Abstract
Macroautophagy (or autophagy) is a conserved degradative pathway that has been implicated in a number of biological processes, including organismal aging, innate immunity, and the progression of human cancers. This pathway was initially identified as a cellular response to nutrient deprivation and is essential for cell survival during these periods of starvation. Autophagy is highly regulated and is under the control of a number of signaling pathways, including the Tor pathway, that coordinate cell growth with nutrient availability. These pathways appear to target a complex of proteins that contains the Atg1 protein kinase. The data here show that autophagy in Saccharomyces cerevisiae is also controlled by the cAMP-dependent protein kinase (PKA) pathway. Elevated levels of PKA activity inhibited autophagy and inactivation of the PKA pathway was sufficient to induce a robust autophagy response. We show that in addition to Atg1, PKA directly phosphorylates Atg13, a conserved regulator of Atg1 kinase activity. This phosphorylation regulates Atg13 localization to the preautophagosomal structure, the nucleation site from which autophagy pathway transport intermediates are formed. Atg13 is also phosphorylated in a Tor-dependent manner, but these modifications appear to occur at positions distinct from the PKA phosphorylation sites identified here. In all, our data indicate that the PKA and Tor pathways function independently to control autophagy in S. cerevisiae, and that the Atg1/Atg13 kinase complex is a key site of signal integration within this degradative pathway.
Keywords: cAMP-dependent protein kinase, macroautophagy, stationary phase, Tor protein kinase
Macroautophagy (hereafter autophagy) is a highly-conserved membrane trafficking pathway that is responsible for the turnover of bulk cytoplasmic protein and organelles (1, 2). This pathway was initially identified as a cellular response to nutrient deprivation (3, 4). However, recent studies indicate that autophagy is involved in a wide variety of physiological processes, including tissue remodeling during development, the removal of protein aggregates, and innate immune responses (5, 6). During autophagy, an isolation membrane emanates from a nucleation site that is known as the preautophagosomal structure (PAS) in Saccharomyces cerevisiae and the phagophore assembly site in mammals (7, 8). This double membrane encapsulates nearby cytoplasm and ultimately targets it to the vacuole/lysosome for degradation. The breakdown products are then recycled to allow for the synthesis of the macromolecules needed for survival during the period of starvation (9). The cellular components mediating autophagy were initially described in S. cerevisiae, and orthologs of many of these Atg proteins have since been identified in other eukaryotes (10, 11).
The flux through the autophagy pathway is tightly controlled by multiple signaling pathways, including the Tor pathway, that are responsible for coordinating cell growth with nutrient availability. One of the key targets of this control appears to be a complex of proteins that contains the Atg1 protein kinase (12–15). Atg1 is specifically recruited to the PAS, and activated, in response to conditions that induce autophagy (7, 16). In contrast, most Atg proteins are constitutively localized to this nucleation structure (17). Recent work suggests that the mammalian Atg1 proteins, ULK1 and ULK2, are also recruited to the phagophore assembly site, and activated, upon nutrient deprivation (18, 19). A key question that remains is how do these signaling pathways work together through Atg1 to ensure the appropriate autophagic response.
In S. cerevisiae, the cAMP-dependent protein kinase (PKA) signaling pathway has also been implicated in the control of autophagy (13, 20). However, the precise nature of this control, and how it is integrated with that of the Tor pathway, is still somewhat controversial (13, 21). In this report, we show that the PKA pathway is critical for the appropriate control of autophagy in S. cerevisiae. Inhibition of PKA signaling is sufficient to induce robust autophagy activity, and this control appears to occur independently, or in parallel to, that exerted by the Tor pathway. For example, both of these pathways target Atg13, a key regulator of Atg1 protein kinase activity, but appear to affect distinct sets of phosphorylation sites on this protein. In all, the data here indicate that both the PKA and Tor pathways are important, but independent, regulators of autophagy, and that the Atg1 protein kinase complex is a key site of signal integration within this pathway.
Results
Inactivation of the PKA Pathway Is Sufficient to Induce Autophagy.
Elevated levels of PKA activity have been shown to inhibit the autophagy process (13). Here, we tested whether the inactivation of this signaling pathway was also sufficient to induce autophagy. To shutdown PKA signaling, we used an inducible form of a dominant negative allele of RAS2, known as RAS2ala22 (22). In S. cerevisiae, the Ras proteins, Ras1 and Ras2, regulate cAMP production, and thus PKA activity, by directly stimulating adenylyl cyclase (23, 24). The RAS2ala22 allele used here was under the control of the promoter from the MET3 gene, a locus that is repressed when methionine is in the growth medium (25). We found that autophagy was efficiently induced upon expression of the RAS2ala22 protein and that the kinetics of induction were similar to that observed with rapamycin treatment (Fig. 1A). Moreover, this RAS2ala22-mediated induction was dependent upon the presence of Atg1 (Fig. 1B). To directly compare the effects of inhibiting the Tor and PKA pathways, we used a concentration of rapamycin that produced a growth arrest similar to that observed in the MET3-RAS2ala22 strain. [Note that rapamycin specifically inhibits the TORC1 complex and that we will be referring to this complex when we discuss Tor signaling in this report (26).] Therefore, a decrease in PKA function that was sufficient to arrest S. cerevisiae growth resulted in a concomitant increase in autophagy activity. These results are consistent with a previous study that used a galactose-inducible form of this RAS2ala22 allele (13).
