 Se atoms were located by SHELX (33) with refinement and phase determination in autoSHARP (34). Density modification and model building with PHENIX (35, 36) placed 408 of 436 residues, and model building was completed using COOT (37). The model was refined using REFMAC (38). Data collection, phasing, and refinement statistics are in Table S1. Structural superpositions were done with SSM (39).
Isolation of SpaA Pili.
Engineered SpaA pili were produced by transformation of the plasmid pAG153, encoding SpaA and SrtA, into a C. diphtheriae strain that lacks spaA and srtA, followed by expression and purification of the polymers (6). These procedures are described more fully in SI Materials and Methods. The use of a His-tagged SpaA construct with a SrtA construct lacking 13 C-terminal residues means that the engineered SpaA pili are secreted into the culture medium and can be purified by nickel-affinity chromatography as previously described (6).
Proteolytic Digestion and Mass Spectral Analyses.
Purified SpaA pili and recombinant SpaA protein were digested and analyzed according to previous protocols (5). Briefly, SDS-PAGE gel bands containing recombinant SpaA or SpaA pili were cut out and incubated with trypsin (Promega) followed by AspN endopeptidase (Roche). Peptides in the m/z range 300–1,600 were analyzed using a Q-STAR XL Hybrid MS/MS system (Applied Biosystems). Searches against SpaA sequence using Mascot search engine version 2.0.05 (Matrix Science) identified linear peptides, and the unmatched peptides were then searched manually to identify those containing noncontiguous peptides crosslinked by isopeptide bonds, either intramolecular or intermolecular. Full details are in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Tom Caradoc-Davies for help with data collection, Martin Middleditch for help with mass spectrometry, and Asis Das for critical insights. This work was supported by the Health Research Council and the Marsden Fund of New Zealand (E.N.B.) and National Institutes of Health Grant AI061381 (to H.T.-T). Data collection was undertaken on the PX1 beamline at the Australian Synchrotron, Victoria, Australia, with support from the New Zealand Synchrotron Group Ltd.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3HR6).
This article contains supporting information online at www.pnas.org/cgi/content/full/0906826106/DCSupplemental.
References
- 1.Ton-That H, Schneewind O. Assembly of pili in Gram-positive bacteria. Trends Microbiol. 2004;12:228–234. doi: 10.1016/j.tim.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Proft T, Baker EN. Pili in Gram-negative and Gram-positive bacteria—structure, assembly and their role in disease. Cell Mol Life Sci. 2009;66:613–635. doi: 10.1007/s00018-008-8477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in gram-positive pathogens. Nat Rev Microbiol. 2006;4:509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- 4.Budzik JM, et al. Amide bonds assemble pili on the surface of bacilli. Proc Natl Acad Sci USA. 2008;105:10215–10220. doi: 10.1073/pnas.0803565105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. Stabilizing isopeptide bonds revealed in Gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- 6.Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 7.Ton-That H, Marraffini LA, Schneewind O. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol Microbiol. 2004;53:251–261. doi: 10.1111/j.1365-2958.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- 8.Gaspar AH, Ton-That H. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J Bacteriol. 2006;188:1526–1533. doi: 10.1128/JB.188.4.1526-1533.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandlik A, Das A, Ton-That H. The molecular switch that activates the cell wall anchoring step of pilus assembly in Gram-positive bacteria. Proc Natl Acad Sci USA. 2008;105:14147–14152. doi: 10.1073/pnas.0806350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swierczynski A, Ton-That H. Type III pilus of corynebacteria: Pilus length is determined by the level of its major pilin subunit. J Bacteriol. 2006;188:6318–6325. doi: 10.1128/JB.00606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swaminathan A, et al. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol Microbiol. 2007;66:961–974. doi: 10.1111/j.1365-2958.2007.05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Budzik JM, Oh SY, Schneewind O. Sortase D forms the covalent bond that links BcpB to the tip of Bacillus cereus pili. J Biol Chem. 2009;284:12989–12997. doi: 10.1074/jbc.M900927200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deivanayagam CC, et al. Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Structure. 2000;8:67–78. doi: 10.1016/s0969-2126(00)00081-2. [DOI] [PubMed] [Google Scholar]
- 14.Symersky J, et al. Structure of the collagen-binding domain from a Staphylococcus aureus adhesin. Nat Struct Biol. 1997;4:833–838. doi: 10.1038/nsb1097-833. [DOI] [PubMed] [Google Scholar]
- 15.Krishnan V, et al. An IgG-like domain in the minor pilin GBS52 of Streptococcus agalactiae mediates lung epithelial cell adhesion. Structure. 2007;15:893–903. doi: 10.1016/j.str.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deivanayagam CC, et al. A novel variant of the immunoglobulin fold in surface adhesins of Staphylococcus aureus: Crystal structure of the fibrinogen-binding MSCRAMM, clumping factor A. EMBO J. 2002;21:6660–6672. doi: 10.1093/emboj/cdf619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q, et al. The Enterococcus faecalis MSCRAMM ACE binds its ligand by the collagen hug model. J Biol Chem. 2007;282:19629–19637. doi: 10.1074/jbc.M611137200. [DOI] [PubMed] [Google Scholar]
- 18.Wikoff WR, et al. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 19.Barocchi MA, et al. A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci USA. 2006;103:2857–2862. doi: 10.1073/pnas.0511017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Budzik JM, Schneewind O. Pili prove pertinent to enterococcal endocarditis. J Clin Invest. 2006;116:2582–2584. doi: 10.1172/JCI30088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lauer P, et al. Genome analysis reveals pili in Group B Streptococcus. Science. 2005;309:105. doi: 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- 22.Patti JM, Allen BL, McGavin MJ, Hook M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 23.Rich RL, et al. Domain structure of the Staphylococcus aureus collagen adhesin. Biochemistry. 1998;37:15423–15433. doi: 10.1021/bi981773r. [DOI] [PubMed] [Google Scholar]
- 24.Kang HJ, Baker EN. Intramolecular isopeptide bonds give thermodynamic and proteolytic stability to the major pilin protein of Streptococcus pyogenes. J Biol Chem. 2009 doi: 10.1074/jbc.M109.014514. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dutton RJ, et al. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci USA. 2008;105:11933–11938. doi: 10.1073/pnas.0804621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heras B, et al. DSB proteins and bacterial pathogenicity. Nat Rev Microbiol. 2009;7:215–225. doi: 10.1038/nrmicro2087. [DOI] [PubMed] [Google Scholar]
- 27.Yeates TO, Clubb RT. How some pili pull. Science. 2007;318:1558–1559. doi: 10.1126/science.1151398. [DOI] [PubMed] [Google Scholar]
- 28.Sokurenko EV, Vogel V, Thomas WE. Catch-bond mechanism of force-enhanced adhesion: counterintuitive, elusive, but…widespread? Cell Host Microbe. 2008;4:314–323. doi: 10.1016/j.chom.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zong Y, et al. A ‘Collagen Hug’ model for Staphylococcus aureus CNA binding to collagen. EMBO J. 2005;24:4224–4236. doi: 10.1038/sj.emboj.7600888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang HJ, Paterson NG, Baker EN. Expression, purification, crystallization and preliminary crystallographic analysis of SpaA, a major pilin from Corynebacterium diphtheriae. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009 doi: 10.1107/S1744309109027596. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 32.Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 33.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 34.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 35.Adams PD, et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Radiat. 2004;11:53–55. doi: 10.1107/s0909049503024130. [DOI] [PubMed] [Google Scholar]
- 36.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr D Biol Crystallogr. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 39.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.