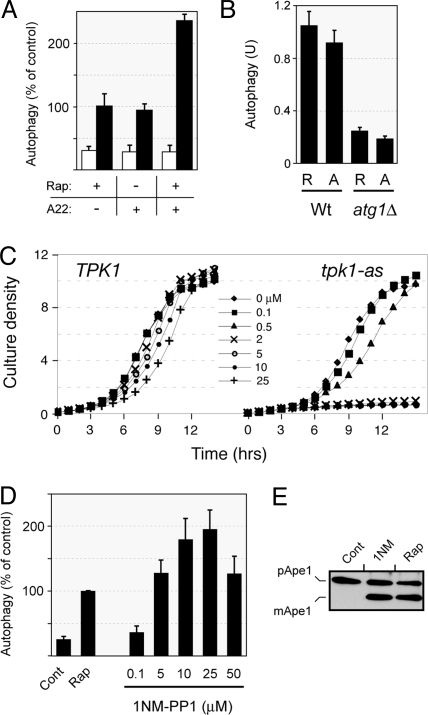
Fig. 1.
Inactivation of the Ras/PKA signaling pathway was sufficient to induce autophagy. (A) Autophagy induction upon rapamycin treatment and/or expression of the dominant-negative RAS2ala22 allele. Wild-type cells (TN125) were grown to mid-log phase and treated with 20 ng/mL rapamycin for 4 h at 30 °C. Cells carrying the MET3-RAS2ala22 allele were transferred to an SC minimal medium lacking methionine for 4 h at 30 °C to induce expression from the MET3 promoter. Autophagy levels were assessed with an alkaline phosphatase (ALP)-based assay as described in the Materials and Methods. The white bars indicate the relative levels of ALP activity in the untreated controls and the black bars the activity following the indicated treatments. (B) The autophagy activity induced upon inactivation of the Ras/PKA pathway was dependent upon the presence of the Atg1 protein. Autophagy levels were assessed with the ALP-based assay in isogenic wild-type and atg1Δ cells after 4 h of rapamycin treatment (R) or exposure to the RAS2ala22 protein (A). (C) Growth curves for isogenic TPK1 and tpk1-as strains in YPAD at 30 °C are shown. The drug 1NM-PP1 was present at the indicated concentrations. (D) Inactivation of PKA signaling in the tpk1-as strain resulted in the induction of autophagy. Autophagy levels were assessed with the ALP-based assay in tpk1-as cells (PHY4710) that were treated for 5 h with either 200 ng/mL rapamycin or the indicated concentrations of 1NM-PP1. (E) Cells over-expressing Ape1 were treated for 4 h with either 200 ng/ml rapamycin or 25 μM 1NM-PP1, and the relative level of Ape1 processing was assessed by Western blotting. Cont, untreated cells.
The second method to inactivate PKA activity made use of an “analog-sensitive” version of this enzyme. This approach for inactivating protein kinases was pioneered by Kevan Shokat and his colleagues, and involves altering a specific residue within the kinase active site (27, 28). Particular substitutions at this “gatekeeper” position render these enzymes sensitive to membrane-soluble inhibitors, like 1NM-PP1 (29). Here, we used a yeast strain that has an “analog-sensitive” allele of TPK1, tpk1-as, as the sole source of PKA activity (27, 28). S. cerevisiae has three functionally redundant PKA catalytic subunits that are encoded by the TPK1–3 genes (30). The tpk1-as strain used here, generously provided by Dr. James Broach, lacks TPK2 and TPK3, and contains an allele of TPK1, tpk1-M164G, that is sensitive to the drug, 1NM-PP1. Interestingly, we found that 1NM-PP1 concentrations that inhibited cell growth in the tpk1-as strain also resulted in a robust induction of the autophagy pathway (Fig. 1 C–E and Figs. S1 and S2). For these experiments, autophagy activity was assessed with three different assays that are described in the Materials and Methods. Therefore, the inhibition of PKA signaling by two different means resulted in an induction of autophagy activity similar to that observed upon inactivation of the Tor pathway. These results are significant because they contradict a recent study that suggested that the loss of PKA activity was not sufficient to induce autophagy (21). This latter study also used a yeast strain that contained analog-sensitive versions of PKA; this strain is in the same genetic background as the tpk1-as strain used here. However, this previous study used a 1NM-PP1 concentration of only 0.1 μM, a concentration that did not have a significant effect upon the growth rate of the tpk1-as strain (Fig. 1C). Therefore, the low level of autophagy observed was likely due to residual PKA activity remaining in the analog-sensitive cells. In all, our data here indicate that the inactivation of the PKA pathway is sufficient to induce autophagy activity in S. cerevisiae cells.
Atg13 Is a Substrate for PKA in Vitro and in Vivo.
The Atg13 protein physically interacts with Atg1 and is required for full Atg1 protein kinase activity in vitro (14, 31–35). Although the mechanistic basis of this activation is not yet understood, the Atg1-Atg13 interaction in S. cerevisiae appears to be regulated by Tor signaling activity. A recent study also identified Atg13 as a candidate substrate for PKA in this budding yeast (20). This identification was based on the presence of evolutionarily-conserved matches in Atg13 to the PKA consensus phosphorylation site. Two sites were very similar to the consensus of R-R-x-S/T-B, where x refers to any amino acid and B to a hydrophobic residue (Sites 2 and 3; Fig. 2A) (36). A third conserved site that deviates more from the consensus was also identified (Site 1). We found that each of these sites was phosphorylated by PKA in vitro, and alteration of all three sites resulted in a greater than 95% decrease in Atg13 phosphorylation (Atg13-AAA; Fig. 2 B and C and Fig. S3). Similar results were observed in vivo with an assay that makes use of an antibody that specifically recognizes phosphorylated PKA sites (Fig. 2D) (37, 38). For these experiments, Atg13 was precipitated from cell extracts and the level of PKA phosphorylation was assessed by Western blotting with this α-substrate antibody. This Atg13 signal was lost upon phosphatase treatment, and was restored by a subsequent incubation with PKA and ATP (Fig. 2D). In addition, this in vivo signal was elevated in cells that possessed elevated levels of PKA activity (Fig. 2E). Finally, Atg13 recognition by this α-substrate antibody was lost following the inactivation of PKA activity (Fig. 2F). In all, these data indicated that Atg13 was a direct substrate for PKA in S. cerevisiae.
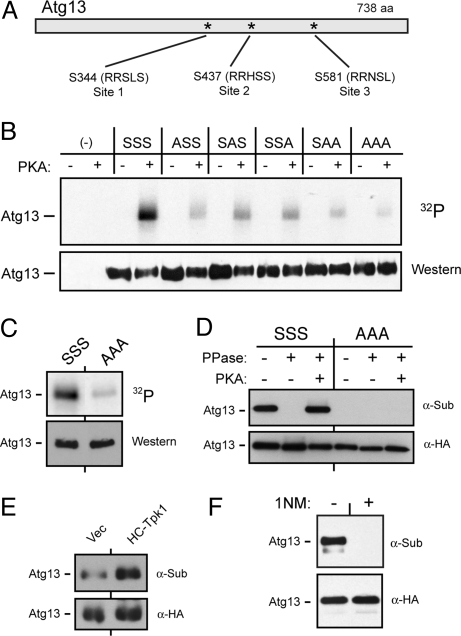
Fig. 2.
The Atg13 protein was a substrate for PKA. (A) The three conserved sites of PKA phosphorylation in Atg13 are shown. (B and C) The in vitro phosphorylation of Atg13 was dependent upon the presence of the above three PKA sites. The indicated Atg13 variants were precipitated from yeast cells and incubated with [γ-32P] ATP and either bovine PKA (bPKA) (in B) or the S. cerevisiae Tpk1 (C). S, serine; A, alanine. (D) The three PKA sites were required for the in vivo phosphorylation of Atg13. The indicated Atg13 proteins were immunoprecipitated from yeast cell extracts with an α-HA antibody and treated with λ phosphatase and then incubated with bPKA and 3 mM ATP, as indicated. The level of PKA phosphorylation was assessed by Western blotting with an α-substrate antibody that recognizes phosphorylated PKA sites. (E) The in vivo level of PKA phosphorylation on Atg13 was elevated in a strain over-expressing Tpk1. (F) Recognition by the α-substrate antibody was lost following inactivation of PKA. The tpk1-as strain was incubated with 10 μM 1NM-PP1 for 4 h and the PKA phosphorylation level of Atg13 was assessed by Western blotting with the α-substrate antibody.
The Loss of PKA Phosphorylation on Atg13 Was Correlated with the Induction of Autophagy.
Atg13 has been shown to be required for Atg1 kinase activity in vitro (14). Here, we tested whether this protein was also required for Atg1 activity in vivo by taking advantage of a previous observation concerning the mobility of this protein in SDS-polyacrylamide gels (14, 20). In particular, autophosphorylation was found to retard the mobility of Atg1 in these gels, and the presence of this slower-migrating band can serve as an indicator of Atg1 kinase activity in vivo (Fig. 3A). We found that the proportion of Atg1 in the slower migrating form increased upon rapamycin treatment (Fig. 3A). This activation of Atg1 did not occur in cells that lacked either Atg13 or Atg17; these mutants were the only atg strains tested that exhibited a significant defect in this Atg1 autophosphorylation. Therefore, Atg13 was required in vivo for Atg1 protein kinase activity.
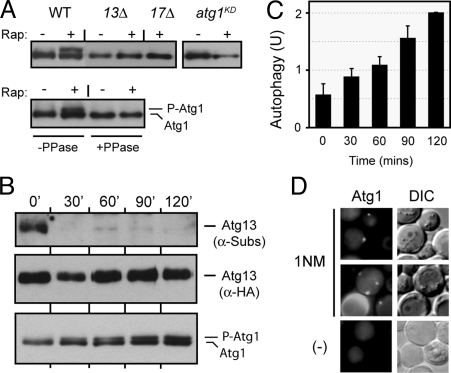
Fig. 3.
The loss of PKA phosphorylation on Atg13 preceded the activation of Atg1 and the induction of autophagy. (A) Atg13 and Atg17 were required for Atg1 autophosphorylation in vivo. Cell extracts were prepared from the indicated yeast strains and the levels of autophosphorylated Atg1 were assessed by Western blotting. The cells were treated with 200 ng/mL rapamycin for 0 or 2 h at 30 °C. The atg1KD strain has a kinase-defective allele of ATG1, atg1-K54A. In the bottom panel, Atg1 was immunoprecipitated from yeast cell extracts and then treated with λ phosphatase, as indicated. (B) The relative levels of Atg13 phosphorylation by PKA (top panel) and of the “activated” form of Atg1 (bottom) were assessed by Western blotting at the indicated times after the addition of 25 μM 1NM-PP1 to a culture of tpk1-as cells. (C) Autophagy activity was assessed with the ALP-based assay at the indicated times after the addition of 25 μM 1NM-PP1 to tpk1-as cells. (D) The localization of an Atg1-YFP protein was assessed by fluorescence microscopy 4 h after the addition of 25 μM 1NM-PP1 to a culture of tpk1-as cells.
We used the tpk1-as strain described above to examine the temporal relationship between the PKA phosphorylation of Atg13, and the induction of both Atg1 kinase activity and autophagy. We found that the PKA-dependent phosphorylation of Atg13 was largely gone after 30 min of treatment with the drug, 1NM-PP1 (Fig. 3B). The first signs of Atg1 kinase activity were apparent at this time, but the fraction of Atg1 in the slower-migrating form continued to increase at later time points (Fig. 3B). Autophagy activity was found to increase in a similar manner (Fig. 3C). Finally, we found that this inactivation of the PKA pathway also resulted in the re-localization of Atg1 from the cytoplasm to the PAS (Fig. 3D). These data therefore indicated that the loss of Atg13 phosphorylation preceded the activation of Atg1 and the induction of the autophagy pathway.
PKA Phosphorylation Regulates the Association of Atg13 with the PAS.
As observed previously, we found that Atg13 was largely cytoplasmic in growing cells and was associated with the PAS upon nutrient limitation (Fig. 4A). Here, we tested whether this localization to the PAS was regulated by PKA phosphorylation. A clear precedent exists as PKA phosphorylation has been shown to inhibit the PAS association of Atg1 (20). We found that the Atg13-AAA variant was localized to the PAS in both growing and nitrogen-starved cells (Fig. 4A and Fig. S4). Moreover, the association of the wild-type Atg13 (Atg13-SSS) with the PAS was inhibited by the presence of the RAS2val19 allele that results in constitutively-elevated levels of PKA activity (39). This inhibition was dependent upon the PKA sites as the Atg13-AAA protein was still constitutively localized to the PAS in RAS2val19 cells (Fig. 4A and Fig. S4). Finally, we found that the wild-type Atg13 protein was recruited to the PAS following the inactivation of the Ras/PKA pathway (Fig. 4B). These data are therefore consistent with PKA phosphorylation regulating the association of Atg13 with the PAS.
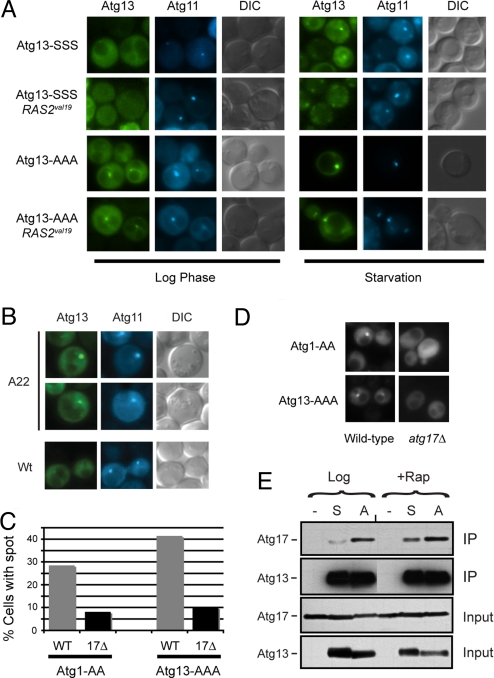
Fig. 4.
The PAS localization of Atg13 was regulated by PKA phosphorylation. (A) Fluorescence microscopy was performed with cells that contained the indicated YFP-Atg13 fusion proteins. The Atg13 proteins had either wild-type (Atg13-SSS) or nonphosphorylatable versions (Atg13-AAA) of the three PKA sites. The CFP-Atg11 fusion protein was present in all cells and served as a marker for the PAS. The RAS2val19 allele was present in the indicated strains. Nitrogen starvation was achieved by transferring the cells from SC glucose minimal medium to the SD-N medium for 1 h at 30 °C. (B) The localization of a wild-type Atg13-YFP protein in the indicated cells was assessed by fluorescence microscopy. A22, RAS2ala22. (C) Atg17 was required for the efficient localization of Atg13-AAA to the PAS in log phase cells. The fraction of cells with Atg1-AA or Atg13-AAA present in a perivacuolar punctate spot in the indicated strains is shown (20). At least 50 cell images were examined for each strain. (D) Representative images for the indicated strains in C. (E) Alteration of the PKA phosphorylation sites in Atg13 influenced the interaction with Atg17. The indicated Atg13 proteins were immunoprecipitated from yeast cell extracts and the relative levels of the associated Atg17 were assessed by Western blotting. The cell extracts were prepared from either mid-log phase cultures (Log) or from cells that were treated with rapamycin (Rap). S, Atg13-SSS; A, Atg13-AAA.
Previous studies have shown that Atg13 interacts with a number of proteins, most notably Atg1 and Atg17 (40–42). Since Atg17 has been suggested to play a role in organizing the PAS, we tested whether the PAS association of Atg13-AAA was dependent upon the presence of Atg17 (43–45). Indeed, we found that the Atg13-AAA variant was less efficiently targeted to the PAS in cells lacking Atg17 (Fig. 4 C and D). In addition, Atg13-AAA exhibited a stronger interaction with Atg17 than the wild-type Atg13 protein and this interaction was no longer influenced by rapamycin treatment (Fig. 4E) (41). Instead, Atg13-AAA appeared to be constitutively associated with Atg17. In contrast, alteration of the PKA sites did not significantly influence the Atg1-Atg13 interaction (Fig. S5A). Finally, Atg17 was found to be associated with the PAS in both growing and nitrogen-starved cells, and this localization was not inhibited by the presence of RAS2val19 (Fig. S5B) (40, 45). Therefore, PKA was apparently regulating the PAS association of Atg13, at least in part, by interfering with its interaction with Atg17.
The PKA and Tor Pathways Independently Target the Atg13 Protein.
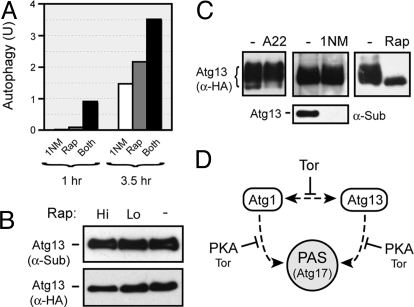
The simultaneous inactivation of both the PKA and Tor pathways produced a more rapid and greater induction of autophagy than was observed with the loss of either pathway alone (Figs. 1A and 5A). This result is consistent with these pathways working independently of each other to control autophagy. To examine this possibility, we tested how shutting down one of these pathways would influence the activity of the other. For these experiments, we used the PKA- and Tor-dependent phosphorylations of Atg13 as reporters for the activity of the respective signaling pathways. We found that the in vivo level of Atg13 phosphorylation by PKA was not diminished upon rapamycin treatment (Fig. 5B). In addition, the inactivation of the Ras/PKA pathway did not result in a loss of the Tor-dependent phosphorylation present on Atg13 (Fig. 5C). This latter phosphorylation causes Atg13 to run as a broad smear on SDS-polyacrylamide gels. This smear rapidly collapses into a tight, faster-migrating band upon rapamycin treatment (Fig. 5C) (14, 46). Finally, the in vitro phosphorylation of Atg13 by PKA did not alter the mobility of this protein in SDS-polyacrylamide gels (see Fig. 2). Therefore, although the precise locations of the Tor-dependent phosphorylation sites on Atg13 have yet to be identified, these positions appear to be distinct from the PKA sites described here. In all, these data are consistent with the Tor and PKA pathways working independently to control autophagy.
Fig. 5.
The PKA and Tor pathways independently target Atg13 to control autophagy activity. (A) Simultaneous inactivation of the PKA and Tor pathways resulted in an elevated autophagy response relative to the loss of either pathway alone. Autophagy levels were assessed with the ALP-based assay in tpk1-as cells that had been treated for the indicated time with either 25 μM 1NM-PP1, 200 ng/mL rapamycin, or both reagents. The data shown are from a single experiment that was representative of at least three independent replicates. (B) The PKA phosphorylation of Atg13 was not diminished upon inactivation of the Tor pathway. Atg13 was immunoprecipitated from cells following a 2 h treatment with either 20 (Lo) or 200 (Hi) ng/mL rapamycin. The level of PKA phosphorylation was subsequently assessed by Western blotting with the α-PKA substrate antibody. (C) Inactivation of the Ras/PKA pathway did not influence the Tor-dependent phosphorylation of Atg13. Cell extracts were prepared from the indicated cells and the level of Tor-dependent phosphorylation of Atg13 was assessed by Western blotting. In the left-hand lanes, cells containing either MET3-RAS2ala22 (A22) or a control plasmid (-) were incubated in methionine-free medium for 6 h to allow for expression from the MET3 promoter. In the middle lanes, the tpk1-as strain, PHY4710, was incubated for 4 h with 0 or 5 μM 1NM-PP1. The bottom panel in the middle lanes shows the level of PKA phosphorylation on Atg13 as assessed by Western blotting with the α-PKA substrate antibody. Note that the relative spread of the Atg13 “smear” is dependent upon the running conditions of the gel. (D) A model depicting the proposed roles of the PKA and Tor pathways in the control of autophagy. The dashed lines indicate the Atg1 and Atg13 interactions with each other or the PAS (and perhaps Atg17), and the solid lines indicate the regulatory effects of the PKA and/or Tor pathways on these interactions. See the text for additional details.
Discussion
Autophagy is a nonspecific degradative process that must be tightly controlled to prevent the inappropriate turnover of material needed for cell growth. The work here adds to our current understanding of this process by demonstrating that the Ras/PKA pathway is an important regulator of autophagy in S. cerevisiae. Inactivation of this signaling pathway by two different methods was sufficient for a robust induction of autophagy activity. This control appears to be exerted through the Atg1 complex as both Atg1 itself and Atg13, a key regulator of this kinase, were shown to be direct substrates for PKA (20) (Fig. 2). For both proteins, this phosphorylation appears to regulate the association with the PAS. Interestingly, the data here indicate that this control by PKA is independent of that exerted by the Tor pathway. For example, although both pathways regulate Atg13 phosphorylation, each appears to control a distinct set of phosphorylation events. In all, this report suggests that the Atg1/Atg13 complex serves as an important point of signal integration within the autophagy pathway. The relevance of this complex as a regulatory control point may be evolutionarily conserved as both Atg1 and Atg13 have been found to be targets for the Tor pathway in other eukaryotes (32, 33, 35, 47).
The autophagy pathway in S. cerevisiae therefore appears to be under the control of at least two independent regulatory inputs. A model that attempts to summarize our current understanding of this regulation is presented in Fig. 5D. In this model, PKA activity specifically inhibits the PAS association of both Atg1 and Atg13. In contrast, the Tor pathway appears to influence both this localization and the Atg1-Atg13 interaction that is necessary for the activation of Atg1 kinase activity (14). Therefore, inactivation of the Tor pathway would result in the formation of an active Atg1-Atg13 complex at the PAS and the ensuing induction of autophagy (1). Upon the loss of PKA activity, however, it is less clear how the Atg1 enzyme is activated. One possibility is that the increased local concentrations of Atg1 and Atg13 at the PAS are sufficient to override the inhibitory effects of Tor activity on the Atg1-Atg13 interaction. Alternatively, Atg13 could be selectively dephosphorylated at the PAS. Although further experimentation is needed to distinguish between these possibilities, the work here shows that the loss of PKA signaling results in increased Atg1 activity and the induction of autophagy. An additional question that arises is why more than one signaling pathway might be needed for the regulation of autophagy. An interesting possibility is that these pathways are responding to distinct nutritional cues and that different types of starvations might elicit distinct autophagy responses in the cell (48). The existence of these multiple inputs would therefore provide the cell with greater flexibility in its response to changing environmental conditions. Determining how these pathways influence this degradative process, and how these signaling activities are coordinated, are therefore important questions for future work.
Materials and Methods
Additional methods, including a description of growth conditions and plasmid construction, are included in SI Text.
Yeast Strain Construction and Growth Conditions.
The yeast strains used in this study were PHY1220 (MATα his3-Δ200 leu2–3,112 lys2–801 trp1–101 ura3–52 suc2-Δ9), PHY1942 (PHY1220 prc1::HIS3 pep4Δ::LEU2 prb1Δ::hisG), TN125 (MATa ade2 his3 leu2 lys2 trp1 ura3 pho8::pho8Δ60), YYK126 (TN125 atg1Δ::LEU2), YYK130 (TN125 atg13Δ::TRP1), PHY3687 (TN125 atg1Δ::LEU2 atg13Δ::kanMX), Y3175 (ade2–1 can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 tpk2::KAN tpk3::TRP1 tpk1-M164G), and PHY4710 (Y3175 pho8::pho8Δ60). This latter strain was generated by integration of a previously described plasmid, PTN9, into Y3175 (49). Strains carrying the MET3-RAS2val19 or MET3-RAS2ala22 alleles were grown in medium containing 500 μM methionine to keep the MET3 promoter in its repressed state. Expression from the MET3 promoter was induced by transferring cells to a medium that lacked methionine. Expression from the CUP1 promoter was induced by the addition of 100 μM CuSO4 to the growth medium. The drug, 1NM-PP1, was generously provided by Kevan Shokat.
Western Blotting and Immunoprecipitations.
Protein extracts for Western blotting were prepared by a glass bead lysis protocol described previously (13). The resulting protein extracts were separated on SDS-polyacrylamide gels and transferred to nitrocellulose membranes (Hybond ECL, Amersham Biosciences) at 4 °C. The membranes were probed with the appropriate primary and secondary antibodies and the Supersignal chemiluminescent substrate (Pierce) was used to illuminate the reactive bands. The immunoprecipitation experiments were performed as described (37, 38).
Autophagy Assays.
The alkaline phosphatase (ALP) assay for autophagy activity was performed as described (13, 49). This assay measures the delivery and subsequent activation of an altered form of the Pho8 phosphatase by the autophagy pathway. Two additional assays assessed the processing of aminopeptidase I (Ape1) and the accumulation of autophagic bodies, and are discussed in SI Text.
PKA Phosphorylation Assays.
In general, the in vitro phosphorylation assays were performed with HA epitope-tagged proteins that were under the control of the yeast CUP1 promoter in the yeast strain, PHY1942. The strains were grown to mid-log phase in selective SC minimal medium containing 2% glucose, and induced with 100 μM copper sulfate for 90 mins. The HA epitope-tagged Atg13 variants were isolated on an α-HA antibody resin (Roche), and incubated with [γ-32P] ATP (PerkinElmer) and 5 U bovine PKA catalytic subunit (Sigma), as described previously (20, 38). A Western immunoblot control was performed with an α-HA antibody (Sigma) to assess the relative amount of Atg13 present in each sample.
The in vivo level of PKA phosphorylation was assessed with an α-PKA substrate antibody (Cell Signaling) as described (37, 38). Briefly, the substrate proteins were immunoprecipitated from yeast cell extracts, separated on SDS-polyacrylamide gels and the relative level of occupancy at the PKA sites was assessed by Western blotting with the α-PKA substrate antibody used at a concentration of 1:2,000.
Fluorescence Microscopy.
The CFP-Atg11, YFP-Atg13, and YFP-Atg17 fusions were under the control of the inducible promoter from the yeast CUP1 gene. Expression of these fusion proteins was induced by the addition of 100 μM CuSO4 for 1 h at 30 °C. The samples were imaged as described (20).
Supplementary Material
Acknowlegements.
We thank James Broach, Daniel Klionsky, Takeshi Noda, Yoshinori Ohsumi, Kevan Shokat, and Jeremy Thorner for reagents used in this study, and members of the Herman lab for helpful discussions and comments on the manuscript. J.S.S. was supported, in part, by a graduate student fellowship from the Jeffrey Seilheimer Lung Cancer Foundation. This work was supported by a grant from the National Institutes of Health (GM65227) to P.K.H.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903316106/DCSupplemental.
References
- 1.Noda T, Suzuki K, Ohsumi Y. Yeast autophagosomes: De novo formation of a membrane structure. Trends Cell Biol. 2002;12:231–235. doi: 10.1016/s0962-8924(02)02278-x. [DOI] [PubMed] [Google Scholar]
- 2.Xie Z, Klionsky DJ. Autophagosome formation: Core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 3.Kopitz J, Kisen GO, Gordon PB, Bohley P, Seglen PO. Nonselective autophagy of cytosolic enzymes by isolated rat hepatocytes. J Cell Biol. 1990;111:941–953. doi: 10.1083/jcb.111.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schworer CM, Mortimore GE. Glucagon-induced autophagy and proteolysis in rat liver: Mediation by selective deprivation of intracellular amino acids. Proc Natl Acad Sci USA. 1979;76:3169–3173. doi: 10.1073/pnas.76.7.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 6.Klionsky DJ. Autophagy: From phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki K, et al. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seglen PO, Gordon PB, Holen I. Non-selective autophagy. Semin Cell Biol. 1990;1:441–448. [PubMed] [Google Scholar]
- 9.Mizushima N, Klionsky DJ. Protein turnover via autophagy: Implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 10.Klionsky DJ, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 11.Meijer WH, van der Klei IJ, Veenhuis M, Kiel JA. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–116. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- 12.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 13.Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem. 2004;279:20663–20671. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamada Y, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephan JS, Herman PK. The regulation of autophagy in eukaryotic cells: Do all roads pass through Atg1? Autophagy. 2006;2:146–148. doi: 10.4161/auto.2.2.2485. [DOI] [PubMed] [Google Scholar]
- 16.Kim J, Huang WP, Stromhaug PE, Klionsky DJ. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 18.Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- 19.Hara T, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Budovskaya YV, Stephan JS, Deminoff SJ, Herman PK. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 2005;102:13933–13938. doi: 10.1073/pnas.0501046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:4180–4189. doi: 10.1091/mbc.E07-05-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powers S, O'Neill K, Wigler M. Dominant yeast and mammalian RAS mutants that interfere with the CDC25- dependent activation of wild-type RAS in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:390–395. doi: 10.1128/mcb.9.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Field J, et al. Mutations of the adenylyl cyclase gene that block RAS function in Saccharomyces cerevisiae. Science. 1990;247:464–467. doi: 10.1126/science.2405488. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki N, et al. Leucine-rich repeats and carboxyl terminus are required for interaction of yeast adenylate cyclase with RAS proteins. Proc Natl Acad Sci USA. 1990;87:8711–8715. doi: 10.1073/pnas.87.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mountain HA, Bystrom AS, Larsen JT, Korch C. Four major transcriptional responses in the methionine/threonine biosynthetic pathway of Saccharomyces cerevisiae. Yeast. 1991;7:781–803. doi: 10.1002/yea.320070804. [DOI] [PubMed] [Google Scholar]
- 26.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 27.Bishop AC, Buzko O, Shokat KM. Magic bullets for protein kinases. Trends Cell Biol. 2001;11:167–172. doi: 10.1016/s0962-8924(01)01928-6. [DOI] [PubMed] [Google Scholar]
- 28.Bishop AC, et al. Design of allele-specific inhibitors to probe protein kinase signaling. Curr Biol. 1998;8:257–266. doi: 10.1016/s0960-9822(98)70198-8. [DOI] [PubMed] [Google Scholar]
- 29.Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 30.Toda T, Cameron S, Sass P, Zoller M, Wigler M. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell. 1987;50:277–287. doi: 10.1016/0092-8674(87)90223-6. [DOI] [PubMed] [Google Scholar]
- 31.Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene. 1997;192:207–213. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- 32.Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganley IG, et al. ULK1. ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol. 2009;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hosokawa N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denis CL, Kemp BE, Zoller MJ. Substrate specificities for yeast and mammalian cAMP-dependent protein kinases are similar but not identical. J Biol Chem. 1991;266:17932–17935. [PubMed] [Google Scholar]
- 37.Chang YW, Howard SC, Herman PK. The Ras/PKA signaling pathway directly targets the Srb9 protein, a component of the general RNA polymerase II transcription apparatus. Mol Cell. 2004;15:107–116. doi: 10.1016/j.molcel.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 38.Deminoff SJ, Howard SC, Hester A, Warner S, Herman PK. Using substrate-binding variants of the cAMP-dependent protein kinase to identify novel targets and a kinase domain important for substrate interactions in Saccharomyces cerevisiae. Genetics. 2006;173:1909–1917. doi: 10.1534/genetics.106.059238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toda T, et al. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985;40:27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- 40.Cheong H, et al. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabeya Y, et al. Atg17 functions in cooperation with atg1 and atg13 in yeast autophagy. Mol Biol Cell. 2005;16:2544–2553. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nair U, Klionsky DJ. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J Biol Chem. 2005;280:41785–41788. doi: 10.1074/jbc.R500016200. [DOI] [PubMed] [Google Scholar]
- 43.Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 45.Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19:2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott SV, et al. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 47.Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20:2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dechant R, Peter M. Nutrient signals driving cell growth. Curr Opin Cell Biol. 2008;20:678–687. doi: 10.1016/j.ceb.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 49.Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